Abstract
Staphylococcus lugdunensis has gained recognition as an atypically virulent pathogen with a unique microbiological and clinical profile. S. lugdunensis is coagulase negative due to the lack of production of secreted coagulase, but a membrane-bound form of the enzyme present in some isolates can result in misidentification of the organism as Staphylococcus aureus in the clinical microbiology laboratory. S. lugdunensis is a skin commensal and an infrequent pathogen compared to S. aureus and S. epidermidis, but clinically, infections caused by this organism resemble those caused by S. aureus rather than those caused by other coagulase-negative staphylococci. S. lugdunensis can cause acute and highly destructive cases of native valve endocarditis that often require surgical treatment in addition to antimicrobial therapy. Other types of S. lugdunensis infections include abscess and wound infection, urinary tract infection, and infection of intravascular catheters and other implanted medical devices. S. lugdunensis is generally susceptible to antimicrobial agents and shares CLSI antimicrobial susceptibility breakpoints with S. aureus. Virulence factors contributing to this organism's heightened pathogenicity remain largely unknown. Those characterized to date suggest that the organism has the ability to bind to and interact with host cells and to form biofilms on host tissues or prosthetic surfaces.
INTRODUCTION
A large number of Staphylococcus species distinct from Staphylococcus aureus comprise the group known as coagulase-negative staphylococci (CNS), so named for their inability to clot plasma due to the lack of production of the secreted enzyme coagulase (5). CNS, which often occur as skin commensals, were historically considered innocuous or, rarely, opportunistic pathogens of low virulence (84). However, the important role of CNS as pathogens, with particular regard to infections associated with indwelling medical devices, is becoming increasingly appreciated (44, 61, 140). CNS are now a leading cause of bacteremia in the United States, Canada, Latin America, and Europe (61, 140), and many of these CNS are resistant to multiple classes of antimicrobial agents (44). Despite this, and in contrast to the case for S. aureus, infections caused by CNS typically manifest as less severe and subacute diseases that are infrequently associated with mortality.
In 1988, Freney et al. described two new coagulase-negative species, Staphylococcus schleiferi and Staphylococcus lugdunensis, isolated from human clinical specimens (68). Since that time, S. lugdunensis, named after Lyon, France, the city where the organism was initially isolated (68), has emerged as an important human pathogen with notable clinical and microbiological characteristics that stand out among those of other CNS. Described previously as “surreptitious” (171) and a “wolf in sheep's clothing,” S. lugdunensis behaves more like S. aureus than other CNS in many respects, including exhibiting an elevated degree of virulence. S. lugdunensis is both a skin commensal and a pathogen responsible for nosocomial and community-acquired infections that may proceed aggressively and with a level of severity reminiscent of that of S. aureus infections. Unlike S. aureus, S. lugdunensis does not possess secreted coagulase. However, some isolates produce a membrane-bound form of the enzyme (clumping factor) that yields a positive result in slide coagulase and/or rapid latex agglutination tests, potentially leading to misidentification of the organism as S. aureus in the clinical laboratory. S. lugdunensis has a propensity to cause native valve endocarditis with a fulminant and highly destructive clinical course that is quite remarkable for a coagulase-negative species, which are otherwise more frequently the etiologic agents of prosthetic valve endocarditis (84). Equally surprising, compared to CNS, most S. lugdunensis isolates remain susceptible to a large number of antimicrobial agents (67).
With little doubt, S. lugdunensis cannot be considered a “typical” coagulase-negative Staphylococcus species, and its successful position as an unusually virulent pathogen deserves attention. In this review, we aim to provide a comprehensive overview of the body of literature on S. lugdunensis since its description 20 years ago, with particular emphasis on the aspects of clinical infection, clinical microbiology, antimicrobial susceptibility, and virulence that are unique to this organism.
CLINICAL DISEASE
In 1989, Etienne et al. described two cases of native valve endocarditis and one case of prosthetic valve endocarditis in which S. lugdunensis was retrospectively identified as being the causative agent (53). In each case, the infection was aggressive, involving valve destruction and abscess formation, and ultimately resulted in the deaths of two of the three patients. The resemblance of the clinical course of S. lugdunensis infections to that of S. aureus was apparent in that early report. The full pathogenic potential of and the wide spectrum of diseases caused by S. lugdunensis are now unmistakable (Table 1). In this section, we review the role of S. lugdunensis as a constituent of the human normal flora as well as a human pathogen.
TABLE 1.
Reported infections caused by Staphylococcus lugdunensis
| Infection type | Reference(s) |
|---|---|
| Cardiovascular infection | |
| Native valve endocarditis | 1, 2, 7, 17, 19, 26, 38, 42, 50, 53, 55, 58, 60, 70, 71, 87, 89, 95, 98, 99, 104-106, 109, 116, 134, 139, 144, 148, 149, 155, 156, 160, 161, 166, 169-171, 180, 183, 187, 189, 193, 195-197 |
| Prosthetic valve endocarditis | 2, 37, 53, 60, 96, 164, 183 |
| Pacemaker-related endocarditis | 2, 15, 101, 162 |
| Myocarditis | 142 |
| Skin and soft tissue infection | 1, 60, 83, 177 |
| Abscesses or wounds | 10, 60, 82, 131, 177, 186 |
| Nonpuerperal breast abscess | 4, 108, 194 |
| Bloodstream infection, sepsis, or septic shock | 25, 51, 60, 178 |
| Toxic shock syndrome | 133 |
| Acute oral infection (abscesses, osteomyelitis) | 200 |
| UTI | 24, 79 |
| Bone and joint infection | 167 |
| Infective arthritis or osteomyelitis | 60, 98, 118, 132, 181 |
| Vertebral osteomyelitis | 77, 98, 121, 198 |
| Disk space infection | 18, 20, 36, 78 |
| Prosthetic joint infection | 154, 158, 182, 198 |
| Central nervous system infection | |
| Brain abscess | 60, 71 |
| Meningitis | 88 |
| Ventriculoperitoneal shunt infection | 52, 157 |
| Peritonitis | 60, 111, 159 |
| Ocular infection | |
| Suppurative keratitis | 141 |
| Postoperative endophthalmitis | 6, 29 |
Characteristics of Colonizing and Infectious Isolates
S. lugdunensis is a constituent of the human normal skin flora and an infrequent, but not rare, human pathogen. Early studies established S. lugdunensis as a skin commensal (82, 183). The organism preferentially colonizes the perineal region (111, 183, 186) and has been rarely found in the anterior nares or nasal cavities of hemodialysis patients (97), cardiothoracic patients (9), and healthy subjects (147). Akiyama et al. reported that five of nine S. lugdunensis isolates (among a total of 162 CNS of colonizing or infective origin) collected from patients with skin lesions in Japan were considered to be colonizing isolates, although the source of isolation was not reported (1). Van der Mee-Marquet et al. (186) cultured the inguinal folds of 140 consecutive patients in an emergency department over three months and found that S. lugdunensis colonized 23% of women and 19% of men, providing supporting data that S. lugdunensis is frequently found in the perineal area.
Herchline and Ayers reported the occurrence of 229 S. lugdunensis isolates, comprising 10% of all non-S. aureus, non-S. epidermidis isolates, during a 63-month study at a U.S. tertiary-care hospital and its associated outpatient clinics (82). The majority of isolates (55%) originated from outpatients. In a review of 50 consecutive cases from which S. lugdunensis was isolated, both colonizing and infectious isolates were identified; infectious isolates derived predominantly from skin-associated infections and, to a lesser extent, from vascular-related infections (82). S. lugdunensis was often found as part of a mixed culture (82). Although S. lugdunensis is commonly described in patients with underlying illnesses or undergoing immunosuppressive therapies (82, 121), it can cause both superficial and serious infections in otherwise healthy people (24, 82, 99, 196).
In addition to Herchline and Ayers' study (82), several groups from multiple countries have reported the frequency of isolation of clinical S. lugdunensis isolates. In contrast to the 10% rate of S. lugdunensis isolation reported by Herchline and Ayers (82), S. lugdunensis accounted for fewer than 3% of non-S. aureus, non-S. epidermidis strains from human clinical sources collected in six Japanese hospitals (91). Overall, in that study, S. lugdunensis accounted for 1.3% of the 1,230 staphylococci (including S. aureus) isolated. S. lugdunensis represented only 1 of 499 (0.2%) CNS recovered from blood cultures in several counties in Denmark during a 2-month period (86). Other rates of isolation of clinically significant S. lugdunensis among CNS range from 0.8% in Korea (165) to 3% in the United States (92) to 6% in Argentina (43).
Bloodstream Infection, Sepsis, and Toxic Shock Syndrome
S. lugdunensis accounted for only 4 of 1,256 (0.3%) CNS causing community-acquired or nosocomially acquired bloodstream infection during a 12-month period in a worldwide network of hospitals, making it the seventh most common coagulase-negative species isolated (140). A retrospective review of the occurrence of clinically significant cases of S. lugdunensis bacteremia at a hospital in the United States over a 12 year period revealed a total of six patients with S. lugdunensis bacteremia (51). In each case, the patient had an underlying disease and the illness manifested as fever. Most patients had intravascular catheters that were reportedly infected. A mecA-positive S. lugdunensis isolate causing catheter-associated bloodstream infection in a premature neonate has been described (178).
Several instances of S. lugdunensis-induced septicemia and septic shock, including one that occurred as a result of receiving contaminated platelets, have been documented (25, 60). A female patient developed S. lugdunensis bacteremia and toxic shock syndrome 48 h after tooth extraction with foam packing (133).
Skin and Soft Tissue Infection
Skin and soft tissue infections account for a prominent number of the total infections caused by S. lugdunensis. In a 63-month study examining the occurrence of S. lugdunensis in consecutive cultures, 55% of the 155 specimens collected from 143 patients originated from wounds, abscesses, or cellulitis (82). Others have reported that S. lugdunensis represents 5 to 6% of CNS isolated from skin lesions (1, 83). The organism is more likely than S. epidermidis or other CNS to be clinically significant when isolated from superficial infections, and it causes suppurative lesions, including furuncles, felons, and sebaceous cysts, at a higher frequency than other CNS (1, 177). Many S. lugdunensis skin infections, particularly abscesses, occur in the perineal, inguinal, or pelvic girdle region (10, 81, 131, 177, 186).
There are three reports describing a total of seven cases of breast abscesses caused by S. lugdunensis in nonlactating women (4, 108, 194). Abscesses developed in two patients shortly after surgical procedures (108), whereas the remainder presented spontaneously.
Oral Infection
S. lugdunensis, as well as S. aureus and other CNS, has been isolated from patients with acute oral infections, including abscesses and osteomyelitis (200). S. lugdunensis, but not S. aureus, was isolated more frequently from patients with oral infection than from healthy subjects, suggesting that S. lugdunensis may be a pathogen when isolated from oral infections (200).
Ocular Infection
S. lugdunensis is an infrequent but significant pathogen in ocular infections. Nearly 6% (31/524) of CNS isolates recovered from the eyelids, anterior chamber fluid, or vitreous fluid of patients with postoperative endophthalmitis were S. lugdunensis, which was second only to S. epidermidis in terms of frequency of isolation (6). All pairs of S. lugdunensis intraocular isolates and eyelid isolates from single patients had identical pulsed-field gel electrophoresis profiles, indicating that the source of infection in these cases was the patient's skin flora (6). Similar results were observed in a more recent study (29), in which there were five cases (5.7%) of S. lugdunensis postoperative endophthalmitis among 87 documented infections. S. lugdunensis endophthalmitis is associated with a high rate of complications and a poor functional prognosis (29). S. lugdunensis has also been reported as a cause of suppurative keratitis (141).
Peritonitis
S. lugdunensis peritonitis after Caesarean section (60) or in patients undergoing continuous ambulatory peritoneal dialysis has been noted (111, 159). S. lugdunensis accounted for 2.3% of CNS recovered from 106 cases of continuous ambulatory peritoneal dialysis-associated peritonitis in 46 patients over a 2-year period (111). S. lugdunensis peritonitis closely resembles peritonitis caused by S. aureus, both of which may involve catheter tunnel abscesses (159).
UTI
Urinary tract infections (UTIs) caused by S. lugdunensis have been observed infrequently in adult and pediatric patients (24, 79). Haile et al. conducted a 3-month prospective study of 4,652 consecutive urine cultures to determine the frequency of detection of S. lugdunensis (79). Of 500 CNS cultured, 31 (6%) grew S. lugdunensis as part of mixed cultures; it was unclear to the authors whether these isolates were uropathogens or contaminants. S. lugdunensis isolated in pure culture was deemed the causative agent of UTI in a child (24).
Infections of the Central Nervous System
S. lugdunensis brain abscesses have been described in patients with dental infection and embolic native valve endocarditis (60, 71). A case of S. lugdunensis meningitis in a child subsequent to ventriculostomy was reported (88). Three blood cultures grew S. lugdunensis, leading the authors to suggest that community-acquired bacteremia may have resulted in the seeding of the cerebrospinal fluid.
Three S. lugdunensis ventriculoperitoneal shunt infections in pediatric and adult patients, of both early and late onset, have been reported (52, 157). In all three cases, the infected shunt was surgically removed. Shunt infections caused by S. lugdunensis may present like a shunt infection caused by S. aureus rather than like one caused by S. epidermidis (157).
Endocarditis
In 1993, Vandenesch et al. reported 11 cases of S. lugdunensis endocarditis, 8 of which involved native mitral and/or aortic valves (183). This report brought the number of documented cases of S. lugdunensis endocarditis in the literature to 20 during the 5 years following the original species description (183). Based on those cases, those authors concluded that S. lugdunensis is an aggressive pathogen when causing endocarditis, based on the findings that most patients had symptoms for less than 3 weeks, that there was a high degree of valve destruction commonly associated with abscess formation, that valve replacement was often required, and that the mortality rate was 70% (14/20 patients died) (183). A further review of several published cases of S. lugdunensis endocarditis also revealed a pattern of embolus formation (95).
In a prospective study of 912 consecutive infective endocarditis cases from a Madrid, Spain, hospital occurring between 1990 and 2003, S. lugdunensis accounted for 1.1% of cases overall or 1.5% of cases excluding endocarditis in intravenous drug users in whom there were no cases of S. lugdunensis endocarditis (2). Four cases of native valve endocarditis (0.8%), two cases of prosthetic valve endocarditis (1.5%), and four cases of pacemaker lead endocarditis (7.8%) were observed (2). Significantly more patients with S. lugdunensis endocarditis underwent surgery than did patients with S. aureus endocarditis (70% versus 37%, respectively; P < 0.04); surgical rates for S. epidermidis endocarditis were similar to those for S. lugdunensis. The mortality rate associated with S. lugdunensis endocarditis (50%) was significantly higher than those associated with S. aureus (14.5%; P < 0.01) and S. epidermidis (20%; P < 0.04). In a univariate analysis of data from 69 cases reported in the literature between 1988 and 2003, a diagnosis of S. lugdunensis endocarditis after 1995 was associated with decreased mortality (2).
More than 80 cases of endocarditis attributable to S. lugdunensis have been reported to date; these are summarized in Table 2. Further characteristics of S. lugdunensis native valve, prosthetic valve, and pacemaker-associated endocarditis are discussed separately below. It is noteworthy that, in addition to endocarditis, S. lugdunensis myocarditis has been described (142).
TABLE 2.
