Abstract
The development and validation of a microsphere immunoassay (MIA) to detect equine antibodies to the major structural proteins of equine arteritis virus (EAV) are described. The assay development process was based on the cloning and expression of genes for full-length individual major structural proteins (GP5 amino acids 1 to 255 [GP51-255], M1-162, and N1-110), as well as partial sequences of these structural proteins (GP51-116, GP575-112, GP555-98, M88-162, and N1-69) that constituted putative antigenic regions. Purified recombinant viral proteins expressed in Escherichia coli were covalently bound to fluorescent polystyrene microspheres and analyzed with the Luminex xMap 100 instrument. Of the eight recombinant proteins, the highest concordance with the virus neutralization test (VNT) results was obtained with the partial GP555-98 protein. The MIA was validated by testing a total of 2,500 equine serum samples previously characterized by the VNT. With the use of an optimal median fluorescence intensity cutoff value of 992, the sensitivity and specificity of the assay were 92.6% and 92.9%, respectively. The GP555-98 MIA and VNT outcomes correlated significantly (r = 0.84; P < 0.0001). Although the GP555-98 MIA is less sensitive than the standard VNT, it has the potential to provide a rapid, convenient, and more economical test for screening equine sera for the presence of antibodies to EAV, with the VNT then being used as a confirmatory assay.
Equine arteritis virus (EAV) is the causative agent of equine viral arteritis (EVA), a respiratory and reproductive disease of horses (61). EAV is a small enveloped virus with a positive-sense, single-stranded RNA genome of 12.7 kb and belongs to the family Arteriviridae (genus Arterivirus, order Nidovirales), which also includes porcine reproductive and respiratory syndrome virus, simian hemorrhagic fever virus, and lactate dehydrogenase-elevating virus of mice (13, 57). The EAV genome includes nine functional open reading frames (ORFs) (55, 57). ORFs 1a and 1b encode two replicase polyproteins (pp1a and pp1ab) (25, 55, 57), and the remaining seven ORFs (2a, 2b, and 3 to 7) encode the structural proteins of the virus. These include four membrane glycoproteins, GP2 (25 kDa), GP3 (36 to 42 kDa), GP4 (28 kDa), and GP5 (30-44 kDa), encoded by ORFs 2b, 3, 4, and 5, respectively; two unglycosylated membrane proteins, E (8 kDa) and M (17 kDa), encoded by ORFs 2a and 6; and the phosphorylated nucleocapsid protein N (14 kDa), encoded by ORF 7 (23, 58, 64). The major envelope glycoprotein GP5 expresses the known neutralization determinants of EAV. The two major envelope proteins GP5 and M form a disulfide-linked heterodimer in the virus particle, and this association is critical for their maturation and for the expression of some of the neutralization epitopes in authentic form (3, 24, 56).
Serological and clinical studies indicate that EAV is widely distributed in equine populations around the world (35, 46, 48). Although there is considerable variation in the sequences of the GP5 proteins of field strains of the virus, there is only one known serotype of EAV, and all strains evaluated thus far are neutralized by polyclonal antiserum raised against the prototype Bucyrus strain of EAV (1, 4, 6, 7, 11, 14, 22, 30). Both natural and experimental infections of horses with either virulent or avirulent strains of EAV result in long-lasting immunity against all strains of the virus (27, 44, 45). EAV infection in horses induces antibodies against the three major viral structural proteins, GP5, M, and N (4, 33, 41). Virus-neutralizing (VN) antibodies are detectable between 1 and 2 weeks after primary infection, and their appearance usually coincides with the onset of clearance of the virus from the circulation (28, 43). However, virus does persist in the male reproductive tract of certain stallions for long periods of time, despite the presence of high titers of virus-specific neutralizing antibodies in their sera.
Serologic diagnosis of acute EAV infection in horses is based on the demonstration of seroconversion or of a ≥4-fold increase in VN antibody titers by comparing paired (acute- and convalescent-phase) serum samples by the virus neutralization test (VNT). The VNT is the principal serological assay used to detect evidence of EAV infection by most laboratories around the world, and it continues to be the current World Organisation for Animal Health (OIE)-prescribed standard test for EVA (20, 40, 50, 54, 61). The assay is used for diagnostic, epidemiological surveillance, international trade, and vaccination-monitoring purposes. The number of equine sera tested annually by a diagnostic laboratory can vary considerably, from several hundred to thousands of samples per year (40, 54, 62). Although the VNT is currently the most highly sensitive and specific serodiagnostic test for EVA infection, it is expensive, labor-intensive, and time-consuming to perform. In addition, results tend to vary among laboratories when adequate attention is not paid to the standardization of both test reagents and procedure. Moreover, serum cytotoxicity caused by anticellular antibodies directed against rabbit kidney 13 (RK-13) cells can be mistaken for a viral cytopathic effect and can give rise to difficulties in test interpretation at lower serum dilutions (20, 29, 40). To overcome these disadvantages, several laboratories have developed and evaluated enzyme-linked immunosorbent assays (ELISAs) to detect antibodies to EAV using whole virus, synthetic peptides, or recombinant viral proteins (e.g., GP5, M, and N) as antigens (7, 14-17, 39, 49, 60, 63). The various studies have shown that the source of antigen as well as the sera evaluated can markedly influence the results obtained with EAV protein-specific ELISAs and competitive ELISAs (4, 34). None of these ELISAs have yet been shown to have sensitivity and specificity equivalent to those of the VNT.
Recent developments in particle array technology have made it possible to perform immunoassays using microspheres (microbeads). The best-established microsphere assay system is the xMap system (Luminex Corp., Austin, TX), which incorporates three well-developed technologies: bioassays, solution-phase microspheres, and flow cytometry. The microsphere assay technology developed by Luminex is ideally suited to a wide range of applications in drug discovery and diagnostics, as well as basic research. Immunoassays based on this particle array technology can overcome the problems associated with the traditional VNT and ELISAs. Some of the distinct advantages of a microsphere immunoassay (MIA) over traditional ELISAs include accuracy; high sensitivity, specificity, and reproducibility; high-throughput sample analysis; and multiplexing capability. MIAs are becoming increasingly popular for the serologic diagnosis of autoimmune and infectious diseases of humans and animals (8, 26, 38, 65, 66). The primary objective of the present study was to develop a reliable immunological assay to detect antibodies to EAV in horses by using Luminex xMap technology and to compare the performance of the assay with that of the VNT. The MIA could be suitable to detect antibodies to EAV in equine sera, and the specificity and sensitivity of this assay should be equivalent to those of the standard VNT.
MATERIALS AND METHODS
Cells.
