Abstract
Nukacin ISK-1 is a lantibiotic produced by Staphylococcus warneri ISK-1. Previous studies have reported that the self-protection system of the nukacin ISK-1 producer involves the cooperative function of the ABC transporter NukFEG and the lantibiotic-binding immunity protein NukH. In this study, the cooperative mechanism between NukFEG and NukH was characterized by using fluorescein-4-isothiocyanate (FITC)-labeled nukacin ISK-1 (FITC-nuk) to clarify the localization of nukacin ISK-1 in the immunity process. Lactococcus lactis recombinants expressing nukFEGH, nukFEG, or nukH showed immunity against FITC-nuk, suggesting that FITC-nuk was recognized by the self-protection system against nukacin ISK-1. Analysis of the interaction between FITC-nuk and energy-deprived cells of the L. lactis recombinants showed that FITC-nuk specifically bound to cells expressing nukH. The interaction between FITC-nuk and nukH-expressing cells was inhibited by the addition of unlabeled nukacin ISK-1 and its derivatives with deletions of the N-terminal tail region, but not by the addition of a synthesized N-terminal tail region. This suggests that the NukH protein recognizes the C-terminal ring region of nukacin ISK-1. The addition of glucose to nukFEGH-expressing cells treated with FITC-nuk resulted in a time-dependent decrease in fluorescence intensity, indicating that FITC-nuk was transported from the cell membrane by the NukFEG protein. These results revealed that after being captured by NukH in an energy-independent manner, nukacin ISK-1 was transported to the extracellular space by NukFEG in an energy-dependent manner.
Lantibiotics are antimicrobial peptides produced by gram-positive bacteria and are characterized by the presence of unusual amino acids such as lanthionine and dehydro amino acid residues (5, 11, 21, 22, 27). Bacterial strains producing antimicrobial compounds that are active against closely related strains possess systems to protect themselves against their products. The self-protection systems of lantibiotic producers comprise two major mechanisms. One is a lantibiotic transport mechanism mediated by the ATP-binding cassette (ABC) transporter LanFEG (1, 3, 6, 14, 24, 25, 26, 30, 31). ABC transporters form one of the largest protein families and are found in all species. They function as molecular pumps and, coupled with ATP hydrolysis, transport various substrates such as nutrients, lipids, and antibiotics (12, 16). Bacterial ABC transporters consist of four domains: two transmembrane domains and two nucleotide-binding domains (12). Sequence similarities and hydrophobicity profiles suggest that LanFEG consists of two transmembrane subunits forming a heterodimer (LanEG) and two nucleotide-binding subunits forming a homodimer (LanF) (5, 25). The expression of lanFEG decreases the amount of cell-associated lantibiotics compared with that in non-lanFEG-expressing cells, suggesting that LanFEG transports cell-associated lantibiotics from the cell membrane to the extracellular space (3, 14, 24, 30, 31). The other self-protection mechanism is a lantibiotic-binding mechanism mediated by LanI (5, 15, 17, 19, 29, 30, 31). NisI (LanI for nisin immunity) is a lipoprotein anchored to the membrane surface via N-terminal lipid-modified cysteine residues (19). Stein and coworkers have reported that heterologous expression of NisI increases the amount of cell-associated nisin and the resistance level (30). Moreover, they have demonstrated that purified His6-NisI interacts with nisin. These results suggest that NisI intercepts nisin before the latter attacks the membrane, thereby preventing pore formation. Interestingly, lantibiotic producers possess either or both of the self-protection systems discussed above: LanFEG for epidermin (24, 25), lacticin 481 (26), mutacin II (6), and mersacidin (1, 14); LanI for lactocin S (29), epicidin 280 (15), and Pep5 (17); and both for nisin and subtilin. Although heterologous expression of either self-protection system imparts partial immunity to nisin and subtilin, the expression of both systems confers full immunity (30, 31). It remains unclear why some lantibiotic producers possess only one self-protection system while others possess two.
Staphylococcus warneri ISK-1 produces nukacin ISK-1, a lacticin 481-type lantibiotic (4, 18, 28). It consists of 27 amino acids, involving 2 molecules of lanthionine, 1 molecule of 3-methyllanthionine, and 1 residue of dehydrobutyrine. The structure of nukacin ISK-1 is believed to be formed by the N-terminal tail region (positions 1 to 7) and the C-terminal ring region, containing unusual amino acids (positions 7 to 27) (2). The self-protection system against nukacin ISK-1 is conferred by the LanFEG-type protein NukFEG and a novel type of lantibiotic-binding immunity protein, NukH (3, 4). Previously, we demonstrated that expression of nukFEG and nukH increases the immunity level of a nukacin ISK-1-sensitive Lactococcus lactis strain and that expression of both is required for full immunity (3). In addition, an in vitro peptide release assay indicated that NukFEG transports cell-associated nukacin ISK-1 and that NukH has binding activity not only against nukacin ISK-1 but also against lacticin 481 (3). Despite the similar functions (lantibiotic binding) of NukH and NisI, their structures are considerably different. NukH is a membrane protein with three transmembrane domains and with the N terminus located on the cytoplasmic side. Evaluation of immunity levels and binding activities of various NukH mutants against nukacin ISK-1 suggested that the whole structure of NukH, except for the N and C termini, is essential for its full immunity and that the third transmembrane helix is dispensable for binding. Importantly, there is no correlation between immunity and the binding activity of NukH. Therefore, we proposed the hypothesis that NukH might inactivate nukacin ISK-1 after binding (23).
