Abstract
The accurate detection and quantitation of cytokines in serum are important in the study of disease mechanisms, pathogenesis, and treatment. Serum cytokines can reflect processes that are occurring at the cellular or tissue level and thus provide a means of indirectly monitoring these processes. Multiplex detection of cytokines allows the simultaneous measurement of multiple cytokines in a sample, increasing the efficiency of measuring the cytokines while reducing the serum sample volumes required for the testing. Two commercially available multiplex platforms were evaluated (Pierce SearchLight and Meso Scale Discovery), using multiplexes capable of simultaneously detecting eight cytokines. The cytokines analyzed in this study were gamma interferon, vascular endothelial growth factor, tumor necrosis factor alpha, interleukin-6 (IL-6), macrophage inflammatory protein 1β, monocyte chemoattractant protein 1, IL-12p40, and IL-4. The range of quantitation of the platforms, the recovery of spiked cytokines, and the detection of the cytokines in serum samples from subjects with ulcerative colitis, Crohn's disease, rheumatoid arthritis, and psoriasis were examined. The findings showed that the detection of the cytokines was highly dependent upon the platform, with the consistency of the detection of cytokines across platforms being dependent upon the cytokine being analyzed. A careful examination of platform assay performance must be made prior to utilizing multiplex platforms in a study. While some cytokines will give similar patterns of results across platforms, others will be highly variable. The use of the same platform within a study or across studies where data will be compared is advised.
The accurate and reproducible measurement of cytokines in serum, plasma, or tissue culture supernatants is important for studies involving disease pathogenesis, treatment, and prognosis (5, 7, 14). Cytokines can be measured by a variety of assay formats, including enzyme-linked immunosorbent assay (ELISA), radioimmunoassay, bioassays, and mass spectrometry (10). In recent years, multiplexed assays have been developed that are capable of simultaneously measuring multiple cytokines in a variety of matrices. These multiplex assays have the advantage of requiring small sample volumes and providing a quick turnaround time for analysis. Multiplex assays have been based primarily on bead-based Luminex-type platforms, which utilize antibodies applied as a coating on microbeads in suspension to detect the target cytokines (3, 16, 17). The development of plate-based multiplex assays (10, 11, 13) is a more recent technology. These plate-based multiplex assays are in concept standard ELISAs, incorporating multiple capture/detection antibodies. Plate-based assays are now being commercially produced and are likely to become more broadly utilized as validated data become available on these platforms.
The plate-based multiplex assays have the advantage of small sample volume, good sensitivity, high throughput, and the potential to measure a large number of cytokines simultaneously (15). However, as with standard ELISA, the multiplex assays are dependent upon the careful choice of the capture/detection antibody pairs, calibration curve standardization, and proper buffering of the sample to minimize matrix interference with the detection of the cytokine.
In the current study, two commercially available multiplex platforms in conjunction with standard ELISAs are compared. Eight commonly measured cytokines were chosen for the comparison: gamma interferon (IFN-γ), vascular endothelial growth factor (VEGF), tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), macrophage inflammatory protein 1β (MIP-1β), monocyte chemoattractant protein 1 (MCP-1), IL-12p40, and IL-4. The range of quantitation of the platforms, the recovery of spiked cytokines, and the detection of the cytokines in serum samples from subjects with ulcerative colitis, Crohn's disease, rheumatoid arthritis (RA), and psoriasis were examined. The findings show that the detection of the cytokines is highly dependent upon the platform, with the consistency of the detection across platforms dependent upon the cytokine being analyzed.
MATERIALS AND METHODS
Samples.
Serum samples from healthy donors and those with inflammatory diseases were obtained from Bioreclaimation (Hicksville, NY). The disease state samples were from subjects with ulcerative colitis (30 to 46 years of age, one male/seven females), Crohn's disease (30 to 69 years, two males/six females), RA (38 to 64 years, two males/six females), and psoriasis (28 to 60 years, one male/seven females).
Whole blood drawn into a dry collection bag was allowed to clot overnight in a refrigerator and spun to serum at 2,800 × g for 20 min in a refrigerated centrifuge (5°C). A prelabeled transfer bag was attached to an available port on the collection bag, and serum was gently expressed into the transfer bag with the use of a plasma extractor device, ensuring that there was no red blood cell contamination.
Multiplex evaluations.
The multiplex assays were manufactured by Meso Scale Discovery (MSD; Gaithersburg, MD) and Pierce Endogen (Pierce Biotechnology, Woburn, MA). Each well of the 96-well plate-based assays contained antibodies to IFN-γ, VEGF, TNF-α, IL-6, MIP-1β, MCP-1, IL-12p40, and IL-4. The manufacturer spotted the antibodies onto the base of the well.