High-passage-number (passage 399 [P399] to P409) RK-13 cells were propagated and maintained in Eagle's minimum essential medium (Mediatech, Herndon, VA) supplemented with 10% fetal calf serum (HyClone, Logan, UT), 100 U of penicillin/ml, 100 μg of streptomycin/ml, 1 μg of amphotericin B/ml, and 0.06% sodium bicarbonate at 37°C (47).
Equine sera.
A total of 2,500 diagnostic equine serum samples were evaluated for the presence of anti-EAV antibodies by the MIA and the VNT. The sera used in the study were selected randomly from those submitted for serological testing to the OIE EVA Reference Laboratory at the Gluck Equine Research Center (n = 1,500) and the Livestock Disease Diagnostic Center (n = 1,000), University of Kentucky, Lexington. Panels of EAV antibody-positive and antibody-negative sera from the Gluck Equine Research Center were selected and used to establish normal ranges of MIA results for negative and positive samples. In addition to these sera, a panel of 192 archived sequential serum samples collected from 18 experimentally infected horses were evaluated with respect to the EAV-specific antibody response by both the VNT and the MIA. The horses were divided into four groups, and each group was inoculated with a different strain of EAV (rVBS, n = 4; 030H, n = 2; KY84, n = 7; and CA95G, n = 5) (2, 9, 10, 51). Blood samples were collected at 0, 2, 4, 6, 8, 10, 12, 14, 21, 28, 35, and 42 days postinfection (dpi) from the EAV rVBS-inoculated horses; at 0, 2, 4, 6, 8, 10, 12, 14, and 21 dpi from the EAV 030H-inoculated horses; at 0, 2, 4, 6, 8, 10, 12, 14, 21, and 28 dpi from the EAV KY84-inoculated horses, and at 0, 2, 4, 7, 9, 14, 21, 28, and 35 dpi from the EAV CA95G-inoculated horses. Sera were aliquoted and stored at −20°C.
PCR amplification, cloning, and sequencing of full-length and truncated versions of ORFs 5, 6, and 7 of EAV.
The oligonucleotide primers for the amplification of the coding sequences of the GP5, M, and N protein genes (ORFs 5, 6, and 7, respectively), as well as the corresponding partial-length genes, were designed according to the published sequence of the virulent Bucyrus strain of EAV (GenBank accession number DQ846750) (9). The nucleotide sequence 5′-CACC-3′ was added at the 5′ end of each primer for directional cloning into the pET TOPO vector (Invitrogen, Carlsbad, CA) (Table 1). The full-length ORFs 5, 6, and 7 (which encode full-length GP5, M, and N proteins, respectively), as well as the coding regions for the amino-terminal ectodomain (amino acids [aa] 1 to 116) and two antigenic regions (aa 55 to 98 and aa 75 to 112) of the GP5 protein (14, 15, 49), the antigenic carboxyl terminus (aa 88 to 162) of the M protein (37), and the antigenic amino terminus (aa 1 to 69) of the N protein (16), were PCR amplified from the plasmid containing the complete genomic sequence of the EAV virulent Bucyrus strain (pEAVrVBS; GenBank accession number DQ846751) (9) by using Pfu DNA polymerase enzyme (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The individual PCR products were concentrated using a Centricon centrifugal filter unit (Ultracel YM-30; Millipore, Billerica, MA) and purified using a commercial kit (QIAGEN, Valencia, CA). The individually amplified cDNA fragments comprising EAV ORFs 5, 6, and 7 and their respective truncated forms were then directly cloned into the pET100 directional TOPO vector by using the Champion pET100/D-TOPO expression kit according to the instructions of the manufacturer (Invitrogen, Carlsbad, CA). The pET100/D-TOPO vector allows the expression of a recombinant protein with an Xpress epitope and a polyhistidine (six-His) tag at the amino terminus. One Shot competent Escherichia coli cells (Invitrogen, Carlsbad, CA) were transformed with plasmids containing individual ORFs. Following transformation and purification, individual plasmids were identified and characterized by restriction enzyme analysis to ensure the correct orientation of the insert. The authenticity of each ORF and of each truncated version was confirmed by automatic sequencing, as previously described (34).
TABLE 1.
Primer pairs used for PCR amplification of full-length and partial-length segments of ORFs 5, 6, and 7
| ORF | Protein name | Primer sense | Primer sequencea | Nucleotide positionb | Size of PCR product (bp) | Recombinant plasmid |
|---|---|---|---|---|---|---|
| 5 | GP51-255 | + | 5′-CACCATGTTATCTATGATTGTATTG-3′ | 11146-11166 | 772 | pET-GP51-255 |
| − | 5′-CTATGGCTCCCATACCTCAG-3′ | 11913-11894 | ||||
| GP51-116 | + | 5′-CACCATGTTATCTATGATTGTATTG-3′ | 11146-11166 | 355 | pET-GP51-116 | |
| − | 5′-CTAGAATTCACGGCCATAGTAA-3′ | 11493-11475 | ||||
| GP555-98 | + | 5′-CACCTACAATTGTTCCGCCAGTAAAAC-3′ | 11308-11330 | 139 | pET-GP555-98 | |
| − | 5′-CTACGGTCCATGCGCCTGTTCC-3′ | 11439-11421 | ||||
| GP575-112 | + | 5′-CACCACGTTTGGAACCGATTG-3′ | 11368-11384 | 121 | pET-GP575-112 | |
| − | 5′-CTAATAGTAAATAAAAGGGGGCATGTC-3′ | 11481-11458 | ||||
| 6 | M1-162 | + | 5′-CACCATGGGAGCCATAGATTC-3′ | 11901-11917 | 493 | pET-M1-162 |
| − | 5′-TCATTGTAGCTTGTAGGCTGTCG-3′ | 12389-12367 | ||||
| M88-162 | + | 5′-CACCATGCCTCGTCTTCGGTCC-3′ | 12162-12179 | 232 | pET-M88-162 | |
| − | 5′-CTATTGTAGCTTGTAGGCTGTCGCCG-3′ | 12386-12364 | ||||
| 7 | N1-110 | + | 5′-CACCATGGCGTCAAGACGATCACG-3′ | 12313-12332 | 337 | pET-N1-110 |
| − | 5′-TTACGGCCCTGCTGGAGGCGC-3′ | 12645-12625 | ||||
| N1-69 | + | 5′-CACCATGGCGTCAAGACGATCACG-3′ | 12313-12332 | 211 | pET-N1-69 | |
| − | 5′-CTAGTTCGACGAAAGGGTGGCGCG-3′ | 12519-12499 |
Expression and purification of full-length GP5, M, and N and the respective partial-length recombinant proteins.