Reported cases of Staphylococcus lugdunensis endocarditis
| Authors, yr | Age (yr)/gendera | Comorbidity(ies) | Valve(s) | Suspected source | Outcome | Reference |
|---|---|---|---|---|---|---|
| Smyth et al., 1988 | 67/F | None | Native aortic | Not stated | Recovered | 169 |
| Etienne et al., 1989 | 72/F | None | Native aortic, mitral, tricuspid | Not stated | Died | 53 |
| 65/F | None | Native mitral | Cutaneous finger lesion | Recovered | ||
| 64/M | None | Prosthetic aortic | Not stated | Died | ||
| Fleurette et al., 1989 | 70/F | None | Not stated | Not stated | Died | 60 |
| 64/M | None | Prosthetic aortic | Not stated | Recovered | ||
| Walsh and Mounsey, 1990 | 32/M | None | Native aortic | Vasectomy 3 mo prior | Recovered | 195 |
| Barker et al., 1991 | 77/M | Congestive cardiac failure; rheumatic fever affecting mitral, aortic, and tricuspid valves | Native mitral | Not stated | Died | 7 |
| Cormican et al., 1992 | 42/F | Mastectomy | Prosthetic aortic | Not stated | Died | 37 |
| Sheppard and Jankowski, 1992 | 59/M | Rheumatic heart disease 13 yr prior | Prosthetic mitral, native aortic | Not stated | Died | 164 |
| Shuttleworth and Colby, 1992 | 60/M | Atherosclerotic disease, aortic aneurysm, end-stage renal disease | Native mitral | Hemodialysis catheter | Died | 166 |
| Schonheyder et al., 1993 | 55/F | Hypertension, mitral valve regurgitation, end-stage renal disease | Native mitral | Not stated | Died | 160 |
| Vandenesch et al., 1993 | 54/M | Not stated | Prosthetic aortic | Cheek infection | Died | 183 |
| 71/F | Not stated | Native mitral | Not stated | Recovered | ||
| 57/F | Not stated | Native mitral, aortic | Arm lymphangitis | Recovered | ||
| 81/F | Not stated | Native mitral | Toe infection | Died | ||
| 66/F | Not stated | Prosthetic aortic | Not stated | Recovered | ||
| 65/M | Not stated | Native mitral | Not stated | Died | ||
| 77/M | Not stated | Native mitral | Not stated | Died | ||
| 23/M | Not stated | Native mitral, aortic | Hickman catheter | Died | ||
| 69/F | Not stated | Native aortic | Not stated | Died | ||
| 37/M | Not stated | Native aortic | Not stated | Died | ||
| 74/M | Not stated | Prosthetic mitral | Pacemaker | Died | ||
| Breen and Karchner, 1994 | 72/F | Coronary artery disease | Native aortic | Coronary angioplasty | Recovered | 17 |
| Costello et al., 1995 | 76/M | Lymphoma | Native mitral | Not stated | Recovered | 38 |
| Kralovic et al., 1995 | 26/M | None | Native aortic | Dental procedure 3 mo prior | Recovered | 99 |
| Dupont et al., 1996 | 88/F | None | Native mitral | Not stated | Died | 50 |
| Koh et al., 1996 | 52/F | None | Native mitral | Not stated | Recovered | 95 |
| Lessing et al., 1996 | 34/M | None | Native aortic | Vasectomy 30 days prior | Recovered | 104 |
| 37/M | None | Native aortic | Vasectomy 21 days prior | Recovered | ||
| 42/M | None | Native aortic | Vasectomy 29 days prior | Recovered | ||
| 45/F | None | Native aortic, tricuspid | Inguinal furuncle 30 days prior | Recovered | ||
| De Hondt et al., 1997 | 33/F | Bicuspid aortic valve | Native aortic, mitral | Not stated | Recovered | 42 |
| Waterer et al., 1997 | 62/M | Rheumatoid arthritis, mitral valve regurgitation | Native mitral | Not stated | Died | 197 |
| Celard et al., 1997 | 41/M | None | Native tricuspid | Cardioverter defibrillator-associated infections for 5 yr | Recovered | 26 |
| Laguno et al., 1998 | 68/F | Pacemaker | Endocarditis on pacemaker leads | Pocket infection 1 year prior | Recovered | 101 |
| Llinares et al., 1998 | 70/F | None | Native mitral | Not stated | Died | 109 |
| 66/M | None | Native mitral | Not stated | Died | ||
| 60/M | None | Native aortic | Not stated | Died | ||
| Sanchis-Bayarri Vaillant et al., 1999 | 65/M | Rheumatic fever affecting mitral valve | Native mitral | Not stated | Recovered | 156 |
| Bobin et al., 1999 | 62/M | Pacemaker | Native tricuspid | Pacemaker insertion 1 mo prior | Recovered | 15 |
| 65/M | Diabetes mellitus, pacemaker | Endocarditis on pacemaker leads | Cutaneous effraction of a toenail 3 days prior | Recovered | ||
| Burgert et al., 1999 | 33/M | Bicuspid aortic valve | Native aortic | Not stated | Recovered | 19 |
| Fervenza et al., 1999 | 39/M | None | Native mitral | Vasectomy 2.5 mo prior | Recovered | 58 |
| Kamaraju et al., 1999 | 65/F | Type 2 diabetes mellitus, hypertension, end-stage renal disease, multiple vascular access infections | Native pulmonary | Not stated | Recovered | 89 |
| Wasserman et al., 1999 | 27/F | Pelvic inflammatory disease | Native mitral | Pelvic inflammatory disease 1 wk prior | Recovered | 196 |
| Kragsbjerg et al., 2000 | 79/M | Psoriasis, hypertension, rheumatoid arthritis, prosthetic knee, aortic and mitral valve regurgitation | Native mitral, aortic | Prosthetic joint infection 1 yr prior | Died | 98 |
| Patel et al., 2000 | 49/M | Renal transplant, Mitral valve prolapse | Native mitral | Not stated | Recovered | 134 |
| 85/F | Total knee arthroplasty | Native mitral | Prosthetic joint infection 16 mo prior | Recovered | ||
| 67/M | Rheumatic heart disease, cryptogenic cirrhosis | Native mitral | Not stated | Recovered | ||
| Polenakovik et al., 2000 | 55/M | Dyslipemia | Not stated | Arteriography via right inguinal area 2 wk prior | Recovered | 144 |
| Renzulli et al., 2000 | 51/M | None | Native aortic, mitral | Not stated | Recovered | 148 |
| Teong et al., 2000 | 22/M | None | Native aortic | Not stated | Recovered | 180 |
| Sanchez et al., 2000 | 71/F | Breast cancer, prosthetic hip failure | Native aortic | Not stated | Recovered | 155 |
| Farrag et al., 2001 | 78/F | None | Native mitral | Not stated | Died | 55 |
| Jones et al., 2002 | 16/M | Congenital aortic stenosis | Native aortic | Not stated | Recovered | 87 |
| Watchler et al., 2002 | 22/F | None | Native mitral | Not stated | Recovered | 193 |
| Sotutu et al., 2002 | 7/M | Congenital aortic stenosis | Native aortic | Not stated | Recovered | 171 |
| Garcia Fernandez et al., 2003 | 65/F | Breast cancer | Native mitral | Not stated | Died | 70 |
| Seenisavan and Yu, 2003 | 36/F | Cocaine abuse | Native mitral | Not stated | Recovered | 161 |
| Rodriguez-Gascon et al., 2003 | 77/F | Hypertension, diabetes mellitus, vulvar carcinoma | Native mitral | Not stated | Recovered | 149 |
| Petzsch et al., 2004 | 68/M | None | Native aortic | Not stated | Recovered | 139 |
| Anguera et al., 2005 | 77/F | Liver cirrhosis | Native mitral | Not stated | Died | 2 |
| 82/F | Ischemic heart disease | Not stated | Not stated | Died | ||
| 68/F | Pacemaker | Endocarditis on pacemaker leads | Not stated | Relapse 1 year later | ||
| 66/M | Pacemaker | Endocarditis on pacemaker leads | Not stated | Died | ||
| 78/M | Pacemaker | Endocarditis on pacemaker leads | Not stated | Recovered | ||
| 70/M | None | Prosthetic aortic | Not stated | Died | ||
| 77/M | None | Prosthetic aortic | Not stated | Recovered | ||
| 43/F | Congenital pulmonary stenosis | Native pulmonary | Not stated | Recovered | ||
| 37/M | None | Native aortic | Not stated | Died | ||
| 63/M | Pacemaker | Endocarditis on pacemaker leads | Not stated | Recovered | ||
| Van Hoovels et al., 2005 | 66/M | Pulmonary lobectomy | Native mitral | Not stated | Recovered | 187 |
| 78/F | Heart failure, gastric ulcers | Native mitral | Not stated | Died | ||
| 19/M | Aortic and mitral valve insufficiency | Native mitral and tricuspid | Not stated | Died | ||
| Seifert et al., 2005 | 61/M | Nephrectomy for cancer, pacemaker | Endocarditis on pacemaker leads | Battery replacement 3 mo prior | Recovered | 162 |
| Gianella et al., 2006 | 49/F | Bicuspid aortic valve | Native aortic | Not stated | Recovered | 71 |
| Sorli Redo et al., 2006 | 66/M | Chronic obstructive pulmonary disease | Native mitral | Not stated | Recovered | 170 |
| Kouberti et al., 2007 | 33/M | Congenital aortic bicuspid valve | Prosthetic aortic | Aortic valve replacement 40 days prior | Died | 96 |
| Viganego et al., 2007 | 75/M | Aortic valve sclerosis, type 2 diabetes mellitus, hypertension | Native aortic | Femoral endoarterectomy and femoral-popliteal bypass 7 mo prior | Died | 189 |
| Matthews et al., 2007 | 77/F | Coronary artery disease | Native mitral | Cardiac catheterization 3 wk prior | Recovered | 116 |
F, female; M, male.
Native valve endocarditis.
S. lugdunensis accounted for nearly 5% of 89 staphylococcal endocarditis isolates recovered from patients at our institution between 1980 and 1999 (134). All S. lugdunensis isolates originated from native valves, comprising 44% of the nine CNS in the collection causing native valve endcoarditis. Upon review of 69 published reports of S. lugdunensis endocarditis from 1988 through 2003, Anguera et al. reported that native valve endocarditis accounted for 77% of all cases (2). Of the native valve cases, the mitral valve was involved 55% of the time. The disease was characterized by acute onset (54% of cases) with cardiac failure, abscess formation, and embolization arising at rates of 45%, 19%, and 30%, respectively. Fifty-one percent of patients underwent surgery, which was associated with a mortality rate of 29%; the overall mortality rate was 42%.
Many reports of S. lugdunensis endocarditis describe cases occurring following surgical procedures or skin trauma in the pelvic region. There have been five reports of S. lugdunensis native valve endocarditis that developed 1 to 3 months following vasectomy in men ranging in age from 32 to 45 years (58, 104, 195). Significant valve damage occurred in all cases. Four patients underwent urgent valve replacement (104, 195), while valvular reconstruction was performed on the fifth patient following successful antimicrobial therapy (58). These cases suggest that S. lugdunensis endocarditis may be a rare complication of vasectomy. In addition, native valve infection has developed in patients following a scrotal wound (134), kidney transplantation (134), and femoral angiography or angioplasty (17, 116, 144, 189).
Three patients on chronic hemodialysis have developed S. lugdunensis endocarditis (89, 160, 166). In each case, the arteriovenous fistula or venous catheter was suspected to be the origin of infection. One patient developed native valve endocarditis of the pulmonary valve (89).
Prosthetic valve endocarditis.
Like other CNS, S. lugdunensis also causes prosthetic valve endocarditis (2, 37, 53, 60, 96, 164, 183). Thirteen percent of 69 cases of S. lugdunensis endocarditis cases between 1988 and 2003 involved prosthetic valves (2). The aortic valve was affected in over three-quarters of cases (2). Over half of patients underwent surgery; S. lugdunensis prosthetic valve endocarditis was associated with a 78% mortality rate (2). Abscess formation, pus, and significant tissue destruction commonly occur in S. lugdunensis prosthetic valve endocarditis (2, 37, 164, 183).
Pacemaker-associated endocarditis.
S. lugdunensis endocarditis due to infected pacemaker systems has been described (2, 15, 101, 162). Pacemaker-associated endocarditis was responsible for 10% of 69 S. lugdunensis endocarditis cases reported in the literature between 1988 to 2003 (2). Symptom onset is reported to be acute, and metastatic infection is common (15, 101). Infection with S. lugdunensis small-colony variants (SCVs) reportedly caused recurrent symptoms in one patient (162). Surgical removal of infected pacemaker systems, in addition to antimicrobial therapy, has been commonly used (2, 15, 101, 162) and is associated with a mortality rate of 14% (2).
Bone and Joint Infection
S. lugdunensis is a noteworthy cause of bone and joint infection. In a 4-year prospective study of the occurrence of CNS in patients undergoing orthopedic surgery for bone and joint infection, S. lugdunensis was isolated at a frequency of 3% in 212 CNS derived from 119 surgeries in 104 patients (167). Another study revealed that during a 40-month period, S. lugdunensis accounted for 1% of the 601 CNS obtained from patients with orthopedic clinical infections, including surgical wounds and infected prostheses (3). Temporal bone osteomyelitis and three cases of S. lugdunensis infective arthritis following surgical procedures have been reported (60, 118, 132, 181).
Vertebral osteomyelitis and disk space infection.
Multiple reports describe vertebral osteomyelitis due to S. lugdunensis (77, 121, 198). In one case, the source of infection was not obvious, although the patient was immunosuppressed (121). Infection in immunocompetent hosts has also been reported (77, 198). S. lugdunensis disk space infection was reported in a patient receiving chemotherapy for multiple myeloma (20), in a patient with osteoarthritis and bacteremia 1 month following foot surgery, and in an immunocompetent patient following disk surgery (78). A patient who developed S. lugdunensis spondylodiscitis several months after the replacement of a pacemaker battery (thought to be the source of infection) was successfully treated with antimicrobial therapy alone (18). Clinical manifestations of S. lugdunensis spine-related infections can be severe (20).
Prosthetic joint infection.
S. lugdunensis should be considered a pathogen when isolated from patients with prosthetic joint infection (63). In a prospective study, S. lugdunensis was the causative agent in 4% (3/79) of all prosthetic joint infection cases from patients undergoing hip or knee arthroplasty revision or resection (182). Four other cases of S. lugdunensis prosthetic joint infection have been communicated in the literature (154, 158, 198). S. lugdunensis prosthetic joint infection has been reported to present from 6 weeks to 4 years after implantation (154). Acute onset occurred in three of the four cases (154, 198).
CLINICAL MICROBIOLOGY
Species Description and Microbiological Characteristics
S. lugdunensis is a gram-positive, nonmotile, catalase-positive coccus (68). Cells are 0.8 to 1.0 μm in diameter and occur singly or in pairs, clusters, or short chains (68). S. lugdunensis is a facultative anaerobe that grows well in rich media at temperatures of between 30 and 45°C. Growth is sustained on 10% or 15% sodium chloride after incubation for 24 or 96 h, respectively (68). Colony morphology, pigmentation, and hemolysis vary among strains (60, 68). The species is coagulase negative (Fig. 1A), but a variable percentage of strains are positive for clumping factor (bound coagulase) when tested in a slide agglutination test with human plasma or various commercial latex agglutination kits (discussed further below) (5, 68). S. lugdunensis does not possess protein A (68, 135). Isolates are positive for acetoin production, nitrate reduction, and ornithine decarboxylase and pyrrolidonyl arylamidase (PYR) activities (Fig. 1B and C, respectively) but negative for oxidase and alkaline phosphatase (5, 68). S. lugdunensis is susceptible to novobiocin and variably resistant to polymyxin B (5, 80). Acid is produced under aerobic growth conditions from d-glucose, d-fructose, d-mannose, maltose, d-trehalose, α-lactose, sucrose, N-acetylglucosamine, and glycerol (5, 68). The cell wall peptidoglycan is composed of l-lysine-glycine5-6, and the cell wall teichoic acids contain glycerol, glucose, and glucosamine (68).
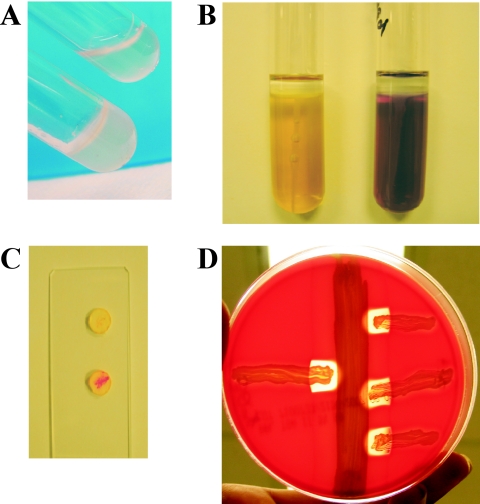
FIG. 1.
Key biochemical characteristics of S. lugdunensis. (A) Tube coagulase (long coagulase) test. S. aureus RN4220 (bottom) clots plasma (BBL rabbit coagulase plasma with EDTA; BD, Franklin Lakes, NJ), whereas S. lugdunensis IDRL-5258 (top) is unable to clot plasma. (B) Ornithine decarboxylase test. S. aureus RN4220 (left) and S. lugdunensis IDRL-5258 (right) were used to inoculate decarboxylase medium agar (BBL Moeller decarboxylase broth base; BD) containing ornithine and overlaid with mineral oil. S. lugdunensis demonstrates ornithine decarboxylase activity, causing a change in the medium's pH indicator (bromocresol purple) to a purple color, signifying a positive result. In contrast, a negative result (yellow) is obtained with S. aureus. (C) Rapid PYR test. S. lugdunensis IDRL-5258 (bottom) produces PYR, exhibiting a positive (pink) reaction in a rapid test (Remel, Lenexa, KS). S. aureus RN4220 (top) is PYR negative. (D) Synergistic hemolysis. The δ-like hemolysin of S. lugdunensis, mediated by three small peptides produced upon expression of the slush locus (see text for details), acts synergistically with the β-hemolysin of S. aureus to produce a zone of complete hemolysis on sheep red blood cells. S. lugdunensis IDRL-5258 (left) and S. lugdunensis IDRL-2414 (right, streaked in triplicate) are streaked perpendicular to, but not touching, S. aureus RN4220, which is streaked vertically. In panels A, B, and D, photographs were taken 18 h after inoculation and incubation in ambient air at 37°C. Panel C was photographed approximately 5 minutes after the rapid test was performed.
To date, the S. lugdunensis genome has not been sequenced. However, the base composition of the genome of the type strain ATCC 43809 (accession no. N860297) has been estimated by thermal denaturation experiments to be 32% G+C (68), which is comparable to the G+C compositions of sequenced S. aureus and S. epidermidis strains (5). Many strains carry one or more plasmids, some of which confer resistance to cadmium (54, 143). Genomic analyses by restriction endonuclease digestion and pulsed-field gel electrophoresis suggest a relatively low level of genomic variability among S. lugdunensis strains (54, 81, 107, 186).
Hemolysis.
S. lugdunensis may demonstrate hemolysis on blood agar containing rabbit erythrocytes and weak hemolysis after 2 days or more on blood agar containing sheep erythrocytes (60, 68). Additionally, a synergistic hemolytic phenotype resembling the activity of the S. aureus δ-hemolysin was observed in 73% of S. lugdunensis strains when streaked perpendicular to a β-hemolysin-producing staphylococcal strain (e.g., Staphylococcus intermedius) on blood agar containing rabbit erythrocytes (185). Similarly, in a different study, 95% of strains demonstrated a zone of complete hemolysis when streaked in proximity with a beta-hemolytic staphylococcal strain on blood agar containing sheep erythrocytes (Fig. 1C) (80).
Colony variation.