The MSD platform used in this study was a 96-well plate-based assay that incorporated electrochemiluminescence as the basis for detection. Following the capture of the cytokine by the spotted antibody, labeled detection antibodies were bound to the antigen. The detection antibodies were coupled to electrochemiluminescent labels that emitted light when electrochemically stimulated via carbon-coated electrodes in the bottom of the array wells. In addition, a detection buffer that was added prior to reading of the array incorporated reactants that enhanced the electrochemical signal. The resulting signal was read using a charge-coupled device (CCD).
In brief the MSD Multi Spot Array assay was run as follows. Calibration curves were prepared in the supplied assay diluent for human serum, with a range of 40,000 pg/ml to 1.2 pg/ml, dependent upon the cytokine. Assay controls consisted of a pool of two healthy human donor serums, spiked with the MSD standard at a high or medium concentration. The acceptance ranges for the controls were set to 100% ± compared to 30% the mean of eight assays. Arrays were preincubated with 25 μl per well of assay diluent for 30 min, with shaking at room temperature. Following the preincubation, 25 μl of sample or calibrator was added in duplicate to the appropriate wells. The array was then incubated at room temperature for 2 h, with shaking. The array was then washed with phosphate-buffered saline plus 0.05% Tween 20, and 25 μl of detection antibody reagent was added. Following a 2-h room-temperature incubation with shaking, the array was washed and the detection buffer was added. The assay results were read using an MSD Sector Imager 6000 incorporating a CCD. Sample cytokine concentrations were determined with Softmax Pro Version 4.6 software, using curve fit models (log-log or four-parameter log-logistic) as suggested by the manufacturer for the specific cytokine.
The Pierce Endogen SearchLight was also a 96-well plate-based assay. The detection for this system utilized ELISA technology. In brief the Pierce SearchLight assay was performed as follows.
Calibration curves were prepared in sample diluent, with a range of 5,000 pg/ml to 0.4 pg/ml, dependent upon the cytokine being analyzed. Assay controls consisted of the sample diluent spiked with the calibration standard at high or medium concentrations. The controls were deemed acceptable if the recovery was within 100% ± 30% of the theoretical concentration. Serum samples were diluted 1:5 in sample diluent. Diluted samples or calibration curves (50 μl) were added in duplicate to the appropriate wells and incubated at room temperature for 1 h with shaking. The plates were then washed with kit wash buffer, and 50 μl of biotinylated antibody detection reagent was added. The plate was then incubated for 30 min at room temperature with shaking. Following a wash step, 50 μl streptavidin-horseradish peroxidase was added to each well and incubated at room temperature for 30 min with shaking. The array was then washed, and 50 μl of substrate solution was added. The assay results were read with the Pierce SearchLight Plus imaging system incorporating a CCD. Sample cytokine concentrations were determined with Softmax Pro 4.6 software, using curve fit models (log-log or four-parameter log-logistic) as suggested by the manufacturer for the specific cytokine.
ELISAs.
Single-analyte analyses of individual cytokines were performed using R&D Systems kits (R&D Systems, Minneapolis, MN). Assays were performed according to the manufacturer's instructions.
Statistical analysis.
The statistical analyses were performed using the Spearman rank correlation coefficient or paired t tests with SigmaStat software (Systat Software Inc., San Jose, CA).
RESULTS
Quantitation ranges.
The manufacturer's reported quantitation range for each cytokine by assay source is listed in Table 1. While it was necessary to confirm the quantitation range of the standard curve, it was also important to validate the quantitation range in the matrix of interest. To this end the lower limit of quantitation (LLOQ) in serum was determined by spiking standard in pooled serum starting at levels several points above the lowest standard. The upper limit of quantitation (ULOQ) was defined as the highest standard on the standard curve. The ranges obtained (Table 1) were based upon the linearity of the calibration curve, and in some cases the endogenous levels of these cytokines were high and the upper range of the assay needed to be extended (resulting in a loss of points on the lower end) to provide an accurate calibration curve for these cytokines For any particular cytokine, the MSD assay gave a broader dynamic range than did the Pierce assay, with the MSD MIP-1β assay having an upper range 50 times higher than that of the Pierce assay. The single-analyte R&D Systems ELISA gave quantitation ranges that generally fell between the ranges for the MSD and the Pierce platforms.
TABLE 1.
Quantitation ranges for multiplex (MSD/Pierce) and single analyte (R&D) assays
| Cytokine | Quantitation range (pg/ml)
|
|||||
|---|---|---|---|---|---|---|
| MSD
|
Pierce
|
R&D
|
||||
| In-house | Vendor | In-house | Vendor | In-house | Vendor | |
| IFN-γ | 4.9-2,500 | 2.4-10,000 | 3.1-400 | 0.8-200 | ||
| IL-6 | 1.2-2,500 | 2.4-10,000 | 3.1-400 | 0.8-200 | 0.2-10.0 | 0.2-10.0 |
| TNF-α | 2.4-2,500 | 2.4-10,000 | 9.4-2,400 | 4.7-1,200 | 1.1-7,000 | 2.2-7,000 |
| IL-4 | 1.2-2,500 | 2.4-10,000 | 6.3-800 | 1.6-400 | ||
| IL-12p40 | 19.5-2,500 | 2.4-10,000 | 2.3-600 | 1.2-300 | 31.2-2,000 | 31.2-2,000 |
| VEGF | 19.5-40,000 | 2.4-10,000 | 39.1-5,000 | 9.8-2,500 | 31.2-2,000 | 31.2-2,000 |
| MCP-1 | 78.1-40,000 | 2.4-10,000 | 25.0-800 | 1.6-400 | 31.2-2,000 | 31.2-2,000 |
| MIP-1β | 78.1-40,000 | 2.4-10,000 | 12.5-800 | 1.6-400 | ||
Spike recovery.