Plasmids pET-GP51-255, pET-GP51-116, pET-GP575-112, pET-GP555-98, pET-M1-162, pET-M88-162, pET-N1-110, and pET-N1-69 were purified and used to transform the BL21 Star(DE3) strain of E. coli for the expression of the recombinant proteins. The E. coli strain BL21 Star(DE3) is specially designed for the expression of genes regulated by the T7 promoter. Following transformation with each expression plasmid, a single colony was grown for approximately 3 h at 37°C in 2 ml of Luria-Bertani liquid medium supplemented with 200 μg of carbenicillin/ml (LBC). Thereafter, 50 μl of the culture was plated onto an LBC agar plate and the plate was incubated overnight at 37°C. On the next day, bacterial colonies on the agar plate were scraped off with a sterile tissue culture scraper and added to a 2-liter flask containing 500 ml of LBC. Cultures were grown to an optical density at 600 nm of 0.5 to 0.7. Protein expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Five hours after induction, bacterial cells were harvested by centrifugation at 3,000 ×g for 15 min at 4°C. Cell pellets were either stored at −80°C or immediately used for further processing.
The recombinant GP5 protein comprising amino acids 1 to 255 (GP51-255) and the recombinant proteins GP51-116, GP575-112, and GP555-98 were purified using a nickel-nitrilotriacetic acid (Ni-NTA) purification system (Invitrogen, Carlsbad, CA) under hybrid conditions. Briefly, individual cell pellets were resuspended in guanidinium lysis buffer (6 M guanidine HCl, 20 mM sodium phosphate, 500 mM NaCl, pH 7.8) containing lysozyme (1 mg/ml) and proteinase inhibitors (Halt proteinase inhibitor cocktail kit; Pierce, Rockford, IL) and slowly mixed on a rocker for 10 min at room temperature. After incubation, cell pellets were lysed by sonication with shortwave pulses of 10 s at high intensity (output, 5). Subsequently, the crude lysates were centrifuged at 3,000 × g for 15 min at 4°C to remove bacterial cellular debris. The supernatant was loaded onto an Ni-NTA agarose column and allowed to bind for 1 h at room temperature. After binding, the agarose-containing column was washed twice with denaturing binding buffer (8 M urea, 20 mM sodium phosphate, 500 mM NaCl, pH 7.8) and twice with denaturing wash buffer (8 M urea, 20 mM sodium phosphate, 500 mM NaCl, pH 6.0), to wash off nonspecifically bound proteins. Subsequently, protein-bound agarose was washed four times with native wash buffer (50 mM NaH2PO4, 0.5 M NaCl, 10 mM imidazole, pH 8.0). Finally, each recombinant protein was eluted from the column with 2-ml fractions of native elution buffer (50 mM NaH2PO4, 0.5 M NaCl, 250 mM imidazole, pH 8.0).
Unlike the GP5 recombinant proteins, the recombinant full-length M1-162, partial-length M88-162, full-length N1-110, and partial-length N1-69 proteins were purified under native conditions. Briefly, individual cell pellets were resuspended in native binding buffer (50 mM NaH2PO4, 0.5 M NaCl, pH 8.0) containing lysozyme (1 mg/ml) and proteinase inhibitors (Halt proteinase inhibitor cocktail kit; Pierce, Rockford, IL) and incubated on ice for 30 min. After incubation, cell pellets were lysed by sonication with shortwave pulses of 10 s at high intensity (output, 5). Subsequently, the crude sonicates were centrifuged at 3,000 × g for 15 min at 4°C to remove cellular debris. The supernatant was loaded onto an Ni-NTA agarose column and allowed to bind for 2 h at 4°C. The recombinant proteins were eluted in native elution buffer (pH 8.0) as described above.
Fractions containing the same recombinant protein were pooled together and centrifuged at 3,000 × g for 45 min with an Amicon Ultra 5K centrifugal filter device (Millipore, Billerica, MA) used to increase the recombinant protein concentration. After concentration, individual proteins were dialyzed against phosphate-buffered saline (PBS; pH 7.4) overnight at 4°C. Final protein concentrations were determined using a bicinchoninic acid protein assay (Pierce, Rockford, IL), and the proteins were stored at −80°C.
Western immunoblotting assay.
The authenticity of the purified recombinant proteins was confirmed by a Western immunoblotting assay as previously described (5, 7). Briefly, proteins were mixed with an equal volume of 2× Laemmli sample buffer (Bio-Rad, Hercules, CA) containing 62.5 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate, 25% glycerol, 0.01% bromophenol blue, and 200 mM dithiothreitol (Pierce, Rockford, IL), and the mixtures were incubated for 15 min at room temperature. Denatured samples were loaded onto sodium dodecyl sulfate-12% polyacrylamide resolving gel and a 5% stacking gel. Following electrophoresis, recombinant proteins were transferred onto a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA). Immunoblotting was performed by using 5% nonfat dried milk powder in Tris-buffered saline with 0.05% Tween 20 (pH 7.6) as the blocking solution. The expression and authenticity of each recombinant protein were confirmed using antibodies against the N-terminal six-His tag, as well as individual EAV protein-specific monoclonal antibodies 6D10 for GP5 (7), 3E2 for N (42), and rabbit anti-EAV M no. 7888 for M protein (41). Bound antibodies were detected by enhanced chemiluminescence, as previously described (7).
VNT.
The neutralizing antibody titers of the test sera were determined as described by Senne et al. and in the OIE standards manual (50, 53). Briefly, serial twofold dilutions of each sample from 1:2 to 1:256 were made in minimal essential medium (Invitrogen, Carlsbad, CA) containing 10% guinea pig complement (Rockland Immunochemicals, Gilbertsville, PA). Each serum sample was tested in duplicate in 96-well plates. A working dilution of virus containing an estimated 200 50% tissue culture infective doses of the modified live-virus strain of EAV (ARVAC; Ft. Dodge Animal Health) in a volume equal to that of the serum sample was added to each well, except those with the serum sample controls. The plates were shaken to ensure mixing of the well contents and then incubated for 1 h at 37°C. A suspension of high-passage-number (P399 to P409) RK-13 cells was added to each well in a volume equivalent to that of the serum-virus mixture, and the plates were incubated at 37°C until a viral cytopathic effect had fully developed in the virus control wells. The titer of a sample was recorded as the reciprocal of the highest serum dilution that provided at least 50% neutralization of the reference virus.
Coupling of recombinant proteins to microspheres.
Aliquots of approximately 50 μg of each recombinant protein were covalently conjugated to carboxyl groups on the surfaces of xMap multianalyte COOH microspheres according to the protocol of the microsphere manufacturer (Luminex Corp, Austin, TX). Each recombinant protein was attached to 6.25 × 106 microspheres, and the microspheres were counted with a hemocytometer so that equivalent quantities of each specific bead set were incorporated into the assay.
Development of MIA.