Variation in colony morphology and pigmentation among S. lugdunensis strains has been described (60, 68, 106, 162). In the original description of the species, colony diameters ranged from 1 to 4 mm (68). Colonies may have yellow to gold pigmentation after 3 to 5 days of incubation or may remain cream-colored or unpigmented (60, 68). Three of the 11 originally described strains exhibited two distinct colony morphotypes, displaying both smooth and glossy or rough and dull morphologies (68). Several colonial morphotypes subcultured from an apparent mixed staphylococcal culture taken from a patient with native valve endocarditis were subsequently all identified as S. lugdunensis (106). Subsequent subculturing of single colonies demonstrated colonial variation through three serial passages (106). In a collection of nine S. lugdunensis isolates, three displayed colony variation after 48 h, as well as after being subcultured and incubated for 24 h (106).
SCVs are a slow-growing, nonpigmented, and nonhemolytic subpopulation of staphylococci, particularly common in S. aureus, that may arise upon culturing of clinical specimens (190). SCVs often grow to only 1/10th of the size expected of a normal colony and present difficulty in identification and susceptibility testing due to their fastidious and auxotrophic growth characteristics (5, 190). S. aureus SCVs may have a defect in the electron transport chain (190). Hemin-auxotrophic SCVs of S. lugdunensis were isolated from thrombotic material scraped from pacemaker leads in a reported case of pacemaker-related endocarditis (162). In addition to the SCVs, the infecting strain produced at least three other colony morphologies that persisted upon serial subculturing (162).
Identification in the Clinical Microbiology Laboratory
CNS are not routinely identified to the species level in most clinical microbiology laboratories. Typically, cultures positive for staphylococci are tested to identify S. aureus, which in many cases can be simply determined with a slide or latex agglutination test for clumping factor and, depending if a commercial kit is used, protein A or other S. aureus-specific cell surface antigens. A negative result in a slide agglutination test may be followed with a tube coagulase test, which can confirm whether the organism is tube coagulase negative and therefore classifiable as a CNS. The enhanced virulence and destructive nature of S. lugdunensis are compelling reasons for the prompt identification of this organism to the species level when it is suspected during infection, especially when isolated from sterile sites. As discussed below, S. lugdunensis is easily identifiable with a relatively few biochemical tests, namely, tests for PYR activity, ornithine decarboxylase activity, and secreted coagulase (via a tube coagulase test). Caution should be exercised when testing staphylococci for clumping factor using rapid methods, as some isolates are clumping factor positive.
Biochemical profile for identification.
S. lugdunensis can be positively differentiated from other CNS by a negative tube coagulase test, a positive PYR reaction, and positive ornithine decarboxylase activity (Fig. 1). S. lugdunensis, along with S. haemolyticus, S. schleiferi, and S. intermedius, is PYR positive (5). While the reference method for staphylococcal PYR testing is with PYR broth, Fig. 1 shows testing of S. lugdunensis and S. aureus for PYR using the disk test that is commonly used for the identification of streptococci. Due to the pigmentation of some staphylococcal species, the PYR disk test may be difficult to interpret. In our experience, however, we have not encountered difficulties when identifying S. lugdunensis with this method. Positive and negative Staphylococcus control strains should be included when performing the PYR disk test.
S. lugdunensis is the only Staphylococcus species for which ≥90% of isolates are positive for ornithine decarboxylase; a positive result can be obtained in as soon as 8 h (5). It should be noted that a small number of S. epidermidis strains reportedly decarboxylate ornithine (5). S. lugdunensis can also be identified by the production of acid from trehalose, mannose, maltose, and sucrose but not from mannitol (5).
Several biochemical identification schemes to differentiate multiple CNS species, including other clumping factor-positive staphylococci, incorporate tests to accurately identify S. lugdunensis (43, 105, 159). Readers are referred to those publications for further information.
Variable presence of clumping factor.
In S. aureus, cell wall-associated clumping factor A is a protein adhesin for fibrinogen that mediates bacterial aggregation upon coming into contact with plasma (5). The initial description of 11 S. lugdunensis strains reported that all were positive for clumping factor when mixed with human plasma (68). However, only 58% were clumping factor positive with human plasma in a subsequent study of 31 isolates (60).
In addition to human plasma, there exist a large number of commercially available latex agglutination kits containing fibrinogen-coated particles to facilitate visible clumping reactions mediated by clumping factor (5). Some commercial kits have improved their sensitivity of detection of S. aureus by incorporating monoclonal antibodies that detect protein A, capsular polysaccharides, or other cell-associated antigens. It has been repeatedly shown that positive results for clumping factor vary significantly, depending on the type of kit used (67, 80, 183). While we detected clumping factor in only 13% (2/15) of S. lugdunensis isolates with the Staphaurex kit (Remel, Lenexa, KS) (67), other investigators have reported positive results ranging from 79% (30/38) with the BBL Staphyloslide kit (BD, Franklin Lakes, NJ) (80) to 82% (9/11) with the Staphyslide kit (bioMérieux, Marcy-l'Etoile, France) (183).
Side-by-side evaluations of S. lugdunensis strain collections in multiple commercial latex agglutination systems have been performed (135, 138). Paulsson et al. tested 11 strains of infectious or colonizing S. lugdunensis with human plasma and three commercial systems (135). Six of 11 strains were positive under all four conditions tested. The remaining five isolates were negative in all tests. A second study evaluated 30 S. lugdunensis isolates in six commercial agglutination kits (138). Positive results ranged from 7% to 60%, depending upon the ability of each kit to detect clumping factor, protein A, and other S. aureus-specific antigens.
The Manual of Clinical Microbiology states that latex agglutination methods for the detection of clumping factor in S. lugdunensis are not as reliable as detection with human plasma (5). However, the use of human plasma in clinical laboratories is discouraged unless it has been determined that it lacks infectious agents and is capable of clotting (5). Overall, this phenotype has varied widely among strains in published studies; results may depend on the testing method used. Collectively, the results of published studies indicate that testing for clumping factor in S. lugdunensis is not a dependable method for the identification of this species.
Identification with commercial kits or automated systems.
Many clinical laboratories employ commercial identification kits or automated instruments that allow quick determination of bacterial species. Numerous systems exist, and manufacturers continuously update their database systems to improve the accuracy of organism identification as new information becomes available. As would be expected, in the years immediately following the description of S. lugdunensis, several identification kits or systems were unable to accurately identify this organism due to incomplete or a general lack of information in the system databases (76, 80, 93, 166). Interestingly, the Microbial Identification System (MIDI, Newark, DE), which identifies organisms based on the analysis of their microbial cellular fatty acid compositions, correctly identified 26/26 S. lugdunensis strains as early as 1994 (172). The Manual of Clinical Microbiology gives the following list of systems that include S. lugdunensis in their databases (23): API-Staph, version 4.0 (bioMérieux); Vitek 2 GP, version 4.01 (bioMérieux) (the older Vitek Legacy did not include S. lugdunensis); MicroScan Conventional Pos ID (LabPro version 1.5) and Rapid Pos ID (LabPro version 1.6) (Dade MicroScan, Inc., West Sacramento, CA); BBL Crystal Gram-Pos ID and Rapid Gram-Pos ID (BD); Microbact Staph 12S (Oxoid); Biolog CP2 version 6.11/6.12 (Biolog, Hayward, CA); MIDI version 5.0 (MIDI); and Phoenix-100 GPID PMIC/II-100 (BD Diagnostics). In addition, the RapID Staph Plus (Remel, Lenexa, KS) correctly identifies S. lugdunensis. The databases of other systems that are not listed here may also include S. lugdunensis; users are advised to consult the manufacturer of their automated identification system for further information on this topic.
Species identification by molecular methods.
Methods to differentiate microorganisms by unique nucleic acid sequences are becoming more commonplace in the clinical microbiology laboratory due in part to increasing technological advances, including real-time PCR and high-throughput DNA sequencing systems. Many efforts particularly focus on the timely identification of pathogenic CNS. Several promising nucleic acid targets that provide accurate identification of S. lugdunensis have been identified and are discussed below.
The sequence diversity of the 16S rRNA genes of staphylococci enables species-level identification. The 16S rRNA gene represents a ubiquitous, highly conserved gene in which certain regions have accumulated changes during the evolution of individual species. By comparing the 16S rRNA sequence of an unknown organism to a sequence repository, such as the GenBank database maintained by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/GenBank/index.html), identification can usually be obtained. Thus, PCR amplification and sequencing of this gene have become an option for molecular identification of pathogenic bacteria in the clinical microbiology laboratory (33), and this technique was used to confirm the identity of a mecA-positive S. lugdunensis isolate upon its isolation from a bloodstream infection in a pediatric patient (178). Real-time PCR assays utilizing fluorescent resonance energy transfer probes that bind to regions of the staphylococcal 16S rRNA gene following amplification with broad-range primers have also been developed (168). Such methods may enable identification of organisms in a shorter time than is needed to amplify and sequence the 16S rRNA gene or other genes.
The internal transcribed spacer (ITS) regions of the prokaryotic rRNA gene locus, which show considerable variability among genera and species, serve to separate the 16S, 23S, and 5S genes and may contain coding sequences for tRNA genes. Staphylococci possess several nonidentical copies of the rRNA locus, and the ITS region of each copy may vary in length and sequence composition. PCR amplification of the ITS regions of a particular species' genome thus yields a polymorphic, species-specific banding pattern. In a study demonstrating the utility of PCR amplification of the ITS region to identify staphylococcal species, two S. lugdunensis strains produced banding patterns that were dissimilar from those of the other 28 staphylococcal species examined (39). In this regard, ITS-PCR provides a method that discriminates S. lugdunensis isolates from other CNS yet does not require sequencing. ITS-PCR banding patterns must be compared to banding patterns from reference strains in order to obtain a species identification, so inclusion of an adequate variety of reference strains is necessary for successful interpretation. Using this method, coupled with microchip gel electrophoresis for rapid analysis of PCR amplification products, S. lugdunensis and other staphylococci were successfully identified from blood culture bottles (69).
Ribotyping, the analysis of rRNA restriction fragment length polymorphisms, provides an alternative method for molecular differentiation of bacterial species. This technique analyzes differences over the entire rRNA locus and may be useful for epidemiological studies. An automated ribotyping system is commercially available (5) and has been used to evaluate collections of CNS isolated from blood or orthopedic prosthesis infections to determine its ability to correctly identify individual species (21, 22). Ribotyping correctly identified all 11 S. lugdunensis strains included in the two published studies (21, 22), indicating the validity of this technique for identification of this species. Interestingly, in both reports, S. lugdunensis strains displayed three unique ribotype patterns.
Sequences of several other genes, including hsp60, sodA, rpoB, and tuf, have proven useful as targets for molecular identification of CNS. A 600-base-pair region of the S. lugdunensis heat shock protein gene, hsp60 (also called groEL), amplified with universal primers, identified S. lugdunensis isolates with 100% accuracy in hybridization experiments (72, 73). Over 40 clinical staphylococcal isolates were tested in these experiments; none of the other species tested were falsely identified as S. lugdunensis (72, 73). Additionally, restriction fragment length polymorphism analysis of groEL following PCR amplification permitted identification of S. lugdunensis and 11 other Staphylococcus species (8). The sodA gene, encoding manganese-dependent superoxide dismutase, is present in and has been sequenced from approximately 40 CNS, including S. lugdunensis (145). The sodA sequences of CNS share less similarity with each other than do their 16S rRNA gene sequences, providing an alternative target for classifying closely related species (145). Sivadon et al. used this target to prospectively identify S. lugdunensis and other CNS causing bone and joint infections (167). Amplification and sequencing of a 751-base-pair region of the RNA polymerase β subunit gene rpoB and an 881-base-pair span of the elongation factor Tu (EF-Tu) gene tuf have also served to differentiate S. lugdunensis from other CNS (47, 113). Sequencing of the rpoB and 16S rRNA genes of several isolates recovered from a pediatric patient with meningitis facilitated identification of S. lugdunensis as the causative organism (88).
ANTIMICROBIAL SUSCEPTIBILITY
Prevalence of Antimicrobial Resistance in S. lugdunensis
S. lugdunensis, unlike most CNS, has remained remarkably susceptible to a wide array of antimicrobial agents. In 2007, we reported that 14 clinical isolates from a variety of sources were susceptible to 10 antistaphylococcal antimicrobial agents representing different drug classes (67). A 15th organism was resistant to trimethoprim-sulfamethoxazole but remained susceptible to the other nine agents tested. The distribution of antimicrobial susceptibilities of these isolates is summarized in Table 3.
TABLE 3.
Distribution of antimicrobial susceptibilities of Staphylococcus lugdunensis clinical isolates
| Antimicrobial agent and parametera | % of isolates with MIC or MBC at or below the following concn (μg/ml)b:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | |
| CFZ | |||||||||||||
| MIC | 20c | 67 | 100 | ||||||||||
| MBC | 7c | 40 | 73 | 93 | 100 | ||||||||
| DAP | |||||||||||||
| MIC | 21 | 50 | 93 | 100 | |||||||||
| MBC | 14 | 29 | 36 | 79 | 100 | ||||||||
| LZD | |||||||||||||
| MIC | 13 | 100 | |||||||||||
| MBC | 100d | ||||||||||||
| MXF | |||||||||||||
| MIC | 87 | 100 | |||||||||||
| MBC | 60 | 80 | 93 | 100 | |||||||||
| NAF | |||||||||||||
| MIC | 47 | 100 | |||||||||||
| MBC | 20 | 73 | 87 | 100 | |||||||||
| Q-D | |||||||||||||
| MIC | 7 | 27 | 60 | 80 | 87 | 100 | |||||||
| MBC | 7 | 13 | 53 | 67 | 73 | 100e | |||||||
| RIF | |||||||||||||
| MIC | 100f | ||||||||||||
| MBC | 20 | 40 | 47 | 53 | 100e | ||||||||
| TET | |||||||||||||
| MIC | 20 | 27 | 67 | 80 | 100 | ||||||||
| MBC | 100e | ||||||||||||
| SXTg | |||||||||||||
| MIC | 47 | 67 | 87 | 93 | 100h | ||||||||
| MBC | 20 | 27 | 40 | 100h | |||||||||
| VAN | |||||||||||||
| MIC | 27 | 87 | 100 | ||||||||||
| MBC | 7 | 20 | 100d | ||||||||||
CFZ, cefazolin; DAP, daptomycin; LZD, linezolid; MXF, moxifloxacin; NAF, nafcillin; RIF, rifampin; Q-D, quinupristin-dalfopristin; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; VAN, vancomycin.
Percentage of clinical S. lugdunensis isolates (n was 15 for all drugs except daptomycin, for which n was 14). S. lugdunensis isolates and the methods used for susceptibility testing were described previously (67).
MIC or MBC ≤ 0.125.
MBC > 128.
MBC > 32.
MIC ≤ 0.03.
SXT susceptibility values correspond to trimethoprim concentrations.
MIC or MBC > 16.
The trend that S. lugdunensis is generally susceptibility to multiple agents, including pencillins, does not change based on the source of infection. Eleven endocarditis isolates were susceptible to the following 19 antimicrobial agents: penicillin, oxacillin, streptomycin, kanamycin, gentamicin, tobramycin, erythromycin, lincomycin, pristinamycin, tetracycline, minocycline, chloramphenicol, perfloxacin, fusidic acid, vancomycin, teicoplanin, fosfomycin, rifampin, and co-trimoxazole (183). Fifteen bloodstream isolates exhibited susceptibility to many common antimicrobials, including penicillin (51). Fifteen S. lugdunensis isolates from diabetic foot infections were susceptible to eight out of nine agents tested, including ceftobiprole, a new broad-spectrum cephalosporin (74).
In a study of non-S. aureus, non-S. epidermidis staphylococci from orthopedic infections, S. hominis, S. capitis, S. haemolyticus, and S. warneri exhibited high rates of resistance (51 to 66% of isolates) to penicillin, ampicillin, cefazolin, and cefamandole (3). In contrast, of eight S. lugdunensis isolates studied, only three were resistant to the same β-lactams. In addition, one S. lugdunensis isolate was resistant to erythromycin. There have also been single reports of S. lugdunensis isolates resistant to streptomycin (86), tetracycline (183), penicillin (183), gentamicin (83), ceftazidime (74), and aminoglycosides and macrolides (60). A single case report described the emergence of resistance to rifampin and ciprofloxacin that correlated with treatment of a persistent S. lugdunensis infection that manifested over 3 years as septic arthritis of both knees, vertebral osteomyelitis, and aortic and mitral valve endocarditis (98).
There has been only one reported observation of S. lugdunensis isolates that are resistant to multiple antimicrobial agents (200). Six S. lugdunensis oral infection isolates collected in Korea were resistant to ampicillin, penicillin, cephalothin, oxacillin, and clindamycin (200). Three of the isolates were also resistant to erythromycin and/or gentamicin, and three harbored plasmids (although it was not determined whether the plasmids contributed to the antimicrobial resistance phenotypes). These organisms are unusual in comparison with other reported collections of S. lugdunensis.
Some S. lugdunensis strains exhibit resistance to the translation-inhibiting antimicrobial agent fusidic acid, which prevents ribosomal release of EF-G. Fusidic acid-resistant S. lugdunensis strains have acquired a chromosomal fusB resistant determinant that encodes an EF-G-binding protein located downstream of groEL in the genome (130).
Frequencies of β-Lactamase and mecA
The frequency of β-lactamase in S. lugdunensis is reported to differ between isolates from North American and European countries. This percentage ranges from 7 to 24% in publications originating from French laboratories (60, 186). In contrast, three separate collections of S. lugdunensis isolates from laboratories in the United States showed rates of 24%, 29%, and 40% (67, 80, 82). A Spanish study and a Swedish study reported rates of 12% and 15% β-lactamase-positive S. lugdunensis isolates, respectively (81). As expected, β-lactamase-positive S. lugdunensis isolates demonstrate resistance to penicillin and other related antimicrobial agents.