The ability of the multiplex and single ELISA platforms to recover antigen spiked into serum was determined. Serum from 10 individual healthy donors was separately spiked with cytokines. The donor samples were not pooled. The concentration of cytokine to be added to the serum was determined by the calibration curve of the individual platforms. The concentrations selected fell within the linear portion of each curve, generally near the upper and lower ends of the curve. The endogenous levels of the cytokine were determined in nonspiked, normal serum and subtracted from the results prior to calculation of the recovery. The range of the endogenous levels of cytokine in the normal samples is given in Table 2.
TABLE 2.
Endogenous levels of cytokines in healthy human serum
| Cytokine | Mean cytokine level (pg/ml) on platforma
|
||
|---|---|---|---|
| MSD | Pierce | R&D | |
| IFN-γ | <LLOQ | <LLOQ | ND |
| IL-4 | <LLOQ | <LLOQ | ND |
| MIP-1 | 248.5 (91.6) | 1,058.7 (1,953.3) | ND |
| MCP-1 | 245.8 (57.5) | 554.4 (302.8) | 169.3 (71.8) |
| IL-6 | 2.3 (1.1) | <LLOQ | 1.5 (0.6) |
| TNF-α | 3.2 (1.4) | <LLOQ | 1.7 (0.7) |
| IL-12p40 | <LLOQ | <LLOQ | 49.2 (28.1) |
| VEGF | 49.3 (32.6) | 257.2 (144.1) | 107.2 (79.5) |
Values in parentheses are standard deviations. ND, not done.
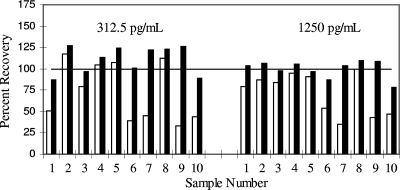
A preliminary evaluation of the spike recovery of cytokine was initially determined for each of the multiplex platforms. During this preliminary evaluation, the recovery of the antigen for a number of the cytokines was variable. In the Pierce platform, VEGF was the cytokine with the poorest recovery, while the MSD platform had recovery problems across a number of cytokines. Typically the recovery of the cytokines did not fall within the acceptable recovery range of 100% ± 30%. The results from the VEGF spiking using the Pierce platform are shown in Fig. 1 as an example. The standard diluent resulted in only 4 of 10 subjects with acceptable recovery at the low spike and 5 of 10 subjects at the high spike. The recovery did not appear to be spike concentration dependent but rather was dependent upon the serum sample being tested. Pooling of the samples reduced the variability (data not shown); however, as individual serum samples will be analyzed during routine testing, it is important to have acceptable recovery in the majority of samples analyzed.
FIG. 1.
Effect of sample diluent on the recovery of spiked samples. Unpooled serum samples (S1 to S10) had VEGF added at a concentration of 1,250 pg/ml or 312 pg/ml. Samples were diluted with either standard sample diluent or modified sample diluent. Samples were tested on the Pierce SearchLight System. Results are calculated as the percentages of the expected values (averages of two separate determinations). Open bars, standard diluent; solid bars, modified diluent.
An investigation into the cause of the inaccuracy was conducted with the manufacturer of each platform. Operator technique, plate washing procedures, sample dilution, and CCD alignment were ruled out as possible causes. Since the inaccuracy was sample dependent, it was surmised that increasing the protein/detergent content of the sample buffer should reduce the variability seen in the sample recovery. This was shown to be the case. Use of the modified diluent improved the accuracy of the spike recovery for both platforms for specific cytokines (Fig. 1 and data not shown). For VEGF 100% of the low-spike samples and 90% of the high-spike samples were within range.
While the exact formulation of the diluents is considered by the manufacturers to be proprietary, the diluents contained additional protein and detergent components known to minimize matrix interference. Further testing of spike recovery used the modified diluent in both platforms.
The recovery of spiked cytokines in the multiplex and single ELISA platforms is given in Table 3. For the multiplexed assays, a recovery of antigen within a 100% ± 30% range of the expected value was considered to be acceptable. With the exception of MCP-1 in both of the multiplexed platforms, incorporating the use of modified sample diluent gave acceptable recovery of the spiked cytokines. The MCP-1 assay overrecovered antigen by more than 30% in both platforms, likely due to differences in the detection of MCP-1 in serum compared to that of the standard that is prepared in assay buffer. The single ELISA platform gave acceptable recovery for all cytokines.