It is very interesting that both nukFEG and nukH are required for the self-protection system of nukacin ISK-1 producers, despite their opposite phenotypes (transport and binding, respectively). Since both immunity systems are localized at the membrane and are essential for full immunity, it could be presumed that there is a functional cooperation between the two systems. In the present study, we used fluorescein-4-isothiocyanate (FITC)-labeled nukacin ISK-1 (FITC-nuk) to determine the cooperative immunity process of NukFEG and NukH. FITC-nuk was found to specifically bind to cells of L. lactis recombinants expressing nukH. We propose that NukH captures the C-terminal ring region of nukacin ISK-1; subsequently, the captured nukacin ISK-1 is transported to the extracellular space by NukFEG.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. warneri ISK-1 was grown in MRS medium (Oxoid, Hampshire, United Kingdom) at 37°C. Lactobacillus sakei subsp. sakei JCM 1157T, Pediococcus pentosaceus JCM 5885, and Leuconostoc mesenteroides subsp. mesenteroides JCM 6124T were grown in MRS medium at 30°C and then used as indicator strains for the immunity assay. L. lactis NZ9000 was grown in M17 medium (Merck, Darmstadt, Germany) supplemented with 0.5% glucose (GM17) at 30°C. Plasmid pNZ8048 was used for the expression of nukF, nukE, nukG, and nukH. The methods of constructing pNZFEGH, pNZFEG, and pNZH, which are pNZ8048 derivatives containing nukFEGH, nukFEG, and nukH, respectively, have been described previously (3). The nukF gene in pNZFEGH was inactivated by digestion with the restriction enzymes BstZ17I and BsaI. After digestion, both ends of the linearized plasmids were blunt ended and were then self-ligated for frameshifts, which resulted in plasmid pNZEGH. For the selection of recombinants carrying pNZ8048 and its derivatives, chloramphenicol was used at a concentration of 10 μg/ml. nukF, nukE, nukG, and nukH were expressed in L. lactis NZ9000 by a procedure based on the nisin-controlled expression system (8, 9).
Preparations of fluorescently labeled peptides.
Nukacin ISK-1 was purified as described previously (3). FITC (Wako, Tokyo, Japan), which has affinity for the amino group (20), was used for labeling nukacin ISK-1. Purified nukacin ISK-1 (0.5 mg/ml) was incubated in 50 mM Tris-HCl (pH 8.5) containing FITC (0.5 mg/ml) at room temperature for 2 h in dark. Subsequently, labeled nukacin ISK-1 was purified by reverse-phase high-performance liquid chromatography (RP-HPLC) using a Resource RPC column (Amersham Pharmacia Biotech, Uppsala, Sweden). FITC-nuk was eluted by a linear gradient of 20 to 60% acetonitrile in water containing 0.1% trifluoroacetic acid for 30 min. The flow rate was 1 ml/min, and the eluates were monitored by absorbance at 220 nm. The molecular mass of each peak obtained was analyzed by electrospray ionization-mass spectrometry (ESI-MS) (Accutof T100LC; JEOL, Tokyo, Japan). After evaporation, purified FITC-nuk was dissolved in 50 mM potassium phosphate (pH 7.0) and stored at −30°C with light shielding. To identify the position of the FITC-labeled residue in nukacin ISK-1, FITC-nuk was cleaved by lysyl endopeptidase (Wako, Osaka, Japan) as described previously (2). Subsequently, the fragments obtained were analyzed by RP-HPLC and ESI-MS. The tail region of nukacin ISK-1 (nukacin1-7) was synthesized chemically by a solid-phase method, as described previously (2). FITC-labeled nukacin1-7 (FITC-nuk1-7) was prepared in the same way as FITC-nuk.
Immunity assay.
For induction of the immunity genes in L. lactis, cells were grown to an A600 of 0.8 and incubated for 3 h in the presence of 10 ng/ml nisin A. After purified nukacin ISK-1 and FITC-nuk (25 μM) were serially diluted with 50 mM potassium phosphate (pH 7.0), 10-μl portions of the solutions were put on plates containing a soft agar (Lactobacilli Agar AOAC; Difco Laboratories, Detroit, MI) that had been seeded with the indicator strains (100-fold dilution). For the recombinants, the soft agar was supplemented with nisin and chloramphenicol at concentrations of 10 ng/ml and 10 μg/ml, respectively, before seeding. After incubation at 30°C for 18 h, the MICs of nukacin ISK-1 and FITC-nuk were defined as the lowest respective concentrations showing a clear visible zone.
Analysis of interaction between L. lactis recombinants and FITC-nuk.
After a 3-h induction of the recombinant L. lactis harboring plasmid pNZ8048 or its derivative containing nukFEGH, nukFEG, or nukH by the addition of nisin A, the cells were harvested and washed twice with 50 mM potassium phosphate (pH 7.0) containing 5 mM MgSO4 (buffer A). To deprive the cells of metabolic energy, the cell suspensions were incubated in 0.5 mM dinitrophenol and washed three times with buffer A. The cell pellets were resuspended to an A600 of 5 in 400 μl of buffer A containing 2.5 μM FITC-nuk and were then incubated for 1 h at 30°C in the presence or absence of 1% sugar (glucose or sorbitol). To investigate the competitive effects of intact nukacin ISK-1, nukacin1-7, and two N-terminal deletion derivatives on the interaction between FITC-nuk and nukH-expressing cells, the unlabeled peptides were individually added to final concentrations of 0.1 to 50 μM. The two N-terminal-deletion derivatives, which lacked 3 lysine residues at positions 1 to 3 (nukacin4-27) or amino acids at positions 1 to 6 (nukacin7-27), were obtained by methods described previously (2). After incubation, the harvested cell pellets were washed twice and resuspended in 400 μl of buffer A. Subsequently, the fluorescence of each suspension was analyzed.