In accordance with the overwhelming antimicrobial susceptibility exhibited by S. lugdunensis isolates, this organism has generally remained susceptible to oxacillin. PCR screening for mecA in large collections of S. lugdunensis has repeatedly yielded negative results (57, 67, 81, 85, 110). In addition to screening S. lugdunensis for oxacillin resistance by mecA PCR, a slide latex agglutination test to detect the presence of PBP2a or Mueller-Hinton agar supplemented with 4% NaCl containing 6 μg/ml oxacillin may also be used (110, 115, 199).
Only two descriptions of mecA in S. lugdunensis presently exist in the English literature (9, 178). A mecA-positive strain identified as S. lugdunensis by conventional phenotypic, automated, and molecular methods was reported as causing a bloodstream infection in a premature neonate with an intravascular catheter in Singapore in 2003 (178). No zone of inhibition was observed with a 5-μg methicillin disk, and the oxacillin Etest MIC was >256 μg/ml. The mecA gene was detected by PCR, and the MRSA-Screen latex agglutination test was positive after induction of expression of the oxacillin resistance gene. The patient was successfully treated with vancomycin. Additionally, Becker et al. reported the isolation of a mecA PCR-positive S. lugdunensis nasal colonizing strain, but no further characterization was performed (9).
Breakpoints for Oxacillin Resistance
In 1999, NCCLS (now the Clinical Laboratory and Standards Institute [CLSI]) lowered the oxacillin resistance breakpoints for CNS from ≥4 μg/ml to ≥0.5 μg/ml in order to improve the sensitivity of identification of mecA-positive isolates as oxacillin resistant (122). Subsequently, Hussain et al. demonstrated that while the revised breakpoints accurately classified several mecA-positive species of CNS (i.e., S. epidermidis, S. hominis, and S. haemolyticus) as oxacillin resistant, S. lugdunensis isolates with MICs of 0.5 to 2 μg/ml but lacking the mecA gene, as detected by PCR, were falsely categorized as being oxacillin resistant (85). A patient with native valve endocarditis caused by an S. lugdunensis mecA-negative isolate, which was classified as oxacillin resistant due to its oxacillin MIC of 1 μg/ml, was successfully treated with ceftriaxone, suggesting that the breakpoints for oxacillin resistance in CNS were not appropriate for S. lugdunensis (134). NCCLS later recommended that S. lugdunensis isolates should be screened for oxacillin resistance by detection of PBP2a by latex agglutination or mecA by PCR (123). Finally, in 2005, the oxacillin breakpoints for S. lugdunensis were revised once more to follow those set for S. aureus, which should more accurately predict mecA-negative S. lugdunensis isolates (34). Currently, S. lugdunensis strains showing oxacillin MICs of ≤2 μg/ml are considered susceptible, whereas those showing oxacillin MICs of ≥4 μg/ml are classified as resistant (34).
Vancomycin Tolerance
Vancomycin, a glycopeptide antimicrobial agent, exerts bactericidal activity against staphylococci. It is generally recommended that vancomycin be reserved for situations in which other antimicrobials are not viable treatment options. A phenomenon known as vancomycin tolerance, in which organisms with MICs indicating susceptibility are refractory to killing in bactericidal killing assays, has been documented in S. aureus (117, 136) and has recently been recognized among S. lugdunensis isolates (16, 67). In our collection of vancomycin-susceptible (MIC range, 0.5 to 2 μg/ml) S. lugdunensis clinical isolates, 93% (n = 15) demonstrated tolerance to vancomycin, as defined by a minimal bactericidal concentration (MBC)/MIC ratio of ≥32 (67). Vancomycin MBCs for 12 isolates were >128 μg/ml (67). Bourgeois et al. reported similar findings when the bactericidal activities of vancomycin and teicoplanin, a related glycopeptide, were tested against clinically significant S. lugdunensis isolates using time-kill curve methodology (16). They found that 6/13 S. lugdunensis isolates were tolerant to vancomycin or teicoplanin, including three organisms that were tolerant to both antimicrobial agents (16). The killing capacities of vancomycin and teicoplanin against the 13 S. lugdunensis isolates were reduced compared to those against 77 other CNS, despite the susceptibility of the isolates to both antimicrobial agents (16).
Optimal treatment of bacterial endocarditis is considered to require a bactericidal antimicrobial regimen. Cases of S. lugdunensis native valve endocarditis have been successfully treated with vancomycin, usually in combination with aminoglycosides or rifampin (2, 161, 183). The general susceptibility of S. lugdunensis to the β-lactams usually precludes the necessity to rely on vancomycin therapy. The observation that S. lugdunensis glycopeptide tolerance may be widespread is concerning and warrants further in vitro and in vivo studies to delineate whether this is a clinically significant finding.
Response of S. lugdunensis Biofilms to Antimicrobial Treatment
Most infections caused by CNS are attributable to the formation of biofilms, surface-associated multicellular communities of microorganisms that surround themselves in a self-produced extracellular polymeric matrix, on host tissues or indwelling medical devices (191). Bacterial biofilms exhibit high levels of resistance to antimicrobial therapies and evade host immune defenses, making biofilm-related infections extremely difficult to treat. Several types of documented S. lugdunensis infection derive from a biofilm etiology, and the clinical features and pathogenesis of these infections are covered elsewhere in this review. Here we discuss our present understanding of the response of S. lugdunensis biofilms to antimicrobial agents.
We investigated the ability of 10 antistaphylococcal agents at concentrations traditionally tested in MIC assays to significantly reduce the number of bacteria recovered from biofilms of 15 S. lugdunensis isolates (67). The biofilm bactericidal concentration (BBC) assay, which measures the amount of regrowth in recovery media following the exposure of biofilms to antimicrobial agents, revealed that the BBC90s for all drugs tested were considered to indicate resistance by the breakpoints for planktonic organisms set forth by the CLSI. Seven drugs had BBCs of ≥128 μg/ml, the highest concentration tested in the assay. Interestingly, the BBC range of the quinolone moxifloxacin was ≤0.125 to 2 μg/ml, and BBCs of 73% of biofilms could be considered to indicate susceptibility by the planktonic breakpoints for this drug.
The activity of sodium metabisulfite, a commonly encountered preservative and antioxidant in intravenously administered pharmaceuticals, was tested against staphylococcal biofilms using an in vitro model of biofilm formation (64). Sodium metabisulfite (0.72 mg/ml) caused only a 1.4 log10 drop in S. lugdunensis viable biofilm cell counts following 24-hour treatment of established biofilms. However, the same concentration of sodium metabisulfite prevented S. lugdunensis biofilm formation in an in vitro microtiter plate biofilm assay, suggesting that sodium metabisulfite may be effective at preventing, but not treating, S. lugdunensis biofilms.
Since subinhibitory concentrations of various antimicrobial agents enhance or impair biofilm formation by S. epidermidis (49, 146), we performed similar studies using a microtiter plate biofilm formation assay to define the effects of 10 antistaphylococcal antimicrobials at subinhibitory concentrations on S. lugdunensis biofilms (67). Tetracycline, which enhances S. epidermidis biofilm formation (146), exerted a negative effect on biofilm formation by 93% of the S. lugdunensis isolates tested. In contrast, the β-lactam nafcillin significantly increased biofilm formation by 93% of the organisms. Considering the widespread susceptibility of S. lugdunensis strains to β-lactam agents, which makes these drugs preferred options for treating S. lugdunensis infections, this result is concerning and deserving of further investigation with in vivo studies. Linezolid also caused a decrease in biofilm formation by 80% of the isolates. The effects of several of the other antimicrobial agents varied among strains, although cefazolin, daptomycin, and rifampin did not substantially affect biofilm formation by most of the isolates.
Cadmium Resistance
Widespread resistance to cadmium and arsenate was reported soon after the description of S. lugdunensis (60). In a collection of 35 S. lugdunensis strains from different European cities, 20 were found to carry plasmids that conferred resistance to cadmium at concentrations of ≥125 μg/ml (143). A 3.2-kb plasmid called pLUG10 is the most frequently occurring plasmid in cadmium-resistant S. lugdunensis strains (28, 143). In S. aureus, the plasmid pOX6 carries a cadmium resistance gene, cadB, which is hypothesized to encode a membrane-associated cadmium-binding protein (137), in association with cadX, which encodes a putative transcriptional activator (28). Probes generated from the S. aureus cadB gene hybridized to pLUG10 DNA in Southern blot analyses, suggesting that cadmium resistance mechanisms were similar in these two species (143). Subsequent sequence analysis of pLUG10 revealed that the 3,117-base-pair plasmid contains two open reading frames with predicted amino acid sequences that share high degrees of identity with CadB and CadX from pOX6 (28). Inactivation of either cadB or cadX in pLUG10 decreases bacterial resistance to cadmium, indicating that both gene products are necessary for full expression of cadmium resistance (28). When transformed into S. aureus strain RN4220, pLUG10 replicates and confers resistance to cadmium at levels commensurate with those when the plasmid is present in S. lugdunensis (28). Interestingly, despite the regions of homologous cadmium resistance between the two plasmids, pLUG10 is a member of the plasmid pT181 family (127) and pOX6 belongs to the distinct family of pC194 plasmids.
The S. aureus plasmid pRW001 also carries a cadmium resistance cassette composed of two genes, cadD and cadX* (41). The predicted CadD protein is 84% identical to CadB from pLUG10, and CadX* represents a truncated peptide that is 86% identical to the positive regulatory protein CadX from pLUG10. The cadmium MIC for S. aureus carrying pRW001 alone was 20 μg/ml, but introduction of cadX from pLUG10 on a second plasmid was found to increase the cadmium MIC to >150 μg/ml, indicating that full-length cadX complemented the nonfunctional cadX* found on pRW001 (41). The cadD-cadX* resistance determinant of pRW001 is located on a 3.9-kb DNA fragment flanked by direct repeats of the insertion sequence element IS257, leading to the hypothesis that this genetic element passed from pLUG10 to pRW001 through a recombination event that resulted in truncation of the C-terminal region of cadX (41).
Although the clinical relevance of cadmium resistance in S. lugdunensis is unclear, in S. aureus, genes for cadmium resistance and β-lactamase have been found together on plasmids (114, 128). In particular, genes related to cadD and cadX of plasmid pRW001 are present in several S. aureus β-lactamase resistance plasmids that demonstrate cadmium resistance (114). In contrast to the case for pRW001, however, this cadX homologue is present as a full-length gene and IS257 sequence elements are absent, suggesting that the recently described family of plasmids acquired the cadmium resistance markers through an alternate evolutionary pathway (114).
PATHOGENESIS AND VIRULENCE FACTORS
Toxins, Hemolysins, and the agr Locus
Efforts to identify toxins in S. lugdunensis similar to those produced by S. aureus, including enterotoxins A to E, toxic shock syndrome toxin 1, and exfoliatin A, have been unsuccessful (60, 133), despite a case of S. lugdunensis toxic shock syndrome having been reported (133).
Hemolytic activity due to α-, β-, or γ-hemolysins has not been detected in S. lugdunensis (185). Further, probes for S. aureus hemolysins did not hybridize to S. lugdunensis genomic DNA in Southern blotting experiments (185). Most S. lugdunensis isolates produce a heat-stable δ-like hemolysin that shares phenotypic properties with the S. aureus delta-toxin, which is encoded by the hld gene (185). In S. aureus, hld is part of the agr locus; hybridization experiments have demonstrated the presence of a sequence similar to hld in S. lugdunensis (185).
agr locus.
The staphylococcal agr (accessory gene regulator) locus is a quorum-sensing system that acts as a global regulator of virulence factors, particularly secreted exoproteins, including enterotoxins, hemolysins, and numerous host protein-modifying enzymes (126). Regions of homology to S. aureus agr have been identified by PCR or Southern blotting in a large number staphylococcal species, including S. lugdunensis (46, 48). agr loci from S. lugdunensis, S. epidermidis, S. saprophyticus, and the coagulase-positive veterinary pathogen S. intermedius, have been characterized in detail (Table 4) (153, 175, 184, 188). The locus is comprised of two divergent transcripts, RNAII and RNAIII, which are expressed from promoters P2 and P3, respectively (126). RNAII encodes four genes, agrB, agrD, agrC, and agrA, whose protein products are the machinery of a two-component signal transduction system (126). The propeptide AgrD is processed into a small peptide that serves as the two-component system autoinducer by the membrane protein AgrB. AgrB secretes the autoinducer peptide (AIP), which in turn binds to AgrC, the transmembrane histidine kinase signal transduction component of the two-component system. Upon AIP binding, AgrC phosphorylates the DNA-binding protein AgrA. Extracellular accumulation of AIP to a critical quorum threshold leads to AgrA activation of expression of promoters P2 and P3. In the S. aureus agr locus, promoter P3 directs transcription of a 517-nucleotide transcript called RNAIII, which acts as an intracellular effector to upregulate transcription of extracellular protein genes and downregulate that of surface protein genes, and contains the hld gene, encoding the 26-amino-acid delta-toxin (126). With the exception of the nonhemolytic S. saprophyticus and S. lugdunensis (discussed below), the hld locus is located within the RNAIII transcription unit (153, 175, 184, 188).
TABLE 4.
Staphylococcus lugdunensis virulence factors
| Virulence factor | Gene name | Description | Homologue(s) in other species | Reference(s) |
|---|---|---|---|---|
| Accessory gene regulator system (agr) and RNAIII | agr locus | Quorum-sensing system that acts as a global regulator of virulence factors | S. aureus, S. epidermidis, S. saprophyticus, S. intermedius agr locus | 11, 46, 48, 126, 153, 175, 184, 188 |
| SLUSH-A, SLUSH-B, SLUSH-C hemolytic peptides | slush locus | Hemolytic peptides with delta-toxin-like activity | S. haemolyticus, S. cohnii subsp. cohnii, S. cohnii subsp. urealyticum, S. caprae, S. xylosus slush-like sequences | 45, 46, 100, 176 |
| OatA peptidoglycan O-acetyltransferase | oatA | Membrane-bound enzyme that confers resistance to lysozyme by O-acetylating cell wall N-acetylmuramic acid and preventing lysozyme binding | S. aureus oatA | 12, 13 |
| vWf-binding protein vWbl | vwbl | Mediates interaction with vWf-expressing host cells, including platelets and endothelial cells; contains an RGD motif | No sequence similarity with known proteins | 125 |
| Fibrinogen-binding protein Fbl | fbl | Facilitates binding to fibrinogen in the host, member of the Sdr (SD repeat) family of Staphylococcus surface proteins | S. aureus clumping factor A (cflA) | 120, 124 |
| Biofilm formation | ||||
| PNAG/PIA extracellular matrix synthesis genes | icaADBC locus | Biosynthetic enzymes of a β-1,6-linked N-acetylglucosamine polysaccharide polymer commonly found in the extracellular matrix of staphylococcal biofilms | S. aureus, S. epidermidis, S. caprae icaADBC | 30, 66 |
| Biofilm extracellular matrix protein(s) | Unknown | Components of the biofilm extracellular matrix | Unknown | 27, 66, 94, 152 |
The S. lugdunensis agr locus is an actively transcribed genomic region that was initially identified by its homology to S. aureus RNAIII in S. lugdunensis isolates demonstrating synergistic hemolytic activity (184). Sequence analysis of the S. lugdunensis agr gene shows it to be ∼63% identical to the S. aureus agr locus, with a similar genomic organization, including promoters P2 and P3, agr-rnaIII, a highly conserved intergenic region, and open reading frames matching agrB, agrD, agrC, and agrA (184) (GenBank accession no. AF173933). Despite the synergistic hemolytic phenotype frequently associated with S. lugdunensis isolates, which was expected to be mediated by a δ-hemolysin homologue, no open reading frames with similarity to the S. aureus hld gene are located in the S. lugdunensis sequence for RNAIII. Further, in contrast to the observation that RNAIII sequences of other CNS are highly homologous with S. aureus RNAIII (179), the homology between the RNAIIIs of S. aureus and S. lugdunensis is rather low, with the highest regions of conservation occurring at the 5′ and 3′ ends (11). Nonetheless, full-length S. lugdunensis RNAIII was shown to be transcribed at a level equivalent to its native background when heterologously expressed in S. aureus from its endogenous promoter (11). This expression pattern was shown to be agr dependent, and S. lugdunensis RNAIII was capable of transcriptional and phenotypic complementation of several agr-regulated exoproteins in an agr-null S. aureus strain (11). These studies indicate that the absence of the hld gene does not impair the ability of S. lugdunensis RNAIII to function as a regulatory molecule.
slush locus.
Following the finding that the S. lugdunensis δ-like hemolysin is not encoded within the agr locus (184), Donvito et al. purified three small exoproteins that exhibited synergistic hemolytic activity with S. aureus on sheep blood agar (45). Sequencing of the peptide fragments following trypsin digestion generated highly related 10- or 23-amino-acid sequences for each of the three peptides. A degenerate DNA probe based on the amino acid sequence of one of the tryptic fragments was hybridized to S. lugdunensis restriction enzyme-digested genomic DNA, and the corresponding hybridizing fragment was subsequently cloned and sequenced in order to determine the genetic locus responsible the synergistic hemolytic peptides. The locus, denoted slush for S. lugdunensis synergistic hemolysin, contains three open reading frames encoding 43-amino-acid peptides of high similarity, called SLUSH-A, SLUSH-B, and SLUSH-C, which are identical to the sequenced hemolytic peptide fragments (Table 4). An additional open reading frame, ORF-X, encoding a predicted 24-amino-acid peptide of unknown function and lacking sequence similarity to the slush open reading frames, lies upstream of slush-A. The slush locus was detected in 14/14 S. lugdunensis strains by Southern blotting, indicating genetic conservation of the locus among strains of this species (46). The same probe also hybridized to DNAs from 4/4 strains of S. haemolyticus, S. cohnii subsp. cohnii, and S. cohnii subsp. urealyticum and to those of some strains of S. caprae (3/4) and S. xylosus (1/4) (46). No sequences bound to the slush probe when hybridized against S. aureus and 11 other CNS species or subspecies (46). Peptides with high degrees of identity to SLUSH-A (∼45 to 65%) are found in the genomes of other pathogenic CNS species, S. saprophyticus and S. haemolyticus (100, 176). Transcriptional regulation of the slush locus may be under control of the agr locus (46).