TABLE 3.
Recovery of spiked cytokines by single and multiplex ELISAs
| Cytokine and kit source (type) | Expected cytokine concn (pg/ml) | Mean observed cytokine concn ± SD (pg/ml) | % Recovery | P value (paired t test)a |
|---|---|---|---|---|
| IFN-γ | ||||
| MSD | 1,250.0 | 1,170.2 ± 84.9 | 108.8 | 0.312 |
| 156.3 | 166.4 ± 8.4 | |||
| Pierce | 100.0 | 103.4 ± 14.8 | 107.5 | 0.197 |
| 25.0 | 27.9 ± 3.6 | |||
| VEGF | ||||
| MSD | 20,000.0 | 21,435.3 ± 1,822.8 | 130.1 | 0.023 |
| 2,500.0 | 2,731.7 ± 268..05 | |||
| Pierce | 1,250.0 | 1,145.5 ± 147.5 | 97.1 | 0.098 |
| 312.5 | 320.6 ± 61.8 | |||
| R&D | 500.0 | 521.9 ± 48.1 | 104.7 | 0.108 |
| 100.0 | 105.1 ± 15.0 | |||
| TNF-α | ||||
| MSD | 1,250 | 1,524.8 ± 149.8 | 124.2 | <0.001 |
| 156.3 | 197.5 ± 14.0 | |||
| Pierce | 600.0 | 699.0 ± 132.6 | 128.9 | 0.003 |
| 150.0 | 212.0 ± 65.3 | |||
| R&D | 2,000 | 2,033.3 ± 72.5 | 108.1 | <0.001 |
| 200 | 228.9 ± 12.6 | |||
| IL-6 | ||||
| MSD | 1,250.0 | 1,288.2 ± 79.3 | 102.6 | 0.070 |
| 156.3 | 159.7 ± 10.2 | |||
| Pierce | 100.0 | 118.8 ± 10.5 | 122.8 | <0.001 |
| 25.0 | 31.7 ± 8.0 | |||
| R&D | 5.0 | 5.0 ± 0.4 | 100.3 | 0.994 |
| 2.0 | 2.0 ± 0.1 | |||
| MIP-1β | ||||
| MSD | 20,000.0 | 23,616.4 ± 1,033.9 | 123.1 | <0.001 |
| 2,500.0 | 2,886.5 ± 201.9 | |||
| Pierce | 200.0 | 203.9 ± 55.2 | 113.4 | 0.006 |
| 50.0 | 62.5 ± 6.3 | |||
| MCP-1 | ||||
| MSD | 20,000.0 | 19,503.6 ± 4,123.3 | 142.5 | 0.648 |
| 2,500.0 | 2,967.3 ± 164.4 | |||
| Pierce | 200.0 | 273.9 ± 30.5 | 161.8 | <0.001 |
| 50.0 | 93.3 ± 23.4 | |||
| R&D | 500.0 | 614.9 ± 29.8 | 117.3 | <0.001 |
| 100.0 | 111.5 ± 8.0 | |||
| IL-12p40 | ||||
| MSD | 1,250.0 | 1,315.6 ± 165.8 | 105.3 | 0.180 |
| 156.3 | 164.7 ± 22.8 | |||
| Pierce | 150.0 | 177.3 ± 5.8 | 124.8 | <0.001 |
| 37.5 | 49.3 ± 3.1 | |||
| R&D | 500 | 564.2 ± 38.6 | 114.9 | <0.001 |
| 100 | 117.0 ± 14.7 | |||
| IL-4 | ||||
| MSD | 1,250.0 | 1,152.3 ± 110.9 | 87.9 | 0.005 |
| 156.3 | 130.6 ± 23.7 | |||
| Pierce | 200.0 | 209.6 ± 15.3 | 118.5 | |
| 50.0 | 63.7 ± 3.8 |
Comparison between all expected values and all observed values per cytokine for each kit.
Disease state samples.
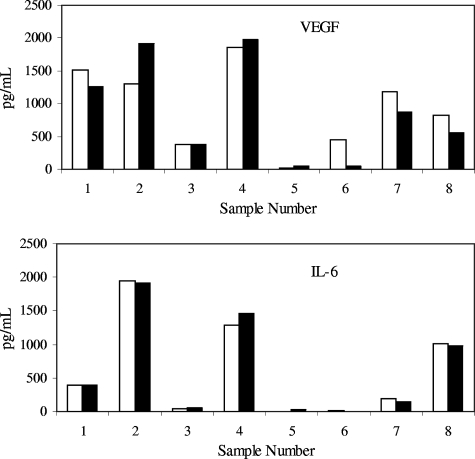
Further testing of the effect of modified sample diluent was performed upon disease state samples. The effect of the sample diluent was not as pronounced as with the spike recovery samples. The detection of VEGF and IL-6 in RA samples is shown as an example (Fig. 2), but the detection of cytokines across analytes and disease states was consistent, regardless of the sample diluent used. The difference is likely related not to disease state serum versus healthy serum but to the detectability of recombinant protein in serum compared to endogenous protein in serum.