Time-dependent transport assays.
The energy-deprived cells of L. lactis recombinants were treated with 2.5 μM FITC-nuk for 1 h at 30°C in the absence of glucose as described above. The washed cells were resuspended to an A600 of 5 in buffer A and preincubated for 5 min at 30°C. The transport assays were initiated by the addition of glucose to a final concentration of 1% to the cell suspensions, and the assay mixtures were further incubated at 30°C. For analysis of the remaining florescence in cells, the transport reactions were stopped by cooling on ice after appropriate incubations. The harvested cell pellets were washed and resuspended in buffer A, and the fluorescence of each cell suspension was analyzed. For real-time monitoring of the transport reaction, the fluorescence of the cell suspensions treated with FITC-nuk was continuously monitored using a fluorimeter. The transport reaction was initiated by the addition of glucose to a final concentration of 1%, and the fluorescence was followed in time until steady state was reached.
Measurement of fluorescence.
The fluorescence of the cells was analyzed by fluorescence microscopy (Eclipse 80i/D-FL; Nikon, Tokyo, Japan) using a Plan Fluor 100× oil objective with a numerical aperture of 1.3 and a working distance of 0.2. The fluorescence of FITC was excited by using a GFP-B filter (460 to 500 nm). Before microscopy, the cells were fixed with 1.6% formaldehyde in buffer A for 1 h on ice. As a control, the cells were observed by difference interference contrast (Nomarski optics). The fluorescence intensities of the cell suspensions (total volume, 400 μl) were measured with a fluorescence spectrophotometer (fluorimeter) (F-7000; Hitachi High-Technologies, Tokyo, Japan), using excitation and emission wavelengths of 480 nm and 530 nm, respectively, and a slit width of 10 nm. The cell suspensions were continuously stirred at 30°C during measurements.
RESULTS
FITC labeling of nukacin ISK-1.
In general, FITC-labeled proteins or peptides are purified by size exclusion chromatography or ion-exchange chromatography (13). However, in our case, purification of the labeled peptide was not successful due to deficient separation of free FITC and FITC-nuk. As the next attempt, we purified FITC-nuk using RP-HPLC. After purification, we obtained three peaks, corresponding to FITC-nuk with 1, 2, and 3 molecules of FITC per molecule of the peptide, respectively (data not shown). Nukacin ISK-1 contains three lysine residues in its N terminus. Therefore, it was suggested that FITC molecules, which have affinity for primary amines, were bound not only to the N terminus but also to the side chains of lysine residues in the N terminus. We used FITC-nuk with 1 molecule of FITC per molecule of the peptide in the subsequent experiments to minimize the effect of labeling. To confirm the position of the labeled residue, FITC-nuk was treated with lysyl endopeptidase and subjected to RP-HPLC and LC-MS analyses, which detected a molecular mass corresponding to unlabeled nukacin4-27 (data not shown). Therefore, we concluded that the FITC-labeled residue is located somewhere between positions 1 and 3 in nukacin ISK-1. Similarly, FITC-nuk1-7 (the FITC-labeled tail region with 1 molecule of FITC per molecule of the peptide) was obtained (data not shown).
Antimicrobial activity of FITC-nuk.
The antimicrobial activities of FITC-nuk against eight indicator strains, including L. lactis NZ9000 recombinants, were determined. FITC-nuk showed lower antimicrobial activities against all tested indicator strains than nukacin ISK-1 (Table 1). The MICs of FITC-nuk against L. sakei subsp. sakei JCM 1157T, P. pentosaceus JCM 5885, and L. lactis NZ9000 harboring pNZ8048 were eightfold higher than those of nukacin ISK-1. We have reported that deletion or alanine substitution of the three lysine residues of nukacin ISK-1 decreases its antimicrobial activity drastically (32-fold decrease) because these lysine residues are important for interaction with the cytoplasmic membrane (2). Importantly, L. lactis expressing the immunity gene nukFEGH, nukFEG, nukEGH, or nukH showed immunity against FITC-nuk even at the highest concentration (25 μM), suggesting that the nukacin ISK-1 self-protection system recognizes FITC-nuk as well as nukacin ISK-1. Admittedly, the addition of FITC to the N terminus of nukacin ISK-1 resulted in a slight decrease in antimicrobial activity, but FITC labeling is not critical for the antibacterial function of nukacin ISK-1.
TABLE 1.
MICs of FITC-nuk and nukacin ISK-1 against Lactococcus lactis NZ9000 recombinants
| Strain | MIC (μM)a of:
|
|
|---|---|---|
| FITC-nuk | Nukacin ISK-1 | |
| Lactobacillus sakei subsp. sakei JCM 1157T | 13 | 1.6 |
| Pediococcus pentosaceus JCM 5885 | 25 | 3.1 |
| Leuconostoc mesenteroides subsp. mesenteroides JCM 6124T | 6.3 | 1.6 |
| Lactococcus lactis NZ9000(pNZ8048) | 6.3 | 0.78 |
| L. lactis NZ9000(pNZFEGH) | ND | ND |
| L. lactis NZ9000(pNZFEG) | ND | 25 |
| L. lactis NZ9000(pNZEGH) | ND | 6.3 |
| L. lactis NZ9000(pNZH) | ND | 6.3 |
Antibacterial activity was determined at least twice for each peptide. ND, not detected at the highest concentration (25 μM).