Resistance to Lysozyme
Lysozyme is an essential enzymatic component of the human innate immune system that defends against microbial infection. As reported in the species description, S. lugdunensis is resistant to 400 mg/ml lysozyme (68). Several other pathogenic staphylococci, including S. aureus, display resistance to high concentrations of lysozyme, whereas nonpathogenic staphylococci remain sensitive or display hypersensitive reactions (12). In S. aureus, lysozyme resistance is mediated by expression of a membrane-bound peptidoglycan O-acetyltransferase (OatA), which O acetylates cell wall N-acetylmuramic acid, preventing binding of lysozyme (13). S. lugdunensis possesses an oatA homologue in its genome and O-acetylated peptidoglycan in its cell wall (Table 4), indicating that the mechanism of lysozyme resistance in S. lugdunensis is likely the same as that in S. aureus (12).
Experimental Animal Studies
Lambe et al. tested the ability of six strains of S. lugdunensis to induce abscess formation in a mouse model of foreign-body infection (102). Pieces of silicone rubber catheter preadhered with each test strain were subcutaneously implanted in mice that subsequently received a subcutaneous injection with the test strain. After 7 days, the six S. lugdunensis strains collectively induced abscess formation in 76% of mice. S. lugdunensis was cultured from 97% of the explanted foreign bodies and/or the surrounding tissues. The results obtained with S. lugdunensis (76% ± 19% abscess formation) did not statistically vary from the collective results obtained from nine S. epidermidis strains (91% ± 13% abscess formation) in the same model, suggesting that the ability of S. lugdunensis to infect foreign bodies is similar to that of S. epidermidis.
To further investigate the relationship of the foreign body to S. lugdunensis virulence, Ferguson et al. used the same mouse model described above but compared abscess formation with and without the presence of the foreign body (56). Abscess formation and positive culture of the surrounding tissues were statistically higher when a foreign body was present for 4/5 S. lugdunensis strains tested. Similar results were obtained with 3/5 S. epidermidis strains examined. S. epidermidis induced abscess formation in the absence of a foreign body at a rate of 11 to 88%, whereas abscess formation in the absence of a foreign body occurred in 8 to 46% of infections induced by the five S. lugdunensis strains, attesting to the ability of S. lugdunensis to cause diseases on par with those of other pathogenic CNS. Virulence was enhanced with most tested strains when a foreign body was involved, suggesting that biofilm formation on implanted foreign devices is a virulence mechanism used by S. lugdunensis.
The pathogenicity of a single S. lugdunensis strain was also tested in a murine model of implant-associated peritoneal infection (151). Although the strain was essentially unable to adhere to the surface of the implant material in vitro, it was found to persist in the peritoneums of normal or neutropenic mice with peritoneal implants at statistically higher numbers than in normal mice without implants. As observed with two strains of S. epidermidis and single strains of S. haemolyticus and S. scheiferi, high numbers of S. lugdunensis organisms disseminated to the liver and kidneys in neutropenic mice with implants, further confirming the generally virulent nature of S. lugdunensis.
Adherence Proteins
The ability of S. lugdunensis to cause endocarditis and foreign-body-associated infections suggests that the organism is proficient at interacting with host tissues and proteins that may coat foreign surfaces following implantation. Paulsson et al. undertook an investigation to identify cell surface factors of S. lugdunensis that facilitate binding to proteins commonly encountered in host tissues (135). Of the soluble proteins tested, S. lugdunensis strains exhibited the highest percentage of binding to purified collagen types I and IV and immunoglobulin G, although less than 40% of the total protein was bound in each case (135). S. lugdunensis strains bound fibrinogen, laminin, vitronectin, fibronectin, thrombospondin, and plasminogen at lower levels (∼8 to 19% bound protein). Strains variably bound to immobilized forms of collagen type I, fibrinogen, and vitronectin. Primers designed to anneal to the S. aureus fibronectin-binding protein gene (fbn) were used to successfully amplify products of the expected size, based on the S. aureus fbn sequence, in three clinical isolates of S. lugdunensis, thereby suggesting the genomic presence of a fibronectin-binding protein gene similar to that found in S. aureus (119).
vWf-binding protein.
A protein that specifically binds von Willebrand factor (vWf) was identified from the affinity selection of a S. lugdunensis phage display library against vWf (Table 4) (125). vWf is a blood plasma glycoprotein produced by platelets and endothelial cells that is involved in several facets of coagulation, including binding to platelets and subendothelial collagen following vascular injury and stabilizing factor VIII of the clotting cascade. The vwbl gene, found in all clinical isolates tested, encodes the 2,060-amino-acid S. lugdunensis vWf-binding protein (vWbl). The protein sequence contains a predicted N-terminal signal peptide that, upon cleavage, results in a 226-kilodalton, 2,013-amino-acid mature protein (125). The 1,255 amino acids following the signal peptide, termed the A region, contain an arginine-glycine-asparagine (RGD) motif. RGD motifs are most often found in integrin-binding proteins, although the physiological relevance of the motif in this protein is unknown. The A region is followed by a series of 10 67-amino-acid repeats that are similar to one another and house the minimal vWf-binding domain of the protein. The C-terminal portion of vWbl likely anchors the protein in the cell membrane. Although S. aureus also possesses a vWf-binding protein (14), vWbl shares no similarity with any other known proteins (125). Interestingly, binding of vWbl to recombinant vWf could be inhibited by the S. aureus vWf-binding protein (125), suggesting that both proteins interact with vWf at the same site.
Fibrinogen-binding protein.
Two concurrent reports described and characterized a fibrinogen-binding protein called Fbl in S. lugdunensis (Table 4) (120, 124). Staphylococcal binding to fibrinogen, the precursor molecule of fibrin, is well documented (62). Fbl is an 881-amino-acid protein encoded by fbl, which was found in all 30 S. lugdunensis strains screened (120, 124). The full-length polypeptide contains an N-terminal signal sequence followed by the N1, N2, and N3 domains that share significant homology (about 60% identity) with S. aureus clumping factor A (ClfA) (120, 124). The fibrinogen-binding region is located within the N2 and N3 domains between residues 40 and 534 (120). Fbl also contains a repetitive SD repeat region between the N3 domain and the C-terminal cell wall anchoring sequence, indicating that Fbl belongs to the large Sdr (SD repeat) family of structurally related staphylococcal surface-associated proteins (62). S. lugdunensis Fbl, whether recombinantly purified from Escherichia coli (124) or expressed on the cell surface of Lactococcus lactis (120), specifically binds fibrinogen, and this interaction can be disrupted with antibodies raised against S. aureus ClfA, confirming the high degree of similarity between ClfA and Fbl and the importance of Fbl in mediating S. lugdunensis binding to fibrinogen. Further, Fbl and ClfA interact with the same general region of fibrinogen, although ClfA binds fibrinogen with a 10-fold-greater affinity than Fbl (120).
Taken together, these studies provide evidence that S. lugdunensis possesses surface-associated proteins that enable bacterial adherence to a variety of host cells and tissues, as well as foreign body surfaces, a critical step in the process of staphylococcal biofilm formation (75). The ability of S. lugdunensis to bind fibrinogen and vWf may contribute to this organism's success in causing endocarditis.
Biofilm Formation
Biofilm formation by CNS, referred to as “slime production” in early reports, has long been associated with virulence (31), although it is now understood that commensal or environmental isolates can also form biofilm. Adherence capabilities of CNS and S. aureus can be quickly and easily assessed in the laboratory by evaluating whether organisms produce a stainable film on the walls of the growth chamber following incubation (31, 32). Initial studies revealed “slime production” on polystyrene conical tubes by 8/31 S. lugdunensis colonizing, infectious, or environmental isolates (60) and on glass tubes by only 4/38 S. lugdunensis clinical isolates (80). Despite the relatively low frequency of in vitro biofilm formation originally observed in these two collections of organisms, these studies indicated that some S. lugdunensis isolates have the potential to adhere to inert surfaces. We confirmed that most organisms in a collection of clinical isolates form in vitro biofilm on polystyrene surfaces (66). Scanning electron microscopy of a 24-h biofilm formed by an S. lugdunensis folliculitis isolate on a silicone elastomer surface revealed a complex architecture of cells that appeared to be embedded in an extracellular substance (Fig. 2A). When these data are coupled with the known types of infections caused by S. lugdunensis and the knowledge that host surface adherence proteins are present in S. lugdunensis, it is reasonable to propose that, like for most other pathogenic staphylococci, biofilm formation is one of the virulence mechanisms employed by this organism (Table 4).
FIG. 2.
Microscopic visualization of S. lugdunensis biofilms. (A) Scanning electron micrograph (magnification, ×11,000) of S. lugdunensis IDRL-2640 biofilm formed on a silicone elastomer disk (Bentec Medical, Woodland, CA) after 24 h. Nonadherent cells were rinsed from the disk following incubation, and the disk was placed in Trumps fixative (4% formaldehyde and 1% glutaraldehyde in phosphate buffer, pH 7.3). Images were acquired on a Hitachi S-4700 instrument (Hitachi Ltd., Tokyo, Japan) by cold-field emission microscopy following critical-point drying and gold-palladium sputter coating. Bar, 5 μm. (B) Confocal scanning laser microscopy image of an S. lugdunensis IDRL-2640 biofilm grown in a chambered coverglass well (Lab-Tek II; Nalge Nunc International, Rochester, NY) for 24 h. Cells (green) were stained with the nucleic acid stain Syto-9 (Molecular Probes, Eugene, OR), and extracellular proteins (red) were visualized with Sypro Ruby (Molecular Probes). Fluorophores were excited with an argon laser at 458 nm (Sypro Ruby) and 488 nm (Syto-9). Fluorescence was detected from Sypro Ruby with an LP 650 filter and from Syto-9 with a BP 505-530 filter. Images were collected on an LSM410 unit equipped with an Axiovert 100 M inverted microscope using a Plan-Apochromat 100×/1.4 NA oil immersion objective (Carl Zeiss, Inc., Thornwood, NY). Bar, 5 μm.
Staphylococcal biofilm formation, characterized in detail for S. epidermidis and S. aureus, proceeds through an ordered series of events in which cells first attach to a surface and begin to form layers as they replicate and accumulate, followed by the production of extracellular matrix components that lead to intercellular adhesion (150). A large number of S. aureus and S. epidermidis adherence proteins mediate primary surface attachment, which is critical to biofilm formation (150). Once established, the elaboration of a polymeric matrix, often composed of polysaccharides, ensues. The matrix serves to connect cells within the structured architecture of the biofilm, as well as to provide protection from harsh conditions encountered in the environment, including interfering with attempts of the host immune system to clear the infection.
Soon after the description of S. lugdunensis, Lambe et al. showed that a glycocalyx, described as a negatively charged polysaccharide matrix surrounding bacterial microcolonies, could be visualized by transmission electron microscopy when cells were stained with ruthenium red, a stain with affinity for polyanionic structures (102, 103). Various amounts of glycocalyx were produced by different S. lugdunensis strains (103). That group further showed that a carbohydrate-containing glycocalyx purified from S. lugdunensis strains effectively stimulated monocyte prostaglandin E2 production, which leads to the inhibition of T-cell proliferation, thus having an immunomodulatory effect on the immune system (173). A later study verified that purified glycocalyx from two distinct S. lugdunensis strains stimulated murine peritoneal macrophages to synthesize prostaglandin E2 and the proinflammatory cytokine interleukin-1. In addition, glycocalyx modulated macrophage production of tumor necrosis factor alpha and nitric oxide (174). This work provides mechanistic evidence that a polymeric carbohydrate-containing material produced by S. lugdunensis protects surface-attached cells from host immune defenses by impeding the normal functions of monocytes and macrophages.
ica locus.
A polysaccharide polymer of β-1,6-linked N-acetylglucosamine residues (PNAG), also referred to as the polysaccharide intercellular adhesin (PIA), is the most well-characterized component comprising the staphylococcal biofilm matrix (75, 129). The genes of the icaADBC locus encode the biosynthetic machinery required to assemble (icaAD), export (icaC), and deacetylate (icaB) PNAG/PIA (75, 192). A negative regulatory protein gene, icaR, is located upstream and in the opposite direction of icaA (35). The genomic organization and the regulation of the icaADBC genes have been described in detail for S. aureus and S. epidermidis (129), and homologues have been detected in several other CNS species, including S. lugdunensis (40, 63). PCR amplification and sequencing of ica genes in S. lugdunensis were first reported by Sandoe and Longshaw in 2001 following characterization of biofilm formation by an isolate causing ventriculoperitoneal shunt infection (157); however, the authors did not submit the sequence to a public sequence repository. We subsequently attempted to amplify icaA from S. lugdunensis isolates causing prosthetic joint infection but were unable to do so with primers designed from conserved areas in the S. aureus and S. epidermidis icaA sequences, suggesting a marked difference in the sequence of the S. lugdunensis ica locus (63). We eventually amplified an icaA homologue using the same primers as Sandoe and Longshaw (157) and used a primer-walking strategy to acquire sequence for the entire region surrounding the S. lugdunensis ica locus (66). More recently, Chokr et al. reported PCR amplification of icaA and/or icaD in only 6/11 S. lugdunensis strains from implant-associated infections (30), although we showed that icaADBC genes were present in all 15 S. lugdunensis isolates that we tested (66). Similar to the case for other staphylococcal ica loci, the S. lugdunensis icaADBC homologues are 30 to 60% identical to their related genes in S. aureus, S. epidermidis, and S. caprae (Table 4). Unexpectedly, icaR was not detected in the genome of S. lugdunensis; rather, a novel open reading frame with putative glycosyl hydrolase activity and which shares a high degree of similarity to the biofilm-degrading enzyme dispersin B from Actinobacillus species was found upstream of icaA. The S. lugdunensis ica genes and the adjacent open reading frame span a region of 4.75 kilobases that is flanked on the 5′ and 3′ ends by homologues of the yycJ and yycI genes, respectively, from other staphylococci.
Composition of the biofilm matrix.
Chokr et al. reported that of 11 S. lugdunensis isolates, only one icaA-positive strain that concomitantly formed biofilm was detected (30). They were unable to detect PIA in biofilm extracts from this strain using an anti-PIA antiserum. PIA was also not detected in static cultures of the remaining five icaA-positive, biofilm-negative S. lugdunensis strains in their collection. Two other organisms formed biofilm but lacked both PIA and the icaA locus, signifying that alternative mechanisms for biofilm formation exist in these strains. In subsequent studies, these investigators showed that biofilms formed by the two ica-negative S. lugdunensis strains contained proteins and extracellular teichoic acids composed of glycerol, glucose, and glucosamine but lacked prominent levels of PNAG (94, 152). Proteinase K and other proteases, but not the carbohydrate-degrading reagent sodium metaperiodate or the biofilm-degrading enzyme dispersin B (90), released preformed ica-negative S. lugdunensis biofilms (27, 94, 152), suggesting that protein components stabilize the PNAG-negative biofilms formed by these two strains (Table 4).
In our own studies with 15 icaADBC-positive S. lugdunensis clinical isolates that form in vitro biofilm to various degrees, we were surprised to find that none of the isolates produced detectable levels of PNAG in biofilms or static-phase cultures (66). Similar to the results described above for the reported ica-negative strains, treatment of ica-positive S. lugdunensis biofilms with proteinase K, trypsin, thermolysin, and chymotrypsin led to the overwhelming detachment of nearly all isolates (66). Visualization of the S. lugdunensis extracellular matrix by fluorescence confocal scanning laser microscopy revealed abundant amounts of extracellular protein (Fig. 2B). Despite the presence of an intact icaADBC locus in all isolates of S. lugdunensis that we examined, the extracellular matrix of in vitro biofilms formed by this species appears to be composed predominantly of proteins (Table 4).
Phenotypic responses of biofilm formation to changing environmental conditions.
The presence of exogenous factors in the growth environment directly influences the staphylococcal biofilm formation phenotype (59). Such changes include accentuation of S. epidermidis and S. aureus biofilm formation in response to increasing glucose concentrations, heightened osmolarity, and ethanol (129). Like for other staphylococci, increasing concentrations of glucose stimulate S. lugdunensis biofilms. However, opposite to what is observed for other staphylococci, the presence of sodium chloride and ethanol in the growth medium prevents most S. lugdunensis isolates tested from adhering to polystyrene (66).
Intravenously administered catecholamine inotropes and heparin also enhance biofilm formation by S. epidermidis (112) and S. aureus (163). When S. lugdunensis isolates were grown in medium containing epinephrine, biofilm production decreased in 47% (7/15) of isolates (65). Dopamine decreased biofilm formation in some isolates but enhanced biofilm formation in the majority of the S. lugdunensis isolates. Dobutamine demonstrated the most pronounced effect on S. lugdunensis biofilms by preventing adherence in 87% of isolates tested. The biofilm phenotype when bacteria were grown in the presence of heparin was identical to the results obtained with dobutamine, which is in direct contrast to the results previously reported for S. aureus (163). Further, treatment of preformed biofilms with concentrations of heparin used clinically to flush catheters resulted in biofilm detachment of 73% of S. lugdunensis isolates, suggesting a mechanism through which organisms colonizing and infecting intravascular catheter surfaces may metastasize to alternate sites in the host.
In nearly all environmental situations examined for biofilms formed by S. lugdunensis isolates thus far, the phenotypic response is opposite to what would be predicted based on prior studies with S. aureus or S. epidermidis. The phenotypic differences observed for S. lugdunensis biofilms under these conditions are likely due to the as-yet-uncharacterized mechanism of PNAG-independent, proteinaceous biofilm formation employed by these isolates.