FIG. 2.
Detection of VEGF and IL-6 in samples obtained from subjects with RA, using either standard diluent or modified diluent. Samples were tested in duplicate on two separate days; the values are the averages of the data points. Open bars, standard diluent; solid bars, modified diluent.
Serum samples from diseased subjects were further tested in each of the multiplex platforms by using modified diluent and in single ELISA formats. For each disease state sample, serum from 10 separate donors was tested in duplicate, on two separate days. The calculated concentration from all four determinations was averaged for comparison purposes.
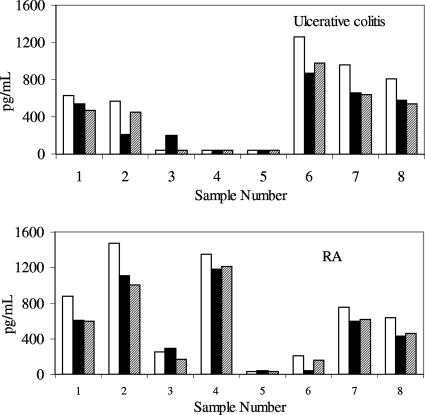
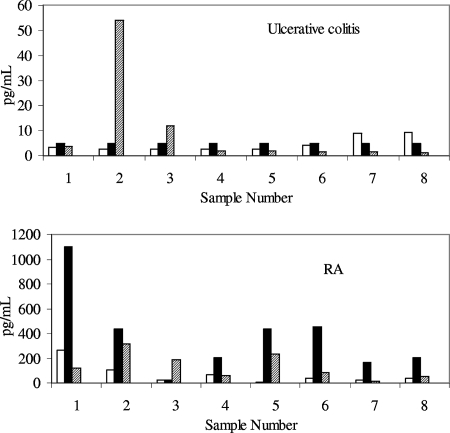
The consistency of cytokine detection across all three platforms varied depending upon the cytokine being tested and the disease state. As an example of platforms giving consistent results across diseases, the recovery seen with VEGF is given in Fig. 3. In both ulcerative colitis and RA, the detection of VEGF was comparable, with similar concentrations being found across platforms.
FIG. 3.
Detection of VEGF in samples obtained from subjects with ulcerative colitis or RA. Samples were tested in duplicate on two separate days; the values are the averages of the data points. Samples were tested on the Pierce SearchLight System. Open bars, MSD; solid bars, Pierce; diagonally striped bars, R&D.
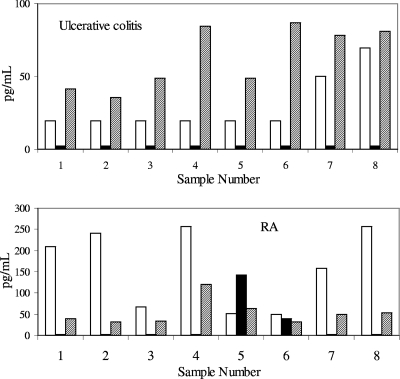
In contrast, the detection of IL-12p40 in samples was highly dependent upon the assay and the disease (Fig. 4). In ulcerative colitis, the Pierce platform did not measure IL-12p40 above the LLOQ of 2.3 pg/ml in any of the samples. The MSD platform gave similar results, with six/eight samples below the LLOQ of 19.5 pg/ml and only two of the samples having detectable analyte. In contrast, the R&D platform detected IL-12p40 in all of the samples with a range of 31.2 to 2,000 pg/ml. The detection of IL-12p40 in RA samples was also platform dependent. In this example, the Pierce and R&D platforms had six/eight samples at less than the LLOQ, while the MSD platform measured detectable IL-12p40 in all samples with a range of 19.5 to 2,500 pg/ml. The detection of TNF-α (Fig. 5) was also platform dependent. In ulcerative colitis, with the MSD and Pierce platforms, the majority of samples were near the LLOQ, while with the R&D assay two of the sample concentrations were well within the quantitation ranges of the assay. In RA samples, the Pierce platform gave much higher TNF-α quantities for seven/eight samples, compared with the MSD and R&D platforms.
FIG. 4.
Detection of IL-12p40 in samples obtained from subjects with ulcerative colitis or RA. Samples were tested in duplicate on two separate days; the values are the averages of the data points. Open bars, MSD; solid bars, Pierce; diagonally striped bars, R&D.
FIG. 5.
Detection of TNF-α in samples obtained from subjects with ulcerative colitis or RA. Samples were tested in duplicate on two separate days; the values are the averages of the data points. Open bars, MSD; solid bars, Pierce; diagonally striped bars, R&D.