A 96-well 1.2-μm-pore-size filter plate was blocked for 2 min with 100 μl of a buffer containing PBS (pH 7.4), 1% bovine serum albumin, and 0.02% sodium azide (PBN buffer) and then washed once with 190 μl of a buffer containing PBS (pH 7.4) and 0.05% Tween 20 (PBS-T). Wells were kept moist by the addition of 20 μl of PBN buffer. Equine serum (50 μl; diluted 1:100 in PBN buffer) was added to each well of the filter plate. Approximately 2,500 recombinant-antigen-conjugated microspheres per protein were added to each well in 50 μl of PBN buffer. The plate was incubated on a shaker in the dark for 30 min at 37°C and then washed three times with PBS-T by using a vacuum manifold. Affinity-purified biotin-labeled goat anti-equine immunoglobulin G (50 μl of a 1:1,000 dilution in PBN buffer) was added, and the plate was incubated on a shaker in the dark for 30 min at 37°C and then washed twice with PBS-T. R-phycoerythrin-conjugated streptavidin (50 μl of a 1:100 dilution in PBN buffer) was added, and the plate was incubated on a shaker in the dark for 30 min at 37°C and then washed twice with PBS-T by using a vacuum manifold. The microspheres were then resuspended in 125 μl of PBN buffer per well, and 75-μl aliquots of the suspensions were transferred into a clear polystyrene 96-well plate. Microspheres were aspirated through the flow cell of a dual-laser Luminex 100 instrument. The median fluorescence intensity (MFI) for 100 microspheres for each specific protein was recorded for each well.
Statistical analysis.
A comparison of the diagnostic performances of the VNT and the MIA using the GP555-98 and N1-110 proteins was conducted using nonparametric density estimation and receiver operating characteristic (ROC) analysis. An ROC curve provides a graphical measure of the accuracy of a continuous diagnostic test. It represents a plot of the true-positive fraction (TPF) versus the false-positive fraction across all possible cutoff values that can be used to dichotomize the data into positive and negative outcomes. A related parameter of interest is the area under the curve (AUC). For continuous tests, the AUC equals the probability that a randomly selected diseased individual will have a test score that is greater than that for a randomly selected nondiseased individual. A diagnostic test that perfectly discriminates between nondiseased and diseased individuals has an ROC curve that is a horizontal line expressed by the following equation: TPF = 1 (y = 1). The corresponding AUC would also be equal to 1. Diagnostic tests that are purely random and, hence, are worthless have an ROC curve that is a 45°-angled line with a corresponding AUC of 0.5 (52).
Cross-sectional data were obtained from a dual-test study design in which both tests were carried out with sera from 2,500 horses and in which the true infection status of each horse was determined using the definitive “gold standard” test, the VNT. Pearson's correlation coefficient was estimated for the GP555-98-based MIA and the VNT. A Bayesian bivariate normal analysis of the log-transformed joint (GP555-98 and N1-110 MIA) outcomes was used to estimate ROC curves and AUCs based on methods developed by Choi et al. (18). Specifically, data derived using sera from infected horses were modeled as a bivariate normal distribution with an unknown mean vector (μGP555-98 for the GP555-98 test and μN1-110 for the N1-110 test) and an unknown covariance matrix that contained three parameters, namely, the variances for the GP555-98 test and for the N1-110 test and the correlation between GP555-98 and N1-110 test values. Independent diffuse normal prior distributions with a mean of 0 and a variance of 100 were used for μGP555-98 and μN1-110, and independent inverse gamma priors with a mean of 1 and a variance of 1,000 were used for the variances. The correlation was assigned a uniform prior ranging from −1 to 1. A bivariate normal distribution was also used to model the data for uninfected horses, with the same prior specification used for infected horses.
Results from the ROC analysis were used to determine an optimal cutoff value for the purpose of classifying horses as EAV antibody positive or negative based on the outcome of the GP555-98 MIA test. The sensitivity and specificity of the GP555-98 MIA test at this cutoff value were then estimated using Bayesian methods. The analysis was performed with the S-Plus (Insightful Corp., Seattle, WA) and WinBUGS (59) programs.
The sensitivity of each recombinant EAV structural protein-based MIA was calculated as follows: [number of true positives/(number of true positives + number of false negatives)] × 100. Test specificities were calculated using the following equation: [number of true negatives/(number of true negatives + number of false positives)] × 100.
RESULTS
Cloning, expression, and purification of recombinant proteins.
Following PCR amplification, all major structural protein genes and their corresponding partial-length ORFs were individually cloned into the pET100/D-TOPO vector. The recombinant protein from each construct was expressed as a bacterial fusion protein with an N-terminal tag containing the Xpress epitope and a six-His tag (Fig. 1A). Various temperatures and IPTG concentrations were evaluated to optimize the conditions for protein expression. With the exception of recombinant GP51-255 and the corresponding partial-length forms (GP51-116, GP575-112, and GP555-98), all the expressed recombinant proteins were present in the soluble fraction of the E. coli lysate. The insoluble GP5 recombinant proteins had to be purified under hybrid conditions instead of native conditions, which were successful for the soluble recombinant proteins (M1-162, M88-162, N1-110, and N1-69).
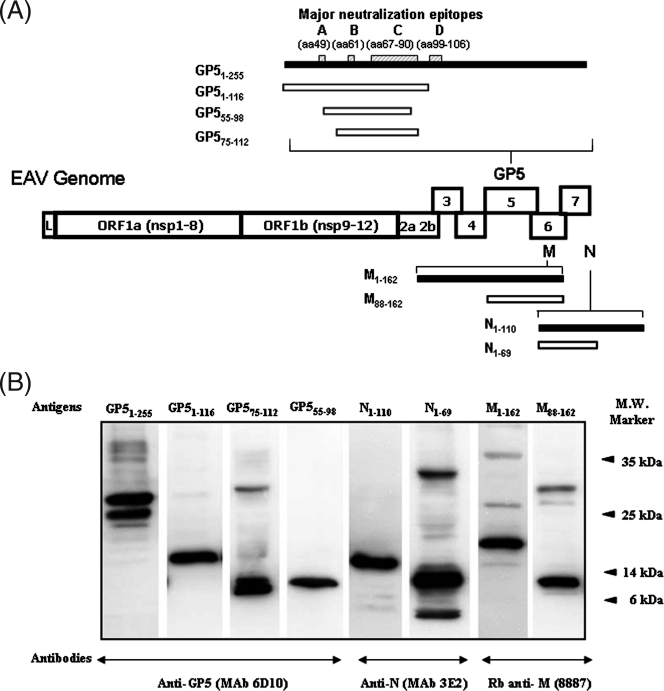
FIG. 1.