CONCLUDING REMARKS
Although S. lugdunensis does not cause infection at the same frequency as S. aureus or S. epidermidis, its pathogenic potential should not be underestimated. In 20 years, the plethora of case reports published on S. lugdunensis (Table 1) reveal the significance of this organism as a pathogen in a large number of infections, including particularly virulent cases of endocarditis. The relative lack of genomic diversity and the prevalent susceptibility to numerous antimicrobial agents suggest that S. lugdunensis has evolved along a different path than the other pathogenic staphylococci. In particular, in comparison with what is known about other staphylococci, the substantial differences in S. lugdunensis biofilm formation phenotypes, the composition of the biofilm matrix, and the genomic organization of the ica locus serve as examples of the distinctive qualities that this organism has acquired during evolution. Further investigations of S. lugdunensis biofilm formation should yield additional insight into other aspects of the properties that contribute to this organism's enhanced virulence. In the present age of readily available genomic sequencing technology, we anticipate that any S. lugdunensis genome sequencing projects that may commence in the future will provide more clarity in understanding the exceptional traits that make this organism such a successful pathogen.
Acknowledgments
We gratefully acknowledge José Miró for sharing his data on S. lugdunensis endocarditis. We thank Prerana Bhatia for photography, Scott Gambe of the Mayo Clinic Electron Microscopy core facility for excellent technical assistance, and Jim Tarara of the Mayo Clinic Flow Cytometry/Optical Morphology core facility for expert confocal microscopy assistance.
This work was supported, in part, by the Anderson Gift.
REFERENCES
- 1.Akiyama, H., H. Kanzaki, J. Tada, and J. Arata. 1998. Coagulase-negative staphylococci isolated from various skin lesions. J. Dermatol. 25:563-568. [DOI] [PubMed] [Google Scholar]
- 2.Anguera, I., A. Del Rio, J. M. Miro, X. Matinez-Lacasa, F. Marco, J. R. Guma, G. Quaglio, X. Claramonte, A. Moreno, C. A. Mestres, E. Mauri, M. Azqueta, N. Benito, C. Garcia-de la Maria, M. Almela, M. J. Jimenez-Exposito, O. Sued, E. De Lazzari, and J. M. Gatell. 2005. Staphylococcus lugdunensis infective endocarditis: description of 10 cases and analysis of native valve, prosthetic valve, and pacemaker lead endocarditis clinical profiles. Heart 91:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arciola, C. R., D. Campoccia, Y. H. An, L. Baldassarri, V. Pirini, M. E. Donati, F. Pegreffi, and L. Montanaro. 2006. Prevalence and antibiotic resistance of 15 minor staphylococcal species colonizing orthopedic implants. Int. J. Artif. Organs. 29:395-401. [DOI] [PubMed] [Google Scholar]
- 4.Asnis, D. S., S. St. John, R. Tickoo, and A. Arora. 2003. Staphylococcus lugdunensis breast abscess: is it real? Clin. Infect. Dis. 36:1348. [DOI] [PubMed] [Google Scholar]
- 5.Bannerman, T. L., and S. J. Peacock. 2007. Staphylococcus, Micrococcus, and other catalase-positive cocci, p. 390-411. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 6.Bannerman, T. L., D. L. Rhoden, S. K. McAllister, J. M. Miller, and L. A. Wilson. 1997. The source of coagulase-negative staphylococci in the Endophthalmitis Vitrectomy Study. A comparison of eyelid and intraocular isolates using pulsed-field gel electrophoresis. Arch. Ophthalmol. 115:357-361. [DOI] [PubMed] [Google Scholar]
- 7.Barker, K. F., J. C. O'Driscoll, and A. Bhargava. 1991. Staphylococcus lugdunensis. J. Clin. Pathol. 44:873-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barros, E. M., N. L. Iorio, M. D. Bastos, K. R. Dos Santos, and M. Giambiagi-Demarval. 2007. Species-level identification of clinical staphylococcal isolates based on polymerase chain reaction-restriction fragment length polymorphism analysis of a partial groEL gene sequence. Diagn. Microbiol. Infect. Dis. 59:251-257. [DOI] [PubMed] [Google Scholar]
- 9.Becker, K., I. Pagnier, B. Schuhen, F. Wenzelburger, A. W. Friedrich, F. Kipp, G. Peters, and C. von Eiff. 2006. Does nasal cocolonization by methicillin-resistant coagulase-negative staphylococci and methicillin-susceptible Staphylococcus aureus strains occur frequently enough to represent a risk of false-positive methicillin-resistant S. aureus determinations by molecular methods? J. Clin. Microbiol. 44:229-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bellamy, R., and T. Barkham. 2002. Staphylococcus lugdunensis infection sites: predominance of abscesses in the pelvic girdle region. Clin. Infect. Dis. 35:E32-E34. [DOI] [PubMed] [Google Scholar]
- 11.Benito, Y., G. Lina, T. Greenland, J. Etienne, and F. Vandenesch. 1998. trans-complementation of a Staphylococcus aureus agr mutant by Staphylococcus lugdunensis agr RNAIII. J. Bacteriol. 180:5780-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bera, A., R. Biswas, S. Herbert, and F. Gotz. 2006. The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun. 74:4598-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bera, A., S. Herbert, A. Jakob, W. Vollmer, and F. Gotz. 2005. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55:778-787. [DOI] [PubMed] [Google Scholar]
- 14.Bjerketorp, J., M. Nilsson, A. Ljungh, J. I. Flock, K. Jacobsson, and L. Frykberg. 2002. A novel von Willebrand factor binding protein expressed by Staphylococcus aureus. Microbiology 148:2037-2044. [DOI] [PubMed] [Google Scholar]
- 15.Bobin, S., A. Durand-Dubief, D. Bouhour, G. Kirkorian, F. Vandenesch, J. Etienne, M. Ballet-Mechain, and D. Peyramond. 1999. Pacemaker endocarditis due to Staphylococcus lugdunensis: report of two cases. Clin. Infect. Dis. 28:404-405. [DOI] [PubMed] [Google Scholar]
- 16.Bourgeois, I., M. Pestel-Caron, J. F. Lemeland, J. L. Pons, and F. Caron. 2007. Tolerance to the glycopeptides vancomycin and teicoplanin in coagulase-negative staphylococci. Antimicrob. Agents Chemother. 51:740-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breen, J. D., and A. W. Karchner. 1994. Usefulness of pulsed-field gel electrophoresis in confirming endocarditis due to Staphylococcus lugdunensis. Clin. Infect. Dis. 19:985-986. [DOI] [PubMed] [Google Scholar]
- 18.Bucher, E., A. Trampuz, L. Donati, and W. Zimmerli. 2000. Spondylodiscitis associated with bacteraemia due to coagulase-negative staphylococci. Eur. J. Clin. Microbiol. Infect. Dis. 19:118-120. [DOI] [PubMed] [Google Scholar]
- 19.Burgert, S. J., M. T. LaRocco, and S. Wilansky. 1999. Destructive native valve endocarditis caused by Staphylococcus lugdunensis. South. Med. J. 92:812-814. [DOI] [PubMed] [Google Scholar]
- 20.Camacho, M., S. Guis, J. P. Mattei, R. Costello, and J. Roudier. 2002. Three-year outcome in a patient with Staphylococcus lugdunensis discitis. Joint Bone Spine. 69:85-87. [DOI] [PubMed] [Google Scholar]
- 21.Campoccia, D., L. Baldassarri, Y. H. An, Q. K. Kang, V. Pirini, S. Gamberini, F. Pegreffi, L. Montanaro, and C. R. Arciola. 2006. Automated ribotyping to distinguish the different nonSau/ nonSep staphylococcal emerging pathogens in orthopedic implant infections. Int. J. Artif. Organs 29:421-429. [DOI] [PubMed] [Google Scholar]
- 22.Carretto, E., D. Barbarini, I. Couto, D. De Vitis, P. Marone, J. Verhoef, H. De Lencastre, and S. Brisse. 2005. Identification of coagulase-negative staphylococci other than Staphylococcus epidermidis by automated ribotyping. Clin. Microbiol. Infect. 11:177-184. [DOI] [PubMed] [Google Scholar]
- 23.Carroll, K. C., and M. P. Weinstein. 2007. Manual and automated systems for detection and identification of microorganisms, p. 192-244. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 24.Casanova-Roman, M., A. Sanchez-Porto, and M. Casanova-Bellido. 2004. Urinary tract infection due to Staphylococcus lugdunensis in a healthy child. Scand. J. Infect. Dis. 36:149-150. [DOI] [PubMed] [Google Scholar]
- 25.Castro, J. G., and L. Dowdy. 1999. Septic shock caused by Staphylococcus lugdunensis. Clin. Infect. Dis. 28:681-682. [DOI] [PubMed] [Google Scholar]
- 26.Celard, M., H. Lelievre, J. F. Obadia, P. Chevalier, F. Forey, F. Vandenesch, and J. Etienne. 1997. Long-standing bacteremia and endocarditis caused by Staphylococcus lugdunensis in a patient with an implantable cardioverter defibrillator. Clin. Microbiol. Infect. 3:387-388. [DOI] [PubMed] [Google Scholar]
- 27.Chaignon, P., I. Sadovskaya, C. Ragunah, N. Ramasubbu, J. B. Kaplan, and S. Jabbouri. 2007. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl. Microbiol. Biotechnol. 75:125-132. [DOI] [PubMed] [Google Scholar]
- 28.Chaouni, L. B., J. Etienne, T. Greenland, and F. Vandenesch. 1996. Nucleic acid sequence and affiliation of pLUG10, a novel cadmium resistance plasmid from Staphylococcus lugdunensis. Plasmid 36:1-8. [DOI] [PubMed] [Google Scholar]
- 29.Chiquet, C., A. Pechinot, C. Creuzot-Garcher, Y. Benito, J. Croize, S. Boisset, J. P. Romanet, G. Lina, and F. Vandenesch. 2007. Acute postoperative endophthalmitis caused by Staphylococcus lugdunensis. J. Clin. Microbiol. 45:1673-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chokr, A., D. Watier, H. Eleaume, B. Pangon, J. C. Ghnassia, D. Mack, and S. Jabbouri. 2006. Correlation between biofilm formation and production of polysaccharide intercellular adhesin in clinical isolates of coagulase-negative staphylococci. Int. J. Med. Microbiol. 296:381-388. [DOI] [PubMed] [Google Scholar]
- 31.Christensen, G. D., W. A. Simpson, A. L. Bisno, and E. H. Beachey. 1982. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37:318-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarridge, J. E., III. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.CLSI. 2005. Performance standards for antimicrobial susceptibility testing; Fifteenth informational supplement. CLSI/NCCLS document M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 35.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooke, R. P., S. E. James, and D. F. Sallomi. 2003. Infective discitis due to Staphylococcus lugdunensis—a case of missed opportunity. Br. J. Biomed. Sci. 60:162-164. [DOI] [PubMed] [Google Scholar]
- 37.Cormican, M. G., K. el Bouri, G. Corbett-Feeney, J. Flynn, and K. Daly. 1992. Staphylococcus lugdunensis endocarditis. J. Infect. 24:335-336. [DOI] [PubMed] [Google Scholar]
- 38.Costello, R., M. Miquel, J. A. Gastaut, and R. Bouabdallah. 1995. Native valve Staphylococcus lugdunensis endocarditis in a patient with lymphoma. Ann. Med. Intern. (Paris). 146:369. [PubMed] [Google Scholar]
- 39.Couto, I., S. Pereira, M. Miragaia, I. S. Sanches, and H. de Lencastre. 2001. Identification of clinical staphylococcal isolates from humans by internal transcribed spacer PCR. J. Clin. Microbiol. 39:3099-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crupper, S. S., V. Worrell, G. C. Stewart, and J. J. Iandolo. 1999. Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J. Bacteriol. 181:4071-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Hondt, G., M. Ieven, C. Vandermersch, and J. Colaert. 1997. Destructive endocarditis caused by Staphylococcus lugdunensis. Case report and review of the literature. Acta Clin. Belg. 52:27-30. [DOI] [PubMed] [Google Scholar]
- 43.De Paulis, A. N., S. C. Predari, C. D. Chazarreta, and J. E. Santoianni. 2003. Five-test simple scheme for species-level identification of clinically significant coagulase-negative staphylococci. J. Clin. Microbiol. 41:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, and M. Beach. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32:S114-132. [DOI] [PubMed] [Google Scholar]
- 45.Donvito, B., J. Etienne, L. Denoroy, T. Greenland, Y. Benito, and F. Vandenesch. 1997. Synergistic hemolytic activity of Staphylococcus lugdunensis is mediated by three peptides encoded by a non-agr genetic locus. Infect. Immun. 65:95-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donvito, B., J. Etienne, T. Greenland, C. Mouren, V. Delorme, and F. Vandenesch. 1997. Distribution of the synergistic haemolysin genes hld and slush with respect to agr in human staphylococci. FEMS Microbiol. Lett. 151:139-144. [DOI] [PubMed] [Google Scholar]
- 47.Drancourt, M., and D. Raoult. 2002. rpoB gene sequence-based identification of Staphylococcus species. J. Clin. Microbiol. 40:1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dufour, P., S. Jarraud, F. Vandenesch, T. Greenland, R. P. Novick, M. Bes, J. Etienne, and G. Lina. 2002. High genetic variability of the agr locus in Staphylococcus species. J. Bacteriol. 184:1180-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunne, W. M., Jr. 1990. Effects of subinhibitory concentrations of vancomycin or cefamandole on biofilm production by coagulase-negative staphylococci. Antimicrob. Agents Chemother. 34:390-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dupont, C., L. Turner, O. Pinguet, V. Leflon, E. Rouveix, M. H. Nicolas-Chanoine, and M. Dorra. 1996. Staphylococcus lugdunensis endocarditis. A new case. Ann. Med. Intern. (Paris) 147:374-375. [PubMed] [Google Scholar]
- 51.Ebright, J. R., N. Penugonda, and W. Brown. 2004. Clinical experience with Staphylococcus lugdunensis bacteremia: a retrospective analysis. Diagn. Microbiol. Infect. Dis. 48:17-21. [DOI] [PubMed] [Google Scholar]
- 52.Elliott, S. P., R. Yogev, and S. T. Shulman. 2001. Staphylococcus lugdunensis: an emerging cause of ventriculoperitoneal shunt infections. Pediatr. Neurosurg. 35:128-130. [DOI] [PubMed] [Google Scholar]
- 53.Etienne, J., B. Pangon, C. Leport, M. Wolff, B. Clair, C. Perronne, Y. Brun, and A. Bure. 1989. Staphylococcus lugdunensis endocarditis. Lancet i:390. [DOI] [PubMed] [Google Scholar]
- 54.Etienne, J., F. Poitevin-Later, F. Renaud, and J. Fleurette. 1990. Plasmid profiles and genomic DNA restriction endonuclease patterns of 30 independent Staphylococcus lugdunensis strains. FEMS Microbiol. Lett. 55:93-97. [DOI] [PubMed] [Google Scholar]
- 55.Farrag, N., P. Lee, R. Gunney, and G. M. Viagappan. 2001. Staphylococcus lugdunensis endocarditis. Postgrad. Med. J. 77:259-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferguson, K. P., D. W. Lambe, Jr., J. L. Keplinger, and J. H. Kalbfleisch. 1991. Comparison of the pathogenicity of three species of coagulase-negative Staphylococcus in a mouse model with and without a foreign body. Can. J. Microbiol. 37:722-724. [DOI] [PubMed] [Google Scholar]
- 57.Ferreira, R. B., N. L. Iorio, K. L. Malvar, A. P. Nunes, L. S. Fonseca, C. C. Bastos, and K. R. Santos. 2003. Coagulase-negative staphylococci: comparison of phenotypic and genotypic oxacillin susceptibility tests and evaluation of the agar screening test by using different concentrations of oxacillin. J. Clin. Microbiol. 41:3609-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fervenza, F. C., G. E. Contreras, K. N. Garratt, and J. M. Steckelberg. 1999. Staphylococcus lugdunensis endocarditis: a complication of vasectomy? Mayo Clin. Proc. 74:1227-1230. [DOI] [PubMed] [Google Scholar]
- 59.Fitzpatrick, F., H. Humphreys, and J. P. O'Gara. 2005. The genetics of staphylococcal biofilm formation—will a greater understanding of pathogenesis lead to better management of device-related infection? Clin. Microbiol. Infect. 11:967-973. [DOI] [PubMed] [Google Scholar]