The correlations between assay results by platform and disease are given in Table 4. In this table, the numbers represent the correlation coefficient resulting from comparing the results of one of the three platforms with the remaining two platforms. An asterisk indicates the statistical significance of the correlation. In this analysis, it can be seen that VEGF gives significant correlations in 11/12 comparisons, MCP-1 has 9/12 comparisons with significance, and MIP-1β has four/four. Thus, for fewer than half of the cytokines, the three platforms gave results that were comparable across the three platforms.
TABLE 4.
Correlation between assay kit results for disease state serum samples
| Cytokine and kit source | Correlation coefficient for disease and kit sourcea
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Ulcerative colitis
|
Crohn's disease
|
RA
|
Psoriasis
|
|||||
| Pierce | R&D | Pierce | R&D | Pierce | R&D | Pierce | R&D | |
| IFN-γ | ||||||||
| MSD | −0.459 | 0.441 | ||||||
| Pierce | ||||||||
| VEGF | ||||||||
| MSD | 0.888** | 0.976** | 0.515** | 0.905** | 0.919** | 0.952** | 0.847** | 0.976** |
| Pierce | 0.976** | 0.524 | 0.994** | 0.905** | ||||
| TNF-α | ||||||||
| MSD | −0.705* | 0.191 | −0.139 | 0.332 | 0.167 | 0.932** | 0.619 | |
| Pierce | −0.515 | 0.167 | 0.561 | |||||
| IL-6 | ||||||||
| MSD | 0.747** | 0.307 | −0.118 | 0.952** | 0.879** | 0.618* | ||
| Pierce | 0.453 | −0.247 | ||||||
| MIP-1β | ||||||||
| MSD | 0.595* | 0.935** | 0.976** | 0.924** | ||||
| Pierce | ||||||||
| MCP-1 | ||||||||
| MSD | 0.400 | 0.929** | 0.918** | 0.929** | 0.682 | 0.976** | 0.820** | 0.934** |
| Pierce | 0.986** | 0.929** | 0.338 | 0.689* | ||||
| IL-12p40 | ||||||||
| MSD | −0.746** | 0.431 | 0.190 | |||||
| Pierce | 0.023 | |||||||
| IL-4 | ||||||||
| MSD | −0.617* | 0.200 | ||||||
| Pierce | ||||||||
*, P ≤ 0.05; **, P ≤ 0.001; n = 10 per disease state, tested in duplicate on two separate days. Empty spaces are due to the results being less than LLOQ or the assay not being performed.
DISCUSSION
This study was intended to evaluate two different solid-phase-based multiplex platforms, based on their range of quantitation, the ability to recover a known quantity of added cytokine, and their ability to detect endogenous cytokine in serum samples obtained from individuals with various inflammatory diseases. Each platform was used to measure the same cytokines, configured by the manufacturer for optimal performance. The results of the multiplex analysis were compared against single ELISA kits that measured the same cytokines. All assays performed well with low intra-assay variability.
The consistency of detection of endogenous cytokine from disease state samples is of critical concern. The measurement of cytokines in serum has a bearing upon mechanism of action studies, pathway analysis, and response to treatment. The results presented in this study demonstrated both the consistency and the variation in measurement between multiplex platforms and single ELISA. Consistent measurements across platforms were seen with the VEGF, MIP-1β, and MCP-1 analysis in the four disease state samples. The remaining five cytokines were not consistent across platforms for the disease state samples. This highlights the need to validate multiplex assays and understand how the data correlate with standard ELISA results.
Having a wide quantitation range is an advantage of the multiplex platforms. The quantitation range of the platforms impacts sample analysis both in terms of LLOQ and in the necessity to repeat those samples that fall beyond the platform's ULOQ. The quantitation range of the MSD platform overall was broader than that for the other two platforms based on calibration curve. However, the MSD platform used undiluted samples for analysis, while the Pierce platform required that the samples be diluted 1:5 prior to testing. Assays to determine the true sensitivity of the platforms in serum showed that the sensitivities of the platforms were similar, although MSD still had a broader dynamic range with ULOQs well above that for the Pierce or the single-analyte ELISAs.
The quantitation assigned to the calibration curve will affect the relative quantity of cytokine detected. The VEGF analysis of ulcerative colitis and RA samples demonstrated this. The three platforms showed the same pattern of detection across samples, but the concentration of the analyte detected showed a consistent bias. The MSD recovery was slightly higher than that with the Pierce and R&D concentrations. This variation was not unexpected across platforms, and the relative difference would not affect the interpretation of results. According to the package inserts accompanying each assay, the three manufacturers used different methods of quantitation of the calibration curves. R&D Systems used in-house, purified recombinant proteins, while MSD and Pierce used vendor-supplied purified recombinant proteins.