A nukH-expressing strain adsorbs FITC-nuk on its surface.
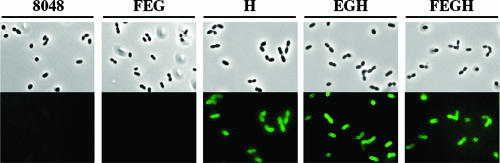
The interaction between FITC-nuk and the energy-deprived cells of L. lactis expressing nukacin ISK-1 immunity genes was investigated. After incubation of the energy-deprived cells with FITC-nuk, the cells were analyzed by fluorescence microscopy. Hardly any visible fluorescence was observed in control (L. lactis harboring pNZ8048) or nukFEG-expressing cells (slight fluorescence was quantitatively determined as described below), suggesting that FITC-nuk has little interaction with these cells (Fig. 1). This result is different from our previous data showing that nukacin ISK-1 bound even to control and nukFEG-expressing cells (4, 23). The weak interactions observed in the present experiment might be explained by a decrease in the antibacterial activity of FITC-nuk compared to that of intact nukacin ISK-1. On the other hand, a green fluorescence emitted from FITC-nuk was detected in nukH-, nukEGH-, and nukFEGH-expressing cells after the incubation (Fig. 1). This indicates that NukH can recognize and capture FITC-nuk despite its decreased antibacterial activity.
FIG. 1.
Detection of FITC-nuk associated with cells of L. lactis recombinants by fluorescent microscopy. After a 3-h induction of recombinant L. lactis harboring plasmid pNZ8048 (8048) or its derivative containing nukFEG (FEG), nukH (H), nukEGH (EGH), or nukFEGH (FEGH) by the addition of nisin A, the cell suspensions were incubated in 0.5 mM dinitrophenol for 30 min at 30°C to deprive them of metabolic energy. The washed cell pellets were incubated with 2.5 μM FITC-nuk for 1 h at 30°C. After being fixed with 1.6% formaldehyde for 1 h on ice, the cells were analyzed by fluorescence microscopy (lower panels). As a control, the cells were observed by difference interference contrast (upper panels).
NukH recognizes the C-terminal ring region of nukacin ISK-1.
The competitive effects of nukacin ISK-1 and its deletion derivatives on the binding of FITC-nuk to nukH-expressing cells were analyzed. After incubation of energy-deprived nukH-expressing cells with FITC-nuk (2.5 μM) in the presence of various concentrations of nukacin ISK-1 or its deletion derivatives, the fluorescence intensities of the cells were measured using a fluorimeter. The fluorescence intensity gradually decreased with an increase in the amount of nukacin ISK-1 and was completely abolished when the concentration of nukacin ISK-1 was fourfold (10 μM) that of FITC-nuk (Fig. 2). This suggested that FITC-nuk bound to nukH-expressing cells in a manner similar to that of nukacin ISK-1. Interestingly, FITC-nuk binding was inhibited by the addition of the derivative nukacin4-27 or nukacin7-27, lacking three lysine residues at positions 1 to 3 or the amino acids at positions 1 to 6, respectively (2). The competitive inhibitory effects of these derivatives were lower than that of nukacin ISK-1; 47% and 28% fluorescence, respectively, was still detected when their concentrations were fourfold that of FITC-nuk (Fig. 2). On the other hand, nukacin1-7 showed no inhibitory effect, even at the highest concentration (50 μM) (Fig. 2). These results indicate that there is no direct interaction between the N-terminal tail region of nukacin ISK-1 and NukH and that NukH recognizes the C-terminal ring region of nukacin ISK-1.
FIG. 2.
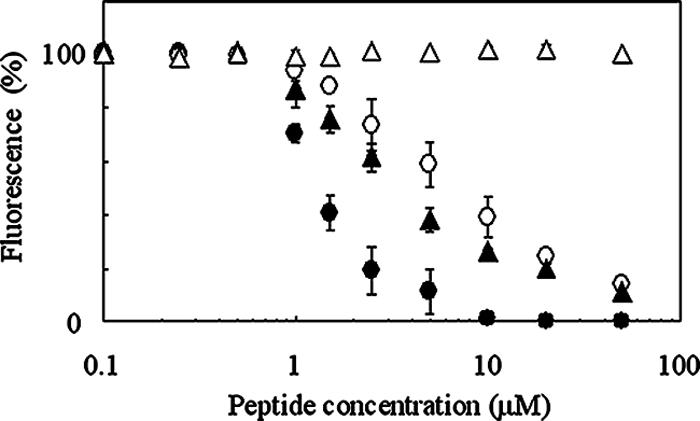
Competitive effects of unlabeled nukacin ISK-1, nukacin4-27, nukacin7-27, and nukacin1-7 on the binding of FITC-nuk to nukH-expressing L. lactis. Energy-deprived nukH-expressing cells of L. lactis NZ9000 were prepared as described in the legend to Fig. 1. The washed cell pellets were incubated with 2.5 μM FITC-nuk in the presence of nukacin ISK-1 (•), nukacin4-27 (○), nukacin7-27 (▴), or nukacin1-7 (▵) at final concentrations of 0.1 to 50 μM for 1 h at 30°C. After a wash with the buffer, the fluorescence intensities of the cell suspensions were measured using a fluorimeter. Relative values calculated on the basis of the fluorescence intensity of the cell suspension incubated without unlabeled peptide are presented. Data are means ± standard errors from two independent experiments.