- 60.Fleurette, J., M. Bes, Y. Brun, J. Freney, F. Forey, M. Coulet, M. E. Reverdy, and J. Etienne. 1989. Clinical isolates of Staphylococcus lugdunensis and S. schleiferi: bacteriological characteristics and susceptibility to antimicrobial agents. Res. Microbiol. 140:107-118. [DOI] [PubMed] [Google Scholar]
- 61.Fluit, A. C., J. Verhoef, and F. J. Schmitz. 2001. Frequency of isolation and antimicrobial resistance of gram-negative and gram-positive bacteria from patients in intensive care units of 25 European university hospitals participating in the European arm of the SENTRY Antimicrobial Surveillance Program 1997-1998. Eur. J. Clin. Microbiol. Infect. Dis. 20:617-625. [DOI] [PubMed] [Google Scholar]
- 62.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 63.Frank, K. L., A. D. Hanssen, and R. Patel. 2004. icaA is not a useful diagnostic marker for prosthetic joint infection. J. Clin. Microbiol. 42:4846-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frank, K. L., and R. Patel. 2007. Activity of sodium metabisulfite against planktonic and biofilm Staphylococcus species. Diagn. Microbiol. Infect. Dis. 57:355-359. [DOI] [PubMed] [Google Scholar]
- 65.Frank, K. L., and R. Patel. 2007. Intravenously administered pharmaceuticals impact biofilm formation and detachment of Staphylococcus lugdunensis and other staphylococci. Diagn. Microbiol. Infect. Dis. doi: 10.1016/j.diagmicrobio.2007.07.008. [DOI] [PubMed]
- 66.Frank, K. L., and R. Patel. 2007. Poly-N-acetylglucosamine is not a major component of the extracellular matrix in biofilms formed by icaADBC-positive Staphylococcus lugdunensis isolates. Infect. Immun. 75:4728-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frank, K. L., E. J. Reichert, K. E. Piper, and R. Patel. 2007. In vitro effects of antimicrobial agents on planktonic and biofilm forms of Staphylococcus lugdunensis clinical isolates. Antimicrob. Agents Chemother. 51:888-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Freney, J., Y. Brun, M. Bes, H. Meugnier, F. Grimont, P. A. D. Grimont, C. Nervi, and J. Fleurette. 1988. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int. J. Syst. Bacteriol. 38:168-172. [Google Scholar]
- 69.Fujita, S., Y. Senda, T. Iwagami, and T. Hashimoto. 2005. Rapid identification of staphylococcal strains from positive-testing blood culture bottles by internal transcribed spacer PCR followed by microchip gel electrophoresis. J. Clin. Microbiol. 43:1149-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia Fernandez, F. J., J. Berjon Reyero, V. Ruiz Quevedo, and E. Arcos Lage El. 2003. Staphylococcus lugdunensis endocarditis. Case report and review of the literature. Rev. Clin. Esp. 203:98-99. [DOI] [PubMed] [Google Scholar]
- 71.Gianella, S., S. Ulrich, B. Huttner, and R. Speich. 2006. Conservative management of a brain abscess in a patient with Staphylococcus lugdunensis endocarditis. Eur. J. Clin. Microbiol. Infect. Dis. 25:476-478. [DOI] [PubMed] [Google Scholar]
- 72.Goh, S. H., S. Potter, J. O. Wood, S. M. Hemmingsen, R. P. Reynolds, and A. W. Chow. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goh, S. H., Z. Santucci, W. E. Kloos, M. Faltyn, C. G. George, D. Driedger, and S. M. Hemmingsen. 1997. Identification of Staphylococcus species and subspecies by the chaperonin 60 gene identification method and reverse checkerboard hybridization. J. Clin. Microbiol. 35:3116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goldstein, E. J., D. M. Citron, C. V. Merriam, Y. A. Warren, K. L. Tyrrell, and H. T. Fernandez. 2006. In vitro activity of ceftobiprole against aerobic and anaerobic strains isolated from diabetic foot infections. Antimicrob. Agents Chemother. 50:3959-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 76.Grant, C. E., D. L. Sewell, M. Pfaller, R. V. Bumgardner, and J. A. Williams. 1994. Evaluation of two commercial systems for identification of coagulase-negative staphylococci to species level. Diagn. Microbiol. Infect. Dis. 18:1-5. [DOI] [PubMed] [Google Scholar]
- 77.Greig, J. M., and M. J. Wood. 2003. Staphylococcus lugdunensis vertebral osteomyelitis. Clin. Microbiol. Infect. 9:1139-1141. [DOI] [PubMed] [Google Scholar]
- 78.Guttmann, G., S. Garazi, and D. Van Linthoudt. 2000. Spondylodiscitis due to Staphylococcus lugdunensis. Clin. Exp. Rheumatol. 18:271-272. [PubMed] [Google Scholar]
- 79.Haile, D. T., J. Hughes, E. Vetter, P. Kohner, R. Snyder, R. Patel, and F. R. Cockerill III. 2002. Frequency of isolation of Staphylococcus lugdunensis in consecutive urine cultures and relationship to urinary tract infection. J. Clin. Microbiol. 40:654-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hébert, G. A. 1990. Hemolysins and other characteristics that help differentiate and biotype Staphylococcus lugdunensis and Staphylococcus schleiferi. J. Clin. Microbiol. 28:2425-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hellbacher, C., E. Tornqvist, and B. Soderquist. 2006. Staphylococcus lugdunensis: clinical spectrum, antibiotic susceptibility, and phenotypic and genotypic patterns of 39 isolates. Clin. Microbiol. Infect. 12:43-49. [DOI] [PubMed] [Google Scholar]
- 82.Herchline, T. E., and L. W. Ayers. 1991. Occurrence of Staphylococcus lugdunensis in consecutive clinical cultures and relationship of isolation to infection. J. Clin. Microbiol. 29:419-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Higaki, S., T. Kitagawa, M. Morohashi, and T. Yamagishi. 1999. Distribution and antimicrobial susceptibility of coagulase-negative staphylococci from skin lesions. J. Int. Med. Res. 27:191-195. [DOI] [PubMed] [Google Scholar]
- 84.Huebner, J., and D. A. Goldmann. 1999. Coagulase-negative staphylococci: role as pathogens. Annu. Rev. Med. 50:223-236. [DOI] [PubMed] [Google Scholar]
- 85.Hussain, Z., L. Stoakes, V. Massey, D. Diagre, V. Fitzgerald, S. El Sayed, and R. Lannigan. 2000. Correlation of oxacillin MIC with mecA gene carriage in coagulase-negative staphylococci. J. Clin. Microbiol. 38:752-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jarlov, J. O., T. Hojbjerg, C. Busch-Sorensen, J. Scheibel, J. K. Moller, H. J. Kolmos, and D. A. Wandall. 1996. Coagulase-negative staphylococci in Danish blood cultures: species distribution and antibiotic susceptibility. J. Hosp. Infect. 32:217-227. [DOI] [PubMed] [Google Scholar]
- 87.Jones, R. M., M. A. Jackson, C. Ong, and G. K. Lofland. 2002. Endocarditis caused by Staphylococcus lugdunensis. Pediatr. Infect. Dis. J. 21:265-268. [DOI] [PubMed] [Google Scholar]
- 88.Kaabia, N., D. Scauarda, G. Lena, and M. Drancourt. 2002. Molecular identification of Staphylococcus lugdunensis in a patient with meningitis. J. Clin. Microbiol. 40:1824-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamaraju, S., K. Nelson, D. N. Williams, W. Ayenew, and K. S. Modi. 1999. Staphylococcus lugdunensis pulmonary valve endocarditis in a patient on chronic hemodialysis. Am. J. Nephrol. 19:605-608. [DOI] [PubMed] [Google Scholar]
- 90.Kaplan, J. B., C. Ragunath, K. Velliyagounder, D. H. Fine, and N. Ramasubbu. 2004. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawamura, Y., X. G. Hou, F. Sultana, K. Hirose, M. Miyake, S. E. Shu, and T. Ezaki. 1998. Distribution of Staphylococcus species among human clinical specimens and emended description of Staphylococcus caprae. J. Clin. Microbiol. 36:2038-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kleeman, K. T., T. L. Bannerman, and W. E. Kloos. 1993. Species distribution of coagulase-negative staphylococcal isolates at a community hospital and implications for selection of staphylococcal identification procedures. J. Clin. Microbiol. 31:1318-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kloos, W. E., and C. G. George. 1991. Identification of Staphylococcus species and subspecies with the MicroScan Pos ID and Rapid Pos ID panel systems. J. Clin. Microbiol. 29:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kogan, G., I. Sadovskaya, P. Chaignon, A. Chokr, and S. Jabbouri. 2006. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol. Lett. 255:11-16. [DOI] [PubMed] [Google Scholar]
- 95.Koh, T. W., S. J. Brecker, and C. A. Layton. 1996. Successful treatment of Staphylococcus lugdunensis endocarditis complicated by multiple emboli: a case report and review of the literature. Int. J. Cardiol. 55:193-197. [DOI] [PubMed] [Google Scholar]
- 96.Kourbeti, I. S., D. E. Alegakis, G. E. Roditakis, and G. Samonis. 2007. Staphylococcus lugdunensis early prosthetic valve endocarditis in a young HIV positive patient. Infection 35:40-42. [DOI] [PubMed] [Google Scholar]
- 97.Koziol-Montewka, M., A. Szczepanik, I. Baranowicz, L. Jozwiak, A. Ksiazek, and D. Kaczor. 2006. The investigation of Staphylococcus aureus and coagulase-negative staphylococci nasal carriage among patients undergoing haemodialysis. Microbiol. Res. 161:281-287. [DOI] [PubMed] [Google Scholar]
- 98.Kragsbjerg, P., J. Bomfim-Loogna, E. Tornqvist, and B. Soderquist. 2000. Development of antimicrobial resistance in Staphylococcus lugdunensis during treatment—report of a case of bacterial arthritis, vertebral osteomyelitis and infective endocarditis. Clin. Microbiol. Infect. 6:496-499. [DOI] [PubMed] [Google Scholar]
- 99.Kralovic, S. M., H. Melin-Aldana, K. K. Smith, and C. C. Linnemann, Jr. 1995. Staphylococcus lugdunensis endocarditis after tooth extraction. Clin. Infect. Dis. 20:715-716. [DOI] [PubMed] [Google Scholar]
- 100.Kuroda, M., A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, K. Matoba, H. Toh, S. Kuhara, M. Hattori, and T. Ohta. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 102:13272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Laguno, M., O. Miro, C. Font, and A. de la Sierra. 1998. Pacemaker-related endocarditis. Report of 7 cases and review of the literature. Cardiology 90:244-248. [DOI] [PubMed] [Google Scholar]
- 102.Lambe, D. W., Jr., K. P. Ferguson, J. L. Keplinger, C. G. Gemmell, and J. H. Kalbfleisch. 1990. Pathogenicity of Staphylococcus lugdunensis, Staphylococcus schleiferi, and three other coagulase-negative staphylococci in a mouse model and possible virulence factors. Can. J. Microbiol. 36:455-463. [DOI] [PubMed] [Google Scholar]
- 103.Lambe, D. W., Jr., C. Jeffery, K. P. Ferguson, and M. D. Cooper. 1994. Examination of the glycocalyx of four species of Staphylococcus by transmission electron microscopy and image analysis. Microbios 78:133-143. [PubMed] [Google Scholar]
- 104.Lessing, M. P., D. W. Crook, I. C. Bowler, and B. Gribbin. 1996. Native-valve endocarditis caused by Staphylococcus lugdunensis. Q. J. Med. 89:855-858. [DOI] [PubMed] [Google Scholar]
- 105.Leung, M. J., N. Nuttall, M. Mazur, T. L. Taddei, M. McComish, and J. W. Pearman. 1999. Case of Staphylococcus schleiferi endocarditis and a simple scheme to identify clumping factor-positive staphylococci. J. Clin. Microbiol. 37:3353-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leung, M. J., N. Nuttall, T. M. Pryce, G. W. Coombs, and J. W. Pearman. 1998. Colony variation in Staphylococcus lugdunensis. J. Clin. Microbiol. 36:3096-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lina, B., F. Vandenesch, J. Etienne, B. Kreiswirth, and J. Fleurette. 1992. Comparison of coagulase-negative staphylococci by pulsed-field electrophoresis. FEMS Microbiol. Lett. 71:133-138. [DOI] [PubMed] [Google Scholar]
- 108.Lina, B., F. Vandenesch, M. E. Reverdy, T. Greenland, J. Fleurette, and J. Etienne. 1994. Non-puerperal breast infections due to Staphylococcus lugdunensis. Eur. J. Clin. Microbiol. Infect. Dis. 13:686-687. [DOI] [PubMed] [Google Scholar]
- 109.Llinares, P., R. Moure, J. Cerqueiro, M. Abalde, E. Miguez, A. Echaniz, and A. Guerrero. 1998. Endocarditis caused by Staphylococcus lugdunensis. Hospital incidence. Enferm. Infecc. Microbiol. Clin. 16:233-236. [PubMed] [Google Scholar]
- 110.Louie, L., A. Majury, J. Goodfellow, M. Louie, and A. E. Simor. 2001. Evaluation of a latex agglutination test (MRSA-Screen) for detection of oxacillin resistance in coagulase-negative staphylococci. J. Clin. Microbiol. 39:4149-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ludlam, H., and I. Phillips. 1989. Staphylococcus lugdunensis peritonitis. Lancet ii:1394. [DOI] [PubMed] [Google Scholar]
- 112.Lyte, M., P. P. Freestone, C. P. Neal, B. A. Olson, R. D. Haigh, R. Bayston, and P. H. Williams. 2003. Stimulation of Staphylococcus epidermidis growth and biofilm formation by catecholamine inotropes. Lancet 361:130-135. [DOI] [PubMed] [Google Scholar]
- 113.Martineau, F., F. J. Picard, D. Ke, S. Paradis, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2001. Development of a PCR assay for identification of staphylococci at genus and species levels. J. Clin. Microbiol. 39:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Massidda, O., M. Mingoia, D. Fadda, M. B. Whalen, M. P. Montanari, and P. E. Varaldo. 2006. Analysis of the beta-lactamase plasmid of borderline methicillin-susceptible Staphylococcus aureus: focus on bla complex genes and cadmium resistance determinants cadD and cadX. Plasmid 55:114-127. [DOI] [PubMed] [Google Scholar]
- 115.Mateo, M., J. R. Maestre, L. Aguilar, F. Cafini, P. Puente, P. Sanchez, L. Alou, M. J. Gimenez, and J. Prieto. 2005. Genotypic versus phenotypic characterization, with respect to susceptibility and identification, of 17 clinical isolates of Staphylococcus lugdunensis. J. Antimicrob. Chemother. 56:287-291. [DOI] [PubMed] [Google Scholar]
- 116.Matthews, P. C., C. G. Missouris, J. Jordaan, and M. P. Lessing. 2007. Staphylococcus lugdunensis endocarditis following cardiac catheterisation. Int. J. Cardiol. doi: 10.1016/j.ijcard.2007.06.068. [DOI] [PubMed]
- 117.May, J., K. Shannon, A. King, and G. French. 1998. Glycopeptide tolerance in Staphylococcus aureus. J. Antimicrob. Chemother. 42:189-197. [DOI] [PubMed] [Google Scholar]
- 118.Mei-Dan, O., G. Mann, G. Steinbacher, S. J. Ballester, R. B. Cugat, and P. D. Alvarez. 2007. Septic arthritis with Staphylococcus lugdunensis following arthroscopic ACL revision with BPTB allograft. Knee Surg. Sports Traumatol. Arthrosc. doi: 10.1007/s00167-007-0379-8. [DOI] [PubMed]
- 119.Minhas, T., H. A. Ludlam, M. Wilks, and S. Tabaqchali. 1995. Detection by PCR and analysis of the distribution of a fibronectin-binding protein gene (fbn) among staphylococcal isolates. J. Med. Microbiol. 42:96-101. [DOI] [PubMed] [Google Scholar]
- 120.Mitchell, J., A. Tristan, and T. J. Foster. 2004. Characterization of the fibrinogen-binding surface protein Fbl of Staphylococcus lugdunensis. Microbiology 150:3831-3841. [DOI] [PubMed] [Google Scholar]
- 121.Murdoch, D. R., R. J. Everts, S. T. Chambers, and I. A. Cowan. 1996. Vertebral osteomyelitis due to Staphylococcus lugdunensis. J. Clin. Microbiol. 34:993-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.NCCLS. 1999. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement M100-S9. NCCLS, Wayne, PA.
- 123.NCCLS. 2002. Performance standards for antimicrobial susceptibility testing. Twelfth informational supplement M100-S12. NCCLS, Wayne, PA.
- 124.Nilsson, M., J. Bjerketorp, B. Guss, and L. Frykberg. 2004. A fibrinogen-binding protein of Staphylococcus lugdunensis. FEMS Microbiol. Lett. 241:87-93. [DOI] [PubMed] [Google Scholar]
- 125.Nilsson, M., J. Bjerketorp, A. Wiebensjo, A. Ljungh, L. Frykberg, and B. Guss. 2004. A von Willebrand factor-binding protein from Staphylococcus lugdunensis. FEMS Microbiol. Lett. 234:155-161. [DOI] [PubMed] [Google Scholar]
- 126.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 127.Novick, R. P. 1989. Staphylococcal plasmids and their replication. Annu. Rev. Microbiol. 43:537-565. [DOI] [PubMed] [Google Scholar]
- 128.O'Brien, F. G., C. I. Botterill, T. G. Endersby, R. L. Lim, W. B. Grubb, and J. E. Gustafson. 1998. Heterogeneous expression of fusidic acid resistance in Staphylococcus aureus with plasmid or chromosomally encoded fusidic acid resistance genes. Pathology 30:299-303. [DOI] [PubMed] [Google Scholar]
- 129.O'Gara, J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270:179-188. [DOI] [PubMed] [Google Scholar]
- 130.O'Neill, A. J., F. McLaws, G. Kahlmeter, A. S. Henriksen, and I. Chopra. 2007. Genetic basis of resistance to fusidic acid in staphylococci. Antimicrob. Agents Chemother. 51:1737-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ortiz de la Tabla, V., F. Gutierrez-Rodero, C. Martin, A. Zorraquino, and I. Belinchon. 1996. Staphylococcus lugdunensis as a cause of abscesses in the perineal area. Eur. J. Clin. Microbiol. Infect. Dis. 15:405-407. [DOI] [PubMed] [Google Scholar]
- 132.Palazzo, E., J. Pierre, and N. Besbes. 1992. Staphylococcus lugdunensis arthritis: a complication of arthroscopy. J. Rheumatol. 19:327-328. [PubMed] [Google Scholar]
- 133.Pareja, J., K. Gupta, and H. Koziel. 1998. The toxic shock syndrome and Staphylococcus lugdunensis bacteremia. Ann. Intern. Med. 128:603-604. [DOI] [PubMed] [Google Scholar]
- 134.Patel, R., K. E. Piper, M. S. Rouse, J. R. Uhl, F. R. Cockerill, 3rd, and J. M. Steckelberg. 2000. Frequency of isolation of Staphylococcus lugdunensis among staphylococcal isolates causing endocarditis: a 20-year experience. J. Clin. Microbiol. 38:4262-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Paulsson, M., A. C. Petersson, and A. Ljungh. 1993. Serum and tissue protein binding and cell surface properties of Staphylococcus lugdunensis. J. Med. Microbiol. 38:96-102. [DOI] [PubMed] [Google Scholar]
- 136.Perry, J. D., A. L. Jones, and F. K. Gould. 1999. Glycopeptide tolerance in bacteria causing endocarditis. J. Antimicrob. Chemother. 44:121-124. [DOI] [PubMed] [Google Scholar]
- 137.Perry, R. D., and S. Silver. 1982. Cadmium and manganese transport in Staphylococcus aureus membrane vesicles. J. Bacteriol. 150:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Personne, P., M. Bes, G. Lina, F. Vandenesch, Y. Brun, and J. Etienne. 1997. Comparative performances of six agglutination kits assessed by using typical and atypical strains of Staphylococcus aureus. J. Clin. Microbiol. 35:1138-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Petzsch, M., W. Leber, B. Westphal, S. Crusius, and E. C. Reisinger. 2004. Progressive Staphylococcus lugdunensis endocarditis despite antibiotic treatment. Wien. Klin. Wochenschr. 116:98-101. [DOI] [PubMed] [Google Scholar]
- 140.Pfaller, M. A., R. N. Jones, G. V. Doern, H. S. Sader, K. C. Kugler, M. L. Beach, et al. 1999. Survey of blood stream infections attributable to gram-positive cocci: frequency of occurrence and antimicrobial susceptibility of isolates collected in 1997 in the United States, Canada, and Latin America from the SENTRY Antimicrobial Surveillance Program. Diagn. Microbiol. Infect. Dis. 33:283-297. [DOI] [PubMed] [Google Scholar]
- 141.Pinna, A., S. Zanetti, M. Sotgiu, L. A. Sechi, G. Fadda, and F. Carta. 1999. Identification and antibiotic susceptibility of coagulase negative staphylococci isolated in corneal/external infections. Br. J. Ophthalmol. 83:771-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pirila, L., K. O. Soderstrom, M. Hietarinta, J. Jalava, V. Kyto, and A. Toivanen. 2006. Fatal myocardial necrosis caused by Staphylococcus lugdunensis and cytomegalovirus in a patient with scleroderma. J. Clin. Microbiol. 44:2295-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Poitevin-Later, F., F. Vandenesch, K. Dyke, J. Fleurette, and J. Etienne. 1992. Cadmium-resistance plasmid in Staphylococcus lugdunensis. FEMS Microbiol. Lett. 78:59-63. [DOI] [PubMed] [Google Scholar]
- 144.Polenakovik, H., T. Herchline, C. Bacheller, and J. Bernstein. 2000. Staphylococcus lugdunensis endocarditis after angiography. Mayo Clin. Proc. 75:656-657. [DOI] [PubMed] [Google Scholar]
- 145.Poyart, C., G. Quesne, C. Boumaila, and P. Trieu-Cuot. 2001. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rachid, S., K. Ohlsen, W. Witte, J. Hacker, and W. Ziebuhr. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:3357-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rasmussen, T. T., L. P. Kirkeby, K. Poulsen, J. Reinholdt, and M. Kilian. 2000. Resident aerobic microbiota of the adult human nasal cavity. APMIS 108:663-675. [DOI] [PubMed] [Google Scholar]
- 148.Renzulli, A., A. Della Corte, M. Torella, G. Dialetto, and M. Cotrufo. 2000. Mitral and aortic valve endocarditis due to Staphylococcus lugdunensis. Tex. Heart Inst. J. 27:67-69. [PMC free article] [PubMed] [Google Scholar]
- 149.Rodriguez-Gascon, M., P. Roig, J. B. Montagud, and J. Merino. 2003. Acute Staphylococcus lugdunensis endocarditis with septic cerebral and pulmonary emboli, showing favorable evolution. Enferm. Infecc. Microbiol. Clin. 21:465-467. [DOI] [PubMed] [Google Scholar]
- 150.Rohde, H., D. Mack, M. Christner, C. Burdelski, G. Franke, and J. K. Knobloch. 2006. Pathogenesis of staphylococcal device-related infections: from basic science to new diagnostic, therpeutic, and prophylactic approaches. Rev. Med. Microbiol. 17:45-54. [Google Scholar]
- 151.Rozalska, B., and A. Ljungh. 1995. Biomaterial-associated staphylococcal peritoneal infections in a neutropaenic mouse model. FEMS Immunol. Med. Microbiol. 11:307-319. [DOI] [PubMed] [Google Scholar]
- 152.Sadovskaya, I., P. Chaignon, G. Kogan, A. Chokr, E. Vinogradov, and S. Jabbouri. 2006. Carbohydrate-containing components of biofilms produced in vitro by some staphylococcal strains related to orthopaedic prosthesis infections. FEMS Immunol. Med. Microbiol. 47:75-82. [DOI] [PubMed] [Google Scholar]
- 153.Sakinc, T., P. Kulczak, K. Henne, and S. G. Gatermann. 2004. Cloning of an agr homologue of Staphylococcus saprophyticus. FEMS Microbiol. Lett. 237:157-161. [DOI] [PubMed] [Google Scholar]
- 154.Sampathkumar, P., D. R. Osmon, and F. R. Cockerill III. 2000. Prosthetic joint infection due to Staphylococcus lugdunensis. Mayo Clin. Proc. 75:511-512. [DOI] [PubMed] [Google Scholar]
- 155.Sanchez, A., I. Martinez, F. Sanz, F. Lopez, and J. M. Aguado. 2000. Aggressive acute endocarditis caused by Staphylococcus lugdunensis complicated with multiple cerebral septic emboli. Enferm. Infecc. Microbiol. Clin. 18:526-527. [PubMed] [Google Scholar]
- 156.Sanchis-Bayarri Vaillant, V., R. Llucian Rambla, and V. Sanchis-Bayarri Bernal. 1999. 7 cases of Staphylococcus lugdunensis infection. An. Med. Interna 16:361-362. [PubMed] [Google Scholar]
- 157.Sandoe, J. A., and C. M. Longshaw. 2001. Ventriculoperitoneal shunt infection caused by Staphylococcus lugdunensis. Clin. Microbiol. Infect. 7:385-387. [DOI] [PubMed] [Google Scholar]
- 158.Sanzéni, L., and H. Ringberg. 2003. Fistulating periprosthetic Staphylococcus lugdunensis hip infection cured by intra-articular teicoplanin injections—a case report. Acta Orthop. Scand. 74:624-625. [DOI] [PubMed] [Google Scholar]
- 159.Schnitzler, N., R. Meilicke, G. Conrads, D. Frank, and G. Haase. 1998. Staphylococcus lugdunensis: report of a case of peritonitis and an easy-to-perform screening strategy. J. Clin. Microbiol. 36:812-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schonheyder, H. C., V. K. Hansen, P. Asschenfeldt, and V. T. Rosdahl. 1993. Staphylococcus lugdunensis: an important cause of endocarditis. A case report. APMIS 101:802-804. [DOI] [PubMed] [Google Scholar]
- 161.Seenivasan, M. H., and V. L. Yu. 2003. Staphylococcus lugdunensis endocarditis—the hidden peril of coagulase-negative staphylococcus in blood cultures. Eur. J. Clin. Microbiol. Infect. Dis. 22:489-491. [DOI] [PubMed] [Google Scholar]
- 162.Seifert, H., D. Oltmanns, K. Becker, H. Wisplinghoff, and C. von Eiff. 2005. Staphylococcus lugdunensis pacemaker-related infection. Emerg. Infect. Dis. 11:1283-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Shanks, R. M., N. P. Donegan, M. L. Graber, S. E. Buckingham, M. E. Zegans, A. L. Cheung, and G. A. O'Toole. 2005. Heparin stimulates Staphylococcus aureus biofilm formation. Infect. Immun. 73:4596-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sheppard, M., and S. Jankowski. 1992. Staphylococcus lugdunensis endocarditis. J. Infect. 25:116-117. [DOI] [PubMed] [Google Scholar]
- 165.Shin, J. H., H. J. Jung, H. R. Lee, J. H. Kim, H. R. Kim, and J. N. Lee. 2007. Prevalence, identification, and antimicrobial susceptibility of Staphylococcus lugdunensis from various clinical specimens in Korea. Jpn. J. Infect. Dis. 60:312-313. [PubMed] [Google Scholar]
- 166.Shuttleworth, R., and W. D. Colby. 1992. Staphylococcus lugdunensis endocarditis. J. Clin. Microbiol. 30:1948-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sivadon, V., M. Rottman, S. Chaverot, J. C. Quincampoix, V. Avettand, P. de Mazancourt, L. Bernard, P. Trieu-Cuot, J. M. Feron, A. Lortat-Jacob, P. Piriou, T. Judet, and J. L. Gaillard. 2005. Use of genotypic identification by sodA sequencing in a prospective study to examine the distribution of coagulase-negative Staphylococcus species among strains recovered during septic orthopedic surgery and evaluate their significance. J. Clin. Microbiol. 43:2952-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Skow, A., K. A. Mangold, M. Tajuddin, A. Huntington, B. Fritz, R. B. Thomson, Jr., and K. L. Kaul. 2005. Species-level identification of staphylococcal isolates by real-time PCR and melt curve analysis. J. Clin. Microbiol. 43:2876-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Smyth, E. G., E. D. Wright, and R. R. Marples. 1988. New type of staphylococcal endocarditis. J. Clin. Pathol. 41:809-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sorli Redo, M. L., A. Aguirre Tejedo, A. Supervia Caparros, and J. L. Echarte Pazos. 2006. Medical treatment in Staphylococcus lugdunensis infective endocarditis with mitral insufficiency. An. Med. Interna 23:395. [DOI] [PubMed] [Google Scholar]
- 171.Sotutu, V., J. Carapetis, J. Wilkinson, A. Davis, and N. Curtis. 2002. The “surreptitious Staphylococcus”: Staphylococcus lugdunensis endocarditis in a child. Pediatr. Infect. Dis. J. 21:984-986. [DOI] [PubMed] [Google Scholar]
- 172.Stoakes, L., M. A. John, R. Lannigan, B. C. Schieven, M. Ramos, D. Harley, and Z. Hussain. 1994. Gas-liquid chromatography of cellular fatty acids for identification of staphylococci. J. Clin. Microbiol. 32:1908-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Stout, R. D., K. P. Ferguson, Y. N. Li, and D. W. Lambe, Jr. 1992. Staphylococcal exopolysaccharides inhibit lymphocyte proliferative responses by activation of monocyte prostaglandin production. Infect. Immun. 60:922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Stout, R. D., Y. Li, A. R. Miller, and D. W. Lambe, Jr. 1994. Staphylococcal glycocalyx activates macrophage prostaglandin E2 and interleukin 1 production and modulates tumor necrosis factor alpha and nitric oxide production. Infect. Immun. 62:4160-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sung, J. M., P. D. Chantler, and D. H. Lloyd. 2006. Accessory gene regulator locus of Staphylococcus intermedius. Infect. Immun. 74:2947-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Takeuchi, F., S. Watanabe, T. Baba, H. Yuzawa, T. Ito, Y. Morimoto, M. Kuroda, L. Cui, M. Takahashi, A. Ankai, S. Baba, S. Fukui, J. C. Lee, and K. Hiramatsu. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292-7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tan, T. Y., S. Y. Ng, and W. X. Ng. 2006. Clinical significance of coagulase-negative staphylococci recovered from nonsterile sites. J. Clin. Microbiol. 44:3413-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tee, W. S., S. Y. Soh, R. Lin, and L. H. Loo. 2003. Staphylococcus lugdunensis carrying the mecA gene causes catheter-associated bloodstream infection in premature neonate. J. Clin. Microbiol. 41:519-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tegmark, K., E. Morfeldt, and S. Arvidson. 1998. Regulation of agr-dependent virulence genes in Staphylococcus aureus by RNAIII from coagulase-negative staphylococci. J. Bacteriol. 180:3181-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Teong, H. H., Y. S. Leo, S. Y. Wong, L. H. Sng, and Z. P. Ding. 2000. Case report of Staphylococcus lugdunensis native valve endocarditis and review of the literature. Ann. Acad. Med. Singap. 29:673-677. [PubMed] [Google Scholar]
- 181.Thomas, S., C. Hoy, and R. Capper. 2006. Osteomyelitis of the ear canal caused by Staphylococcus lugdunensis. J. Infect. 53:e227-229. [DOI] [PubMed] [Google Scholar]
- 182.Trampuz, A., K. E. Piper, M. J. Jacobson, A. D. Hanssen, K. K. Unni, D. R. Osmon, J. N. Mandrekar, F. R. Cockerill, J. M. Steckelberg, J. F. Greenleaf, and R. Patel. 2007. Sonication of removed hip and knee prostheses for diagnosis of infection. N. Engl. J. Med. 357:654-663. [DOI] [PubMed] [Google Scholar]
- 183.Vandenesch, F., J. Etienne, M. E. Reverdy, and S. J. Eykyn. 1993. Endocarditis due to Staphylococcus lugdunensis: report of 11 cases and review. Clin. Infect. Dis. 17:871-876. [DOI] [PubMed] [Google Scholar]
- 184.Vandenesch, F., S. J. Projan, B. Kreiswirth, J. Etienne, and R. P. Novick. 1993. agr-related sequences in Staphylococcus lugdunensis. FEMS Microbiol. Lett. 111:115-122. [DOI] [PubMed] [Google Scholar]
- 185.Vandenesch, F., M. J. Storrs, F. Poitevin-Later, J. Etienne, P. Courvalin, and J. Fleurette. 1991. Delta-like haemolysin produced by Staphylococcus lugdunensis. FEMS Microbiol. Lett. 62:65-68. [DOI] [PubMed] [Google Scholar]
- 186.van der Mee-Marquet, N., A. Achard, L. Mereghetti, A. Danton, M. Minier, and R. Quentin. 2003. Staphylococcus lugdunensis infections: high frequency of inguinal area carriage. J. Clin. Microbiol. 41:1404-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Van Hoovels, L., P. De Munter, J. Colaert, I. Surmont, E. Van Wijngaerden, W. E. Peetermans, and J. Verhaegen. 2005. Three cases of destructive native valve endocarditis caused by Staphylococcus lugdunensis. Eur. J. Clin. Microbiol. Infect. Dis. 24:149-152. [DOI] [PubMed] [Google Scholar]
- 188.Van Wamel, W. J., G. van Rossum, J. Verhoef, C. M. Vandenbroucke-Grauls, and A. C. Fluit. 1998. Cloning and characterization of an accessory gene regulator (agr)-like locus from Staphylococcus epidermidis. FEMS Microbiol. Lett. 163:1-9. [DOI] [PubMed] [Google Scholar]
- 189.Viganego, F., T. M. Nazir, C. Y. Lee, J. P. Hafner, and R. Smith. 2007. Staphylococcus lugdunensis endocarditis following lower extremity revascularization. Int. J. Cardiol. 116:e4-6. [DOI] [PubMed] [Google Scholar]
- 190.von Eiff, C., G. Peters, and K. Becker. 2006. The small colony variant (SCV) concept—the role of staphylococcal SCVs in persistent infections. Injury 37:S26-33. [DOI] [PubMed] [Google Scholar]
- 191.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2:677-685. [DOI] [PubMed] [Google Scholar]
- 192.Vuong, C., S. Kocianova, J. M. Voyich, Y. Yao, E. R. Fischer, F. R. Deleo, and M. Otto. 2004. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 279:54881-54886. [DOI] [PubMed] [Google Scholar]
- 193.Wachtler, M., E. Strobel, U. Koch, M. Overbeck, W. Witte, W. Doering, and D. Eichenlaub. 2002. Native mitral valve endocarditis caused by Staphylococcus lugdunensis in a 22-year-old woman. Infection 30:251-253. [DOI] [PubMed] [Google Scholar]
- 194.Waghorn, D. J. 1994. Staphylococcus lugdunensis as a cause of breast abscess. Clin. Infect. Dis. 19:814-815. [DOI] [PubMed] [Google Scholar]
- 195.Walsh, B., and J. P. Mounsey. 1990. Staphylococcus lugdunensis and endocarditis. J. Clin. Pathol. 43:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wasserman, E., L. Lombard, and G. Walzl. 1999. Staphylococcus lugdunensis endocarditis in a young, previously healthy female. Eur. J. Clin. Microbiol. Infect. Dis. 18:289-291. [DOI] [PubMed] [Google Scholar]
- 197.Waterer, G., R. Wilson, S. Dimmitt, and M. Watson. 1997. Staphylococcus lugdunensis endocarditis. Aust. N. Z. J. Med. 27:84-85. [DOI] [PubMed] [Google Scholar]
- 198.Weightman, N. C., K. E. Allerton, and J. France. 2000. Bone and prosthetic joint infection with Staphylococcus lugdunensis. J. Infect. 40:98-99. [DOI] [PubMed] [Google Scholar]
- 199.Yamazumi, T., I. Furuta, D. J. Diekema, M. A. Pfaller, and R. N. Jones. 2001. Comparison of the Vitek gram-positive susceptibility 106 card, the MRSA-Screen latex agglutination test, and mecA analysis for detecting oxacillin resistance in a geographically diverse collection of clinical isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 39:3633-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.You, Y. O., K. J. Kim, B. M. Min, and C. P. Chung. 1999. Staphylococcus lugdunensis—a potential pathogen in oral infection. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 88:297-302. [DOI] [PubMed] [Google Scholar]