The source of variation across platforms has been attributed to matrix effects (6), antibody cross-reactivity with other cytokines (1), the accuracy of the quantitation of the calibration curve (9), the source of the antibodies used for capture and detection (2), and the use of serum or plasma for sample collection (19). Matrix effects were shown in this study to have an effect upon the recovery of spiked cytokine in healthy donor samples and were corrected by modifying the sample diluent. However, analysis of disease samples with the modified diluent showed that the accuracy of detection was not as affected by use of the modified diluent. In the healthy donor serum, the unspiked samples had analyte concentrations near the LLOQ for both of the multiplex platforms.
Antibody cross-reactivity with similar cytokines is a potential source of variation as well. However, the package inserts and review of the cross-reactivity data with the manufacturers showed cross-reactivity of the cytokine detection in the products to be well assessed and controlled.
Another significant source of assay variation across platforms is the antibodies used for the capture and detection of antigen (6, 16). The multiplex manufacturers used a mix of in-house sources and external vendors for their antibody pairs, the source of which is held as proprietary by the manufacturer. Should the antibodies recognize the same epitopes on the target protein, then the platforms should give similar results. This is likely to be the case in the VEGF, MIP-1β, and MCP-1 assays. The remaining five antibodies in the platforms probably do not recognize the same epitopes. As an example, the IL-12p40 detection varies so significantly across platforms that the capture/detection sites of the antibodies are very likely to be distinct epitopic sites or possibly on fragmented or precursor forms of the cytokine (2).
It is known that the sample source can affect results. For example the use of plasma or serum can affect cytokine detection (19). In single ELISA, the prognostic ability of VEGF concentrations was greater in serum than it was in plasma. Differences in the performance of the platforms with regard to sample matrix are an important consideration in choosing a platform but were beyond the scope of this study, which focused on human serum analyses.
Heterophilic antibodies can be found in chronic inflammatory conditions such as RA. These antibodies can produce false-positive results or interfere in detection when serum is being analyzed by antigen-capture ELISA methods (4, 8, 12). The variable response seen in the cytokine detection in some of the disease samples could be due to the presence of heterophilic antibodies. Methods to reduce the nonspecific binding of these antibodies are recommended when testing disease samples with poor spike recovery on multiplex platforms. The preincubation of serum with mouse antibody or the depletion of heterophilic antibodies with protein L (4) has been shown to be effective.
In a comparison of multiplex bead arrays Khan et al. (9) found that although the cytokine levels follow similar patterns, the absolute levels of the cytokines measured varied by manufacturer. It was suggested that the quantitative values obtained from within a specific platform were reproducible but that across platforms there was enough variation that comparison of results would be difficult.
Urbanowska et al. (18) developed a plate-based multiplex consisting of six cytokines. The development of the assay included measures of the accuracy and precision and the LLOQ and ULOQ and a comparison with single ELISA technology. The correlation between multiplex and single ELISA was found to be good, with the correlation coefficients above 0.9 for all but one of the cytokines. Pooled serum was used for the development work, which tends to give more consistent results than does unpooled serum but is not reflective of the intended use of the assay for individual serum analyses.
We have found in this study that the solid-phase multiplex assays can perform reliably within a platform. However, certain steps must be taken when using these assays to obtain optimal data. It is important to work closely with the manufacturer of the platform chosen for the work. The manufacturers should, prior to supplying the multiplex array, test the possible combinations of antibodies on their array. This will reduce the incidence of cross-reactions and will combine analytes that require similar serum dilutions to be placed upon the same array.
The multiplex array should be validated in-house prior to study use. Additional validation steps should include spike recovery, dilutional linearity, stability of samples, the inter- and intraobserver variability, and measurement of the endogenous analyte. While assay validation is a multistep procedure, the recovery of spiked sample should be within predetermined ranges, typically within 100% ± 30% of the expected value. Pooled samples typically give better recovery than single samples. However, while pooled serum is useful for certain validation procedures, using pooled samples can lead to erroneous conclusions regarding the recovery of spiked antigen. It is important to understand the individual variation in recovery in normal sera as well as in available sera from the patient population(s) of interest. Through the use of single samples, we found that the recovery of antigen could be significantly affected due to inherent variability between subjects and that this could be improved by modifying the sample diluent.
The use of multiplex platforms for sample analysis has been increasing. The advantages of multiplex analysis, such as small sample volume, time savings, and the ability to simultaneously obtain data from a large number of cytokines, have been a great advancement in immunoassays. Due to the differences in the detection of cytokines across different platforms, a careful examination of the platform assay performance must be made prior to comparing results across platforms. While some cytokines will give similar patterns of results across platforms, others will be highly variable. Therefore, the use of the same platform across a study should be considered. While the assay results may be biased compared with other platforms, internally the results should be consistent. As the use of these assays increases and the volume of data in serum and plasma increases, the interpretation of data across platforms will be important and as such looking at relative change rather than absolute quantitation may be necessary. The expression of results as the percent change from a baseline level may be required for reliable interpretation.
Acknowledgments
This work was supported by Centocor R&D. All of the authors are employees of Centocor.
Footnotes
Published ahead of print on 14 November 2007.