(A) Schematic representation of various EAV structural proteins expressed in E. coli. The recombinant full-length proteins derived from EAV ORFs 5, 6, and 7 and their respective truncated versions are depicted. The black bars denote full-length proteins and the white bars denote partial-length proteins. The putative major neutralization sites A (aa 49), B (aa 61), C (aa 67 to 90), and D (aa 99 to 106) located in the N-terminal ectodomain of the GP5 protein are indicated by hatched boxes (1, 4, 6). L, leader sequence; nsp, nonstructural proteins. (B) Western blot analysis of purified bacterial recombinant proteins with EAV protein-specific monoclonal antibodies (MAb) and rabbit (Rb) antisera. Monoclonal or rabbit EAV-specific antibodies used to detect each recombinant protein are indicated at the bottom of the figure. The molecular mass (M.W.) marker is indicated on the right. The molecular masses of recombinant GP51-255, GP51-116, GP575-112, GP555-98, N1-110, N1-69, M1-162, and M88-162 proteins were 32, 17, 7, 8, 15.3, 10.6, 20.6, and 11 kDa, respectively.
The expression and authenticity of the recombinant proteins were confirmed by Western immunoblotting assays. EAV structural protein-specific antibodies strongly reacted with the respective recombinant proteins (Fig. 1B). In addition, the attachment of six-His tags to the expressed proteins was demonstrated by Western immunoblotting with horseradish peroxidase-labeled anti-HisG antibody (data not shown).
VNT results.
Of the 2,500 equine serum samples tested, 1,750 were negative for VN antibodies to EAV (titers were <1:4). A total of 750 sera were positive for antibodies to EAV, with titers ranging from 1:4 to ≥1:512.
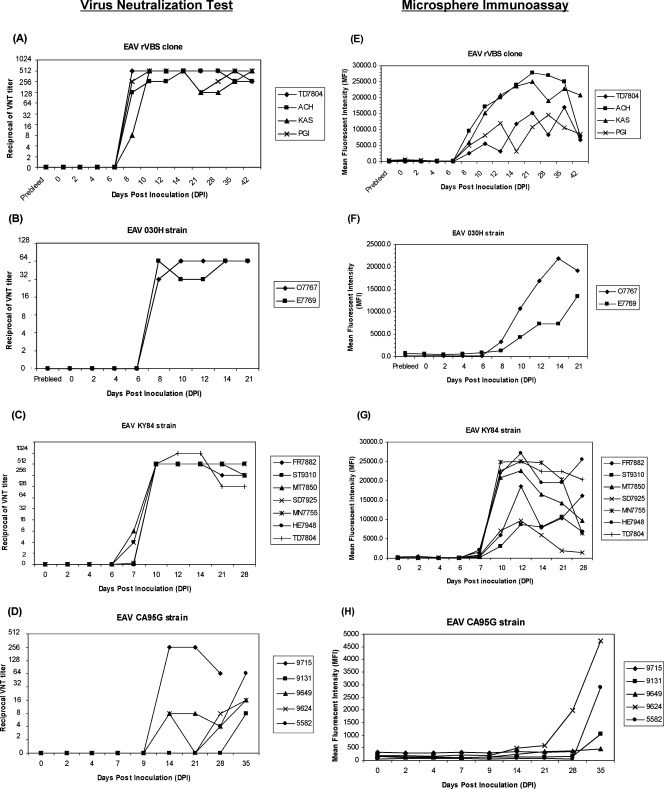
In addition to the diagnostic samples, a panel of 192 archived sequential sera from 18 horses experimentally infected with different strains of EAV was evaluated using the VNT. The horses experimentally inoculated with EAV rVBS, 030H, and KY84 strains developed VN antibodies to EAV 7 to 10 days postexposure (Fig. 2A, B, and C). Sera from horses inoculated with EAV rVBS and KY84 had VN antibody titers ranging from 1:128 to 1:1,024. The majority of horses in these two groups maintained a titer of ≥1:512 throughout the study. Horses infected with the EAV 030H laboratory strain developed lower antibody titers ranging from 1:32 to 1:64. In contrast, horses inoculated with the avirulent CA95G strain developed detectable serum antibody titers from 14 to 35 dpi (Fig. 2D). Neutralizing antibody titers in sera from the CA95G-inoculated horses were generally lower than those in sera from horses inoculated with the other virus strains, ranging from only 1:4 to 1:16. Only one horse in this group developed a high antibody titer (1:256).
FIG. 2.
Comparison of antibody responses to EAV as determined by GP555-98 MIA and VNT using sequential serum samples from experimentally infected horses (n = 18). The reciprocal VN titers and MFI values for horses inoculated with different EAV strains, rVBS, 030H, KY84, and CA95G, are plotted against dpi. (A, B, C, and D) VN antibody responses in horses experimentally infected with EAV rVBS (n = 4), EAV 030H (n = 2), EAV KY84 (n = 7), and EAV CA95G (n = 5) as measured by the standard VNT. (E, F, G, and H) VN antibody responses in horses experimentally infected with EAV rVBS (n = 4), EAV 030H (n = 2), EAV KY84 (n = 7), and EAV CA95G (n = 5) as measured by the GP555-98 MIA. Designations at the right of each graph represent individual identities of the horses from which samples were obtained.
Development of EAV recombinant structural protein-based MIAs.
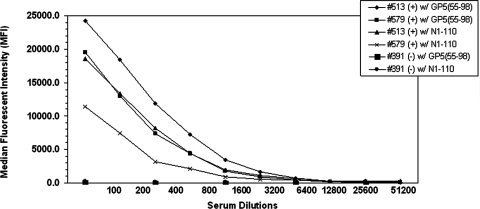
Eight expressed recombinant proteins were used to coat polystyrene microbeads in the development of the MIAs. The optimal dilution of the test sera to be used in the MIAs was predetermined by the use of known strong-positive (titer, ≥1:512) and negative (titer, <1:4) equine sera. The optimal working dilution was considered to be the serum dilution with the lowest level of background activity from nonspecific reactions. At a serum dilution of 1:100, the maximal dynamic range for all the recombinant proteins was attained; this dilution was defined as the optimal serum dilution for each protein (Fig. 3). To establish cutoff levels for the MIAs specific for each EAV recombinant protein, 20 equine sera from known EAV-seronegative horses were used. The MIA positive cutoff for each protein was established at 3 standard deviations (SD) above the microsphere MFI for the known VNT-negative sera. The MFI for GP51-255 was 279.5 (SD, 345), with an assay cutoff of 1,315; the MFI for GP51-116 was 237.9 (SD, 321), with a cutoff of 1,202; the MFI for GP575-112 was 220.8 (SD, 160.4), with a cutoff of 702; the MFI for GP555-98 was 263.1 (SD, 245), with a cutoff of 998; the MFI for M1-162 was 344.7 (SD, 209.4), with a cutoff of 973; the MFI for M88-162 was 377.3 (SD, 228.4), with a cutoff of 1,063; the MFI for N1-110 was 432.1 (SD, 161.7), with a cutoff of 917; and the MFI for N1-69 was 216.6 (SD, 166.5), with a cutoff of 716. Samples with MFI values above the cutoff value were designated as positive, and those with values below the cutoff value were considered to be negative. The subsequent analysis of 125 positive sera (with VNT titers ranging from 1:4 to >1:512) and 125 neutralizing antibody-negative sera tested in parallel by the VNT allowed the evaluation of the performance of each recombinant protein (for 250 samples) in the MIA. MIAs based on recombinant GP5 proteins had high specificity. The specificity of GP51-255-, GP51-116-, and GP575-112-based MIAs was 98.4%, with only 2 false-positive results among 125 VNT-negative samples. The GP555-98-based MIA had slightly lower specificity, 97.6%, than the other GP5-based MIAs. The sensitivities of the GP51-255- and GP51-116-based MIAs were the lowest among all those of the protein-based MIAs (3.2% and 5.6%, respectively), as these assays detected only 4 and 7 of 125 VNT-positive samples. The sensitivities of the recombinant GP575-112- and GP555-98-based MIAs were higher (41.6% and 77.6%, respectively), with the detection of 52 and 97 of 125 VNT-positive sera, respectively. Both MIAs based on M proteins (M1-162 and the partial-length M88-162) had high specificity (97.6% and 98.4%, respectively), and these assays correctly detected 122 and 123 of 125 VNT-negative samples. However, both had very low sensitivity (6.4%) and detected anti-EAV antibodies in only 8 of 125 VNT-positive sera. The sensitivity of the recombinant N1-69-based MIA was also very poor; this assay detected anti-EAV antibodies in only 10 (8.0%) of 125 VNT-positive sera. In contrast, the MIA incorporating full-length N1-110 detected anti-EAV antibodies in 77 of 125 VNT-positive sera, resulting in significantly higher sensitivity (61.6%). In summary, these results confirm that MIAs based on GP51-255, GP51-116, GP575-112, N1-69, M1-162, and M88-162 were markedly less sensitive than the N1-110 and GP555-98 MIAs (61.6% and 77.6%, respectively) in detecting anti-EAV antibodies in equine sera as determined by comparison to the results of the VNT (Table 2). Accordingly, the MIAs based on recombinant proteins that showed poor sensitivity were excluded from further study, and only GP555-98 and N1-110 protein-based MIAs were selected for further development and validation.
FIG. 3.
GP555-98 and N1-110 MIA analysis of serially diluted serum samples. Sera from horses with EAV-neutralizing antibodies (+) and negative control equine sera (−) were serially diluted and evaluated in the GP555-98 and N1-110 MIAs. The MFI for each dilution of the standard was determined. Results are reported as MFI per 100 microspheres. Samples are identified by number in the upper right corner. w/, with.
TABLE 2.
Sensitivities and specificities of MIAs using individual recombinant EAV structural proteins as antigens based on testing of 125 VNT-positive and 125 VNT-negative serum samples
| Recombinant EAV structural protein | Sensitivity (%) | Specificity (%) |
|---|---|---|
| GP51-255 | 3.2 | 98.4 |
| GP51-116 | 5.6 | 98.4 |
| GP575-112 | 41.6 | 98.4 |
| GP555-98 | 77.6 | 97.6 |
| M1-162 | 6.4 | 97.6 |
| M88-162 | 6.4 | 98.4 |
| N1-110 | 61.6 | 91.2 |
| N1-69 | 8 | 97.6 |
Validation of the GP555-98- and N1-110-based MIAs.
The same panel of 2,500 diagnostic equine sera evaluated by the VNT was analyzed for the presence of EAV-specific antibodies using GP555-98- and N1-110-based MIAs. Of the 750 VNT-positive sera, only 698 were determined to be positive by the GP555-98 MIA. The GP555-98 MIA failed to detect anti-EAV antibodies in 52 serum samples that were confirmed to be positive by the VNT (giving 52 false-negative results). Furthermore, only 1,644 of the 1,750 VNT-negative sera gave negative results with the GP555-98 MIA assay. This finding indicated that GP555-98 gave 106 false-positive results compared to the VNT. The overall sensitivity and specificity of the GP555-98 MIA were 93.1% and 93.9%, respectively. Similarly, the N1-110 MIA gave 256 false-negative results (only 494 of 750 VNT-positive samples were identified as positive) and 374 false-positive results (1,376 of 1,750 VNT-negative samples were identified as negative) compared to the VNT. The sensitivity and specificity of the N1-110 MIA were 65.9% and 78.6%, respectively.
As mentioned previously, the cutoff values were assigned on the basis of 3 SD above the MFI for 20 EAV antibody-negative equine sera. However, the cutoff value calculated according to this method varied slightly for each of the protein-coated bead sets; this variation may have been a factor contributing to the differences in sensitivity observed between the MIA and the standard VNT. Moreover, a statistical approach using the negative reference population assumes a normal distribution of the test variable and does not consider the sensitivity of the assay (12). Both sensitivity and specificity are important parameters when a diagnostic test is applied to a given population, and an alternative approach to the determination of the cutoff value is to consider both the negative and positive reference populations (21, 31, 32). This approach was adopted in this study by using nonparametric ROC analysis to define the cutoff value with the entire equine serum data set derived from the study. Such an approach to the tabulation of results is recommended in comparative studies of this type, in order to eliminate variability in the day-to-day performance of the assay (36). By using the ROC analysis, the optimal cutoff value to classify sera as positive or negative was recalculated to be an MFI of 992. With this cutoff, the GP555-98 MIA identified 1,694 of 1,750 VNT-negative samples as negative and determined 806 of the total of 2,500 samples to be positive. Given these results, the sensitivity and specificity of the GP555-98 MIA as calculated by Bayesian analysis were 92.6% (95% credible interval [the Bayesian analog of a confidence interval], 90.1 to 94.3%) and 92.9% (95% credible interval, 91.7 to 94.1%), respectively, compared to the VNT. The cutoff value that maximized the tradeoff between the sensitivity and the specificity of the N 1-110-based MIA was 1,261.43. At this cutoff, the estimated sensitivity and specificity were 88.3% and 54.7%, respectively.
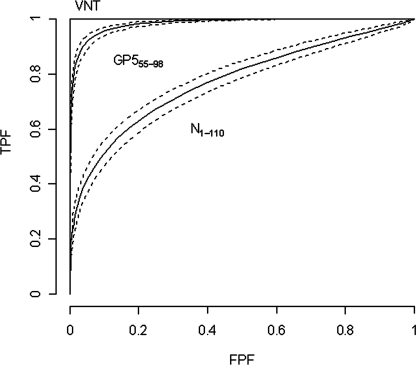
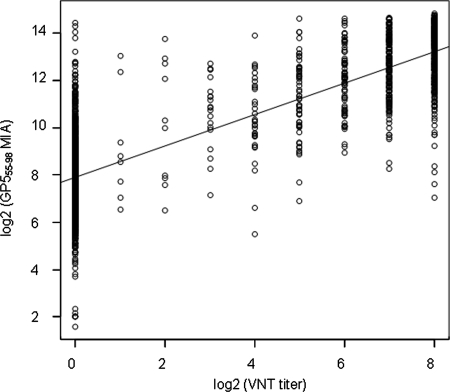
The results from both recombinant protein-based MIAs were subjected to Bayesian analysis. The areas under the ROC curves for both GP555-98 and N1-110 MIAs were compared to that for the VNT. The AUC for the GP555-98 MIA was higher (0.983, with a 95% credible interval of 0.979 to 0.986) than the AUC for the N1-110 MIA (0.772, with a 95% credible interval of 0.751 to 0.793) (Fig. 4). This result confirms that the diagnostic accuracy of the GP555-98-based MIA is significantly higher than that of the N1-110-based MIA; the GP555-98-based MIA can reliably detect anti-EAV antibodies in equine sera. An analysis of the correlation between the results of the GP555-98 MIA and those of the VNT was performed, and it indicated significant correlation between the outcomes of the GP555-98 MIA and the standard VNT (r = 0.84; P < 0.0001) (Fig. 5).
FIG. 4.
ROC curves depicting the sensitivities and specificities of the GP555-98 and N1-110 MIAs compared to the VNT. An ROC curve is a plot of the TPF versus the false-positive fraction (FPF) across all possible cutoff values that can be used to dichotomize the data into positive and negative outcomes. A related parameter of interest is the AUC. Estimated ROC curves (solid lines) with pointwise 95%-credible-interval bands (dashed lines) for the GP555-98 MIA and the N1-110 MIA are indicated.
FIG. 5.
Analysis of the correlation between the GP555-98 MIA and the VNT for the detection of anti-EAV antibodies in equine sera. A scatter plot of GP555-98 MIA results (MFI values) versus VNT titers determined using 2,500 equine sera is depicted on the log2 scale. The VNT data were plotted on the x axis, and the MIA data were plotted on the y axis.
Comparison of the sensitivities of the GP555-98 MIA and the VNT using sequential serum samples from experimentally inoculated horses.
A panel of 192 archived sequential serum samples from 18 horses in four groups, each inoculated with a different strain of EAV, was used to compare the antibody responses detected by the GP555-98 MIA and the VNT (2, 9, 10, 51).
Sera collected from EAV rVBS (n = 4)- and EAV 030H (n = 2)-inoculated horses gave reactive signals in the GP555-98 MIA from day 8 postinfection. The MFI values for sera collected from days 8 to 42 were positive, with values for EAV strains rVBS and 030H ranging from 2,541 to 27,736 (Fig. 2E) and from 1,295 to 21,874 (Fig. 2F), respectively. Seroconversions detected by the GP555-98 MIA were in full agreement with the results of the VNT, which also detected neutralizing antibodies from day 8 (Fig. 2A and B). Similarly, reactive signals in sera from horses inoculated with EAV KY84 (n = 7) were detected from 7 dpi (two horses) and 10 dpi (other horses). The MFI values for sera from horses inoculated with EAV rVBS, 030H, and KY84 ranged from 1,648 to 27,142. The only discrepancy between the results of the GP555-98 MIA and the VNT involved serum from one horse (SD7925), which had a threshold neutralizing antibody titer (1:4) on day 7 (Fig. 2C); this sample was not positive by the MIA (Fig. 2G). In general, the pattern of antibody development determined with the GP555-98 MIA correlated very well with the pattern found using the standard VNT. Both assays detected EAV-specific antibodies in samples collected more than 10 dpi. If consideration is given to the fact that the two tests measure antibodies on different scales, there was greater variability in the results from the MIA than in those from the VNT. Despite this finding, the sensitivity of the MIA was not affected by this variability.
In contrast, when 44 sequential serum samples from five horses inoculated with the highly attenuated EAV CA95G strain were used to compare the GP555-98 MIA and the VNT, the GP555-98 MIA failed to detect consistent positive reactive signals (Fig. 2H). The earliest reactive signal was detected by the GP555-98 MIA on day 28 postinfection for horse 9624, whereas samples from this horse had low levels of neutralizing antibodies detectable by the VNT from 14 dpi (Fig. 2D). Similarly, horse 5582 had a threshold neutralizing antibody titer (1:4) detected on day 28 by the VNT; this titer was not detected by the GP555-98 MIA, which detected reactive signals only from 35 dpi. Horse 9131 developed a low neutralizing antibody titer (1:8) by 35 dpi, which was detected by the GP555-98 MIA. Sera from horses 9715 and 9649 did not show reactive signals at all, whereas VN antibodies were detected at low levels from 14 dpi. The MFI values for the antibody-positive sera from CA95G-inoculated horses ranged from 1,055 to 4,688.
DISCUSSION
This study evaluated a newly developed MIA based on GP555-98 as an alternative diagnostic procedure for the detection of antibodies to EAV. The evaluation of various forms of recombinant viral structural proteins (full-length proteins and partial-length proteins containing the antigenic regions) allowed the identification of the best recombinant protein antigen for use in the MIA. Previous reports have shown that two bacterial fusion proteins (aa 1 to 116 and 55 to 98) and a synthetic peptide (aa 81 to 106), both based on GP5 of EAV, can be used as effective diagnostic antigens in ELISAs (15, 49). It has been demonstrated previously that horses immunized with portions of GP5 either expressed in bacteria (aa 55 to 98) or constructed as a synthetic oligopeptide (aa 75 to 97) developed EAV-neutralizing antibodies (14, 49). Chirnside et al. used a series of recombinant bacterial fusion proteins derived from EAV ORF 7 to define the immunoreactive region of the viral nucleocapsid (N) protein (16). These studies indicated that the major N protein epitope that reacts with anti-EAV equine sera is located within the amino acid residue segment from 1 to 69 in the terminal region of the protein. Similarly, it has been demonstrated previously that only the carboxyl-terminal sequence (aa 88 to 162) of the M protein is necessary to identify equine serum antibodies specific to the EAV M protein; it was further suggested that this region should be useful for the serodetection of EAV-infected horses (37). On the basis of the foregoing data, it was decided to express the full-length GP51-255, M1-162, and N1-110 proteins, various antigenic regions of GP5 (aa 1 to 116, 75 to 112, and 55 to 98), and the M88-162 and N1-69 proteins as bacterial fusion proteins for use in MIAs to detect antibodies to EAV. When the sensitivities and specificities of these different MIAs were determined by comparing their results to those of the VNT, all of the MIAs, with the exception of the MIA based on the GP555-98 bacterial fusion protein, had very low specificities and sensitivities; accordingly, they were considered unsuitable as serodiagnostic assays for EVA infection.
The low immunoreactivity of MIAs incorporating three of the GP5 proteins (GP51-255, the N-terminal ectodomain segment GP51-116, and GP575-112), as well as the M (M1-162 and M88-162) and N (N1-110 and N1-69) proteins, may result from the misfolding and aggregation of the E. coli-expressed recombinant proteins. Despite the fact that the GP5 N-terminal ectodomain contains the major neutralization epitopes of EAV, the three MIAs incorporating GP5 proteins, the full-length GP51-255, the N-terminal ectodomain segment GP51-116, and GP575-112, had markedly low sensitivities. It has been reported previously that in the absence of the M protein, the recombinant GP5 protein tends to form large protein aggregates that are neither immunogenic nor immunoreactive (4, 33). Similarly, it has been shown that the GP5 protein, when expressed by baculovirus or mammalian expression systems, is not processed properly and is easily misfolded (2). As the length of the recombinant GP5 was reduced, the sensitivity of the MIA increased significantly; this finding would indicate that a reduction in length allows the exposure of some linear epitopes present in the antigenic region of the recombinant GP5 protein.
There was very good agreement between the results of the GP555-98-based MIA and those of the VNT. However, the GP555-98 MIA was found to have lower sensitivity and specificity than the VNT. Many factors may adversely affect the sensitivity of the GP555-98 MIA. (i) The short length of the GP5 protein incorporated into the assay may have targeted antibodies that were specific only for linear epitopes present in the region of aa 55 to 98 and not for any conformational epitopes. In addition, the failure of the GP555-98 MIA to detect some sera that possessed VN antibodies to EAV may have been due to the lack of two major neutralization sites located outside of the GP555-98 region, neutralization sites A (aa 49) and D (aa 99 to 106), and/or the lack of conformational interaction of the four neutralization sites (A to D) in the N-terminal ectodomain of GP5 as shown previously (1). (ii) Recombinant proteins expressed in E. coli differ from the virion-associated GP5 protein, due to the lack of secondary-structural folding and posttranslational modifications. (iii) The VNT putatively detects antibodies to the GP5 major envelope glycoprotein, which carries the known neutralization determinants of EAV (4). Although there is only one known serotype of EAV, the GP5 protein has been shown to be the most variable of the EAV structural proteins among field strains of the virus (5, 11). Antigenic variation among EAV field strains may reduce the sensitivity of the assay. It should be noted that the cloned GP5 amino acid sequence (aa 55 to 98) was derived from the virulent Bucyrus strain of EAV and field strains of EAV have been shown to differ significantly in this region; this variation may result in significant differences in neutralization phenotype among viral isolates as determined using polyclonal equine sera. (iv) The lower sensitivity of the GP555-98 MIA may also be associated with the dilution of the test samples. Previously, it has been documented that recombinant proteins expressed by bacteria can exhibit high levels of background reactivity with equine serum (15, 16, 49). In the present study, we determined 1:100 to be the optimal serum dilution for the minimization of nonspecific binding of antibodies to the bacterial recombinant proteins. At this dilution, the majority of sera with low antibody titers (1:4 to 1:8) detected by the VNT gave false-negative results in the GP555-98 MIA (Table 3). It should be emphasized, therefore, that the MIA can reliably detect only sera with moderate to high titers (≥1:16) of antibodies to EAV.
TABLE 3.
Distribution of numbers of samples with false-negative results from GP555-98 MIAa
| VN titer | No. of samples with false-negative MIA results/no. of VNT-positive samples (%) |
|---|---|
| 1:4 | 5/7 (71.4) |
| 1:8 | 4/10 (40.0) |
| 1:16 | 6/21 (28.6) |
| 1:32 | 10/34 (29.4) |
| 1:64 | 11/58 (18.9) |
| 1:128 | 4/76 (5.3) |
| 1:256 | 5/162 (3.1) |
| ≥1:512 | 6/382 (1.6) |
A total of 750 samples were positive by VNT.
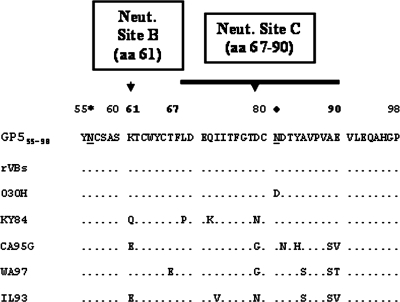
Upon the testing of sera from horses experimentally exposed to the rVBS, 030H, and KY84 strains of EAV, there was excellent concordance between the GP555-98 MIA and the VNT in the detection of the antibody response to the virus. While the 030H and KY84 strains of EAV had one and four substitutions, respectively, between aa 55 and 98 compared to the cloned GP555-98, sera from horses exposed to these virus strains did not give any false-negative reactions in the GP555-98 MIA. In contrast, sera from the EAV CA95G strain-inoculated horses failed to give a consistent positive signal in the GP555-98 MIA. Upon comparative amino acid sequence analysis, the EAV CA95G virus strain exhibited six amino acid substitutions in the GP555-98 region, five of which were located between aa 79 and 90 (Fig. 6). This finding would indicate that amino acid substitutions located in the region of aa 79 to 90 may have a significant effect on the sensitivity of the GP555-98 MIA. Furthermore, a comparative amino acid analysis of GP5 showed that some other field strains of EAV (e.g., WA97 and IL93) had multiple amino acid substitutions between aa 79 and 90 (Fig. 6). Accordingly, sera from horses exposed to EAV strains that have significant numbers of amino acid substitutions in this region may well give false-negative results in the GP555-98 MIA. To maximize the sensitivity of a GP5-based MIA, it may be necessary to include a cocktail of GP555-98 proteins comprising multiple sequences representative of strains of EAV known to differ phenotypically rather than depend on a single GP555-98 sequence from one strain.
FIG. 6.
Aligned deduced amino acid sequences of GP555-98 proteins from various laboratory (rVBS and 030H) and field (KY84, CA95G, WA97, and IL93) strains of EAV. Dots indicate the same amino acid as that in the sequence at the top. Letters indicate the amino acid substitution at each site. The predicted N-linked glycosylation sites are underlined. The respective conserved and variable glycosylation sites are indicated by * and ⧫. Neut., neutralization.
ELISAs targeting whole virus or recombinant GP5, M, and N proteins have been investigated for the serological diagnosis of EAV infection (15-17, 33, 39, 41, 49, 60, 63). An ELISA procedure incorporating an