Energy-dependent transport of FITC-nuk captured by NukH in nukFEGH-expressing cells.
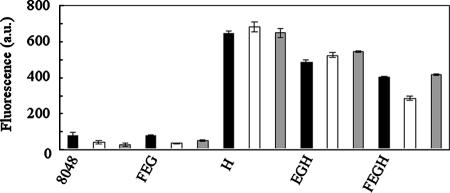
Using FITC-nuk, we investigated the effect of glucose on the transport activity of nukFEGH-expressing cells. ABC transporters transport substrates by using energy obtained from ATP hydrolysis as a driving force. Therefore, the addition of glucose as a source of metabolic energy to suspensions of energy-deprived cells presumably activates NukFEG, thereby leading to transport of FITC-nuk from the cell surface. We determined the fluorescence intensities remaining in the cells of recombinant L. lactis by using a fluorimeter after incubations with or without glucose followed by removal of the supernatants. As a result, the fluorescence intensities of nukFEGH-expressing cells were high and then decreased after the addition of glucose (a decrease of 113 arbitrary units), while no decrease was observed for the nukH- or nukEGH-expressing cells (Fig. 3). Furthermore, the addition of sorbitol, which is not metabolized by the L. lactis recombinants, had no effect on the fluorescence intensities of nukH-, nukEGH-, and nukFEGH-expressing cells (Fig. 3). These results indicate that the transport reaction requires metabolic energy and that the binding and hydrolysis of ATP by NukF is essential for the functioning of NukFEG as a transporter. It should be emphasized that only a small amount of FITC-nuk was bound to the control (L. lactis harboring pNZ8048) and nukFEG-expressing strains (Fig. 3). These results show that most of the FITC-nuk attached to nukH-, nukEGH-, or nukFEGH-expressing cells was captured specifically by NukH in an energy-independent manner. Additionally, FITC-nuk1-7 did not bind to control or nukH-expressing cells at all (data not shown), indicating that the C-terminal region of FITC-nuk, but not the N-terminal tail region and FITC molecule, is responsible for interaction with cells. Taken together, it was proposed that NukFEG transports the FITC-nuk captured by NukH to the outsides of cells.
FIG. 3.
Quantitative analyses of cell-associated FITC-labeled peptides using a fluorimeter. Energy-deprived cells of recombinant L. lactis harboring plasmid pNZ8048 (8048) or its derivative containing nukFEG (FEG), nukH (H), nukEGH (EGH), or nukFEGH (FEGH) were prepared as described in the legend to Fig. 1. The cell pellets were incubated with 2.5 μM FITC-nuk for 1 h at 30°C in the absence of sugar (solid bars) or in the presence of 1% glucose (open bars) or sorbitol (shaded bars). After a wash with the buffer, the fluorescence intensity of the cell suspension was measured using a fluorimeter. Data are means ± standard errors from two independent experiments. a.u., arbitrary units.
Time-dependent transport of FITC-nuk in nukFEGH-expressing cells.
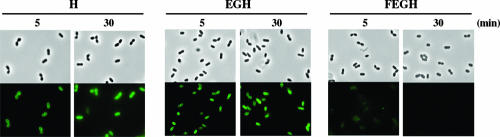
After the addition of glucose to suspensions of energy-deprived cells treated with FITC-nuk, their fluorescence was analyzed at appropriate time intervals by fluorescence microscopy. In nukH- and nukEGH-expressing cells, fluorescence was stable for at least 30 min after the addition of glucose (Fig. 4). In contrast, a significant decrease in fluorescence was observed in nukFEGH-expressing cells after the addition of glucose. Slight but visible fluorescence was observed after a 5-min incubation, and it became invisible after a 30-min incubation (Fig. 4). The fluorescence intensities of FITC-nuk remaining in nukFEGH-expressing cells were measured using a fluorimeter after appropriate incubations in the buffer with glucose, followed by removal of the supernatants containing free FITC-nuk. As a result, the fluorescence intensities (cell-associated FITC-nuk) were rapidly decreased during a 5-min incubation (Fig. 5A). The fluorescence intensity after a 30-min incubation was approximately identical to those of control (pNZ8048) and nukFEG-expressing cells shown in Fig. 3, suggesting that most of the FITC-nuk captured by NukH was time-dependently transported to the outsides of nukFEGH-expressing cells.
FIG. 4.
Effects of glucose on the transport of FITC-nuk in nukH-, nukEGH-, and nukFEGH-expressing L. lactis. Energy-deprived cells of L. lactis NZ9000 expressing nukH (H), nukEGH (EGH), or nukFEGH (FEGH) were prepared and incubated with FITC-nuk as described in the legend to Fig. 1. After the cells treated with FITC-nuk were washed and resuspended, transport reactions were initiated by the addition of glucose to final concentrations of 1%, and the reaction mixtures were further incubated at 30°C. After 5-min and 30-min incubations, the cell suspensions were cooled down on ice, and washed cells were fixed with 1.6% formaldehyde for 1 h on ice. The cells were analyzed by fluorescence microscopy (lower panels) and were observed by difference interference contrast as a control (upper panels).
FIG. 5.
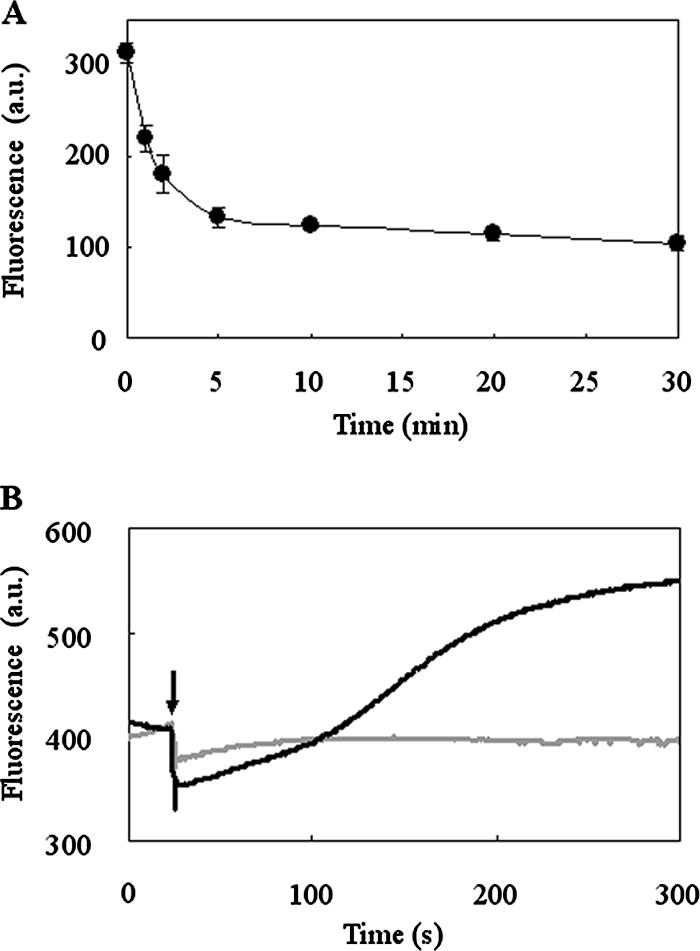
Time-dependent transport of FITC-nuk in nukFEGH-expressing L. lactis. (A) The transport reaction of FITC-nuk in L. lactis NZ9000 expressing nukFEGH was performed as described in the legend to Fig. 4. After incubation for 1, 2, 5, 10, 20, or 30 min, the cell suspensions were cooled on ice. After a wash with the buffer, the fluorescence intensity of the cell suspension was measured using a fluorimeter. Data are means ± standard errors from two independent experiments. a.u., arbitrary units. (B) After the addition of glucose to a final concentration of 1% to the cell suspension of L. lactis expressing nukFEGH (indicated by the arrow), the fluorescence was followed in time until steady state was reached (black line). As a control experiment, the same volume of buffer was added instead of the glucose solution (gray line).
Next, we monitored the transport reaction in real time. In this assay, in contrast to the experiment for which results are shown in Fig. 5A, the change in fluorescence after the addition of glucose was followed continuously by using a fluorimeter without removal of the supernatants. After the addition of glucose to the suspension of nukFEGH-expressing cells treated with FITC-nuk, a significant increase in the fluorescence was observed (the temporary collapse of the fluorescence after the addition of glucose is due to the dilution of the reaction mixture) (Fig. 5B). Notably, the increasing levels of fluorescence intensity showed good agreement with the decreasing levels of cell-associated FITC-nuk shown in Fig. 5A, i.e., they almost reached steady state after 5 min of incubation. Therefore, we concluded that an increase in fluorescence was caused by the transport of FITC-nuk by NukFEG. It is interesting that the increase in fluorescence was observed after the addition of glucose even though the total amount of FITC-nuk in the cell suspension was constant during the analysis. A possible explanation for this phenomenon is that the fluorescence of FITC-nuk had been weakened in some degree after its capture by NukH into the lipid bilayer and was subsequently restored after transport by NukFEG to the extracellular space. This is the first report on the real-time transport of the cell-associated lantibiotic by the lantibiotic-immunity ABC transporter.
DISCUSSION
In this report, we have described the functional analysis of the self-protection system of nukacin ISK-1 producers by using fluorescently labeled nukacin ISK-1. In the bioassay, FITC-nuk showed a decrease in antibacterial activity compared with that of nukacin ISK-1 (Table 1). The most likely explanation for the lower activity of FITC-nuk is that the attachment of FITC, which is a large hydrophobic molecule, decreased the electrostatic interaction between the lysine residues and the membrane. The interaction between FITC-nuk and L. lactis cells expressing nukFEGH, nukFEG, nukEGH, or nukH was analyzed by fluorescence microscopy using a fluorimeter. Despite its lower antibacterial activity, FITC-nuk bound to nukH-, nukEGH-, and nukFEGH-expressing cells (Fig. 2 and 3), and it bound with different efficiencies in the absence of sugar (Fig. 3). This might be due to the fact that the expression levels of NukH in each expressing cell type are different and/or to the fact that the NukF, -E, and -G subunits affect the interaction between NukH and FITC-nuk. Importantly, the binding of FITC-nuk was competitively inhibited by the addition not only of nukacin ISK-1 but also of nukacin4-27 and nukacin7-27 (Fig. 2). Furthermore, nukacin1-7 did not inhibit the binding of FITC-nuk (Fig. 2). These results strongly suggest that NukH captures nukacin ISK-1 by recognizing its C-terminal region. The capture of the C-terminal region of nukacin ISK-1 might be attributed to the effective inactivation mechanism of NukH, because nukacin ISK-1 contains unusual amino acids, which are deduced to be essential for antimicrobial activity, in its C-terminal region. Nukacin7-27 inhibited the binding of FITC-nuk better than nukacin4-27. This suggested that the ring region of nukacin ISK-1 is a minimum unit for the recognition by NukH and that the remaining tail region after the digestion of three lysine residues is somewhat unfavorable for the recognition by NukH. The highly competitive inhibition of nukacin ISK-1 compared to that of nukacin4-27 and nukacin7-27 suggests that the interaction of nukacin ISK-1 with the cytoplasmic membrane plays a role in the binding of nukacin ISK-1 by NukH. One putative mechanism to explain the hypothesis is that the interaction between the three lysine residues in the N terminus of nukacin ISK-1 and the cytoplasmic membrane gives the membrane-localized NukH access to nukacin ISK-1, thereby increasing the capturing efficiency of NukH.
In a previous study on the functional analysis of LanFEG, it was proposed that LanFEG scavenges and exports lantibiotics from the membrane to the extracellular space in order to maintain the amounts of cell-associated lantibiotic below a critical level, thereby contributing to the tolerance of the host cell (3, 14, 24, 30, 31). This proposal appears to be plausible, because pore formation concomitant with membrane insertion is the best-known mode of action of lantibiotics. However, ABC transporters that have been identified with respect to their substrates are believed to catalyze only the transmembrane movements of substances. An exception that has been reported previously is the Lol system, which releases lipoproteins from the inner membrane to the outer membrane in gram-negative bacteria (32). In that sense, the proposed function of the lantibiotic immunity-associated ABC transporter appears to be unique in the ABC transporter family. It should be noted that this is the first report to demonstrate the proposed function of the ABC transporter involved in the self-protection system as a scavenger by visualizing the localization of the lantibiotic in the immunity process. FITC-nuk bound to nukFEGH-expressing cells was transported in an energy-dependent manner; on the other hand, no transport was observed in nukH- and nukEGH-expressing cells (Fig. 3 and 4). This result showed that NukFEG recognizes and transports FITC-nuk captured by NukH in nukFEGH-expressing cells. Additionally, we could observe time-dependent transport of FITC-nuk in nukFEGH-expressing cells (Fig. 4 and 5). The increase in fluorescence shown in Fig. 5B suggested that an interaction between FITC-nuk and NukH, concomitant with the immersing of FITC-nuk in the lipid bilayer, has an influence on the fluorescence intensity of FITC-nuk. Taking all the evidence together, we propose the hypothesis that after being captured by NukH, FITC-nuk is exported to the extracellular space by NukFEG. According to investigative studies on the structure of ABC transporters, it is presumed that conformational changes in transmembrane domains caused by dimerization of nucleotide-binding domains drive the transport of substrates (7, 10). However, a pathway for the transmembrane movement of substances is not required for the proposed mechanism of the lantibiotic self-protection system. Therefore, our proposed mechanism of NukFEG and NukH cooperation in lantibiotic transport seems to be reasonable.
The hypothesized function of NukH is similar to that of substrate-binding proteins (SBPs) found in general ABC transporter systems (16). It is proposed that SBPs impart high affinity and specificity to the transport systems for efficient transport. In gram-negative bacteria, SBPs located at the periplasm (periplasmic binding proteins) bind the respective substrates and deliver them to the ABC transporters. In gram-positive bacteria, which lack a periplasm, SBPs are anchored to the membrane surface via a lipid moiety (16). Before now, there was no report on SBPs that localize and function in the membrane; however, the phenotype of NukH is consistent with that of SBPs in terms of high affinity and specificity for its substrate and cooperation with the transporter. Therefore, it appears that NukH functions as a novel type of SBP in the lantibiotic self-protection system.
Acknowledgments
This work was partially supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS), the Japan Science Society, the Novartis Foundation (Japan) for the Promotion of Science, the Novozymes Japan Research Fund, and the Nagase Science and Technology Foundation and by JSPS research fellowships.
Footnotes
Published ahead of print on 19 October 2007.
REFERENCES
- 1.Altena, K., A. Guder, C. Cramer, and G. Bierbaum. 2000. Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl. Environ. Microbiol. 662565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asaduzzaman, S. M., J. Nagao, Y. Aso, J. Nakayama, and K. Sonomoto. 2006. Lysine-oriented charges trigger the membrane binding and activity of nukacin ISK-1. Appl. Environ. Microbiol. 726012-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aso, Y., K. Okuda, J. Nagao, Y. Kanemasa, N. T. B. Phuong, K. Shioya, T. Sashihara, J. Nakayama, and K. Sonomoto. 2005. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci. Biotechnol. Biochem. 691403-1410. [DOI] [PubMed] [Google Scholar]
- 4.Aso, Y., T. Sashihara, J. Nagao, Y. Kanemasa, H. Koga, T. Hashimoto, T. Higuchi, A. Adachi, H. Nomiyama, A. Ishizaki, J. Nakayama, and K. Sonomoto. 2004. Characterization of a gene cluster of Staphylococcus warneri ISK-1 encoding the biosynthesis of and immunity to the lantibiotic, nukacin ISK-1. Biosci. Biotechnol. Biochem. 681663-1671. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee, C., M. Paul, L. Xie, and W. A. van der Donk. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105633-684. [DOI] [PubMed] [Google Scholar]
- 6.Chen, P., F. Qi, J. Novak, and P. W. Caufield. 1999. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl. Environ. Microbiol. 651356-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson, R. J., and K. P. Locher. 2006. Structure of a bacterial multidrug ABC transporter. Nature 443180-185. [DOI] [PubMed] [Google Scholar]
- 8.de Ruyter, P. G., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 1783434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 623662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, J., G. Yang, and H. S. Mchaourab. 2005. Structural basis of energy transduction in the transport cycle of MsbA. Science 3081023-1028. [DOI] [PubMed] [Google Scholar]
- 11.Dufour, A., T. Hindre, D. Haras, and J. P. Le Pennec. 2007. The biology of lantibiotics from the lacticin 481 group is coming of age. FEMS Microbiol. Rev. 31134-167. [DOI] [PubMed] [Google Scholar]
- 12.Fath, M. J., and R. Kolter. 1993. ABC transporters: bacterial exporters. Microbiol. Rev. 57995-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsum, U. 1973. Characterization of FITC-labelled F(ab′)2 fragments of IgG and a rapid technique for the separation of optimally labelled fragments. J. Immunol. Methods 2183-195. [DOI] [PubMed] [Google Scholar]
- 14.Guder, A., T. Schmitter, I. Wiedemann, H.-G. Sahl, and G. Bierbaum. 2002. Role of the single regulator MrsR1 and the two-component system MrsR2/K2 in the regulation of mersacidin production and immunity. Appl. Environ. Microbiol. 68106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heidrich, C., U. Pag, M. Josten, J. W. Metzger, R. W. Jack, G. Bierbaum, G. Jung, and H.-G. Sahl. 1998. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl. Environ. Microbiol. 643140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins, C. F. 2001. ABC transporters: physiology, structure and mechanism—an overview. Res. Microbiol. 152205-210. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann, A., T. Schneider, U. Pag, and H.-G. Sahl. 2004. Localization and functional analysis of PepI, the immunity peptide of Pep5-producing Staphylococcus epidermidis strain 5. Appl. Environ. Microbiol. 703263-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura, H., H. Matsusaki, T. Sashihara, K. Sonomoto, and A. Ishizaki. 1998. Purification and partial identification of bacteriocin ISK-1, a new lantibiotic produced by Pediococcus sp. ISK-1. Biosci. Biotechnol. Biochem. 622341-2345. [DOI] [PubMed] [Google Scholar]
- 19.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216281-291. [DOI] [PubMed] [Google Scholar]
- 20.Maeda, H., N. Ishida, H. Kawauchi, and K. Tsujimura. 1969. Reaction of fluorescein-isothiocyanate with proteins and amino acids. I. Covalent and non-covalent binding of fluorescein-isothiocyanate and fluorescein to proteins. J. Biochem. (Tokyo) 65777-783. [DOI] [PubMed] [Google Scholar]
- 21.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25285-308. [DOI] [PubMed] [Google Scholar]
- 22.Nagao, J., S. M. Asaduzzaman, Y. Aso, K. Okuda, J. Nakayama, and K. Sonomoto. 2006. Lantibiotics: insight and foresight for new paradigm. J. Biosci. Bioeng. 102139-149. [DOI] [PubMed] [Google Scholar]
- 23.Okuda, K., Y. Aso, J. Nagao, K. Shioya, Y. Kanemasa, J. Nakayama, and K. Sonomoto. 2005. Characterization of functional domains of lantibiotic-binding immunity protein, NukH, from Staphylococcus warneri ISK-1. FEMS Microbiol. Lett. 25019-25. [DOI] [PubMed] [Google Scholar]
- 24.Otto, M., A. Peschel, and F. Götz. 1998. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tü3298. FEMS Microbiol. Lett. 166203-211. [DOI] [PubMed] [Google Scholar]
- 25.Peschel, A., and F. Götz. 1996. Analysis of the Staphylococcus epidermidis genes epi F, -E, and -G involved in epidermin immunity. J. Bacteriol. 178531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rincé, A., A. Dufour, P. Uguen, J.-P. Le Pennec, and D. Haras. 1997. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 634252-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sahl, H.-G., R. W. Jack, and G. Bierbaum. 1995. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur. J. Biochem. 230827-853. [DOI] [PubMed] [Google Scholar]
- 28.Sashihara, T., H. Kimura, T. Higuchi, A. Adachi, H. Matsusaki, K. Sonomoto, and A. Ishizaki. 2000. A novel lantibiotic, nukacin ISK-1, of Staphylococcus warneri ISK-1: cloning of the structural gene and elucidation of the structure. Biosci. Biotechnol. Biochem. 642420-2428. [DOI] [PubMed] [Google Scholar]
- 29.Skaugen, M., C. I. Abildgaard, and I. F. Nes. 1997. Organization and ex-pression of a gene cluster involved in the biosynthesis of the lantibiotic lactocin S. Mol. Gen. Genet. 253674-686. [DOI] [PubMed] [Google Scholar]
- 30.Stein, T., S. Heinzmann, I. Solovieva, and K.-D. Entian. 2003. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 27889-94. [DOI] [PubMed] [Google Scholar]
- 31.Stein, T., S. Heinzmann, S. Dusterhus, S. Borchert, and K.-D. Entian. 2005. Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J. Bacteriol. 187822-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2212-218. [DOI] [PubMed] [Google Scholar]