REFERENCES
- 1.Aziz, N., P. Nishanian, R. Mitsuyasu, R. Detels, and J. L. Fahey. 1999. Variables that affect assays for plasma cytokines and soluble activation markers. Clin. Diagn. Lab. Immunol. 6:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Jager, W., and G. T. Rijkers. 2006. Solid-phase and bead-based cytokine immunoassay: a comparison. Methods 38:294-303. [DOI] [PubMed] [Google Scholar]
- 3.de Jager, W., H. te Velthuis, B. J. Prakken, W. Kuis, and G. T. Rijkers. 2003. Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells. Clin. Diagn. Lab. Immunol. 9:133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Jager, W., B. J. Prakken, J. W. J. Bijlsma, W. Kuis, and G. T. Rijkers. 2005. Improved multiplex immunoassay performance in human plasma and synovial fluid following removal of interfering heterophilic antibodies. J. Immunol. Methods 300:124-135. [DOI] [PubMed] [Google Scholar]
- 5.Dinarello, C. A. 2000. Proinflammatory cytokines. Chest 118:503-508. [DOI] [PubMed] [Google Scholar]
- 6.Elshal, M. F., and J. P. McCoy. 2006. Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods 38:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahey, J. L., J. M. G. Taylor, B. Manna, P. Nishanian, N. Aziz, J. V. Giorgi, and R. Detels. 1998. Prognostic significance of plasma markers of immune activation, HIV viral load and CD4 T-cell measurements. AIDS 12:1581-1590. [DOI] [PubMed] [Google Scholar]
- 8.Hennig, C., L. Rink, U. Fagin, W. J. Jabs, and H. Kirchner. 2000. The influence of naturally occurring heterophilic anti-immunoglobulin antibodies on direct measurement of serum proteins using sandwich ELISAs. J. Immunol. Methods 235:71-80. [DOI] [PubMed] [Google Scholar]
- 9.Khan, S. S., M. S. Smith, D. Reda, A. F. Suffredini, and J. P. McCoy. 2004. Multiplex bead array assays for the detection of soluble cytokines: comparison of sensitivity and quantitative values among kits from multiple manufacturers. Cytometry B Clin. Cytom. 618:35-39. [DOI] [PubMed] [Google Scholar]
- 10.Kingsmore, S. F. 2006. Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat. Rev. Drug Discov. 5:310-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lash, G. E., P. J. Scaife, B. A. Innes, H. A. Otun, S. C. Robson, R. F. Searle, and J. N. Bulmer. 2006. Comparison of three multiplex cytokine analysis systems: Luminex, SearchLight and FASTQuant. J. Immunol. Methods 309:205-208. [DOI] [PubMed] [Google Scholar]
- 12.Martins, T. B., B. M. Pasi, C. M. Litwin, and H. R. Hill. 2004. Heterophile antibody interference in a multiplexed fluorescent microsphere immunoassay for quantitation of cytokines in human serum. Clin. Diagn. Lab. Immunol. 11:325-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendoza, L. G., P. McQuary, A. Mongan, R. Gangadharan, S. Brignac, and M. Eggers. 1999. High-throughput microarray-based enzyme-linked immunosorbent assay (ELISA). BioTechniques 27:778-788. [DOI] [PubMed] [Google Scholar]
- 14.Moser, B., and K. Willimann. 2004. Chemokines: role in inflammation and immune surveillance. Ann. Rheum. Dis. 63(Suppl. II):ii84-ii89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen, U. L., and B. H. Geierstanger. 2004. Multiplexed sandwich assays in microarray format. J. Immunol. Methods 290:107-120. [DOI] [PubMed] [Google Scholar]
- 16.Prabhakar, U., E. Eirikis, and H. M. Davis. 2002. Simultaneous quantification of proinflammatory cytokines in human plasma using the LabMAP assay. J. Immunol. Methods 260:207-218. [DOI] [PubMed] [Google Scholar]
- 17.Prabhakar, U., E. Eirikis, M. Reddy, E. Silvestro, S. Spitz, C. Pendley, H. M. Davis, and B. E. Miller. 2004. Validation and comparative analysis of a multiplexed assay for the simultaneous quantitative measurement of Th1/Th2 cytokines in human serum and human peripheral blood mononuclear cell culture supernatants. J. Immunol. Methods 291:27-38. [DOI] [PubMed] [Google Scholar]
- 18.Urbanowska, T., S. Mangialaio, C. Zickler, S. Cheevapruk, P. Hasler, S. Regenass, and F. Legay. 2006. Protein microarray platform for the multiplex analysis of biomarkers in human serum. J. Immunol. Methods 316:1-7. [DOI] [PubMed] [Google Scholar]
- 19.Werther, K., I. J. Christensen, H. J. Nielsen, and the Danish RANX05 Colorectal Cancer Study Group. 2002. Prognostic impact of matched preoperative plasma and serum VEGF in patients with primary colorectal carcinoma. Br. J. Cancer 86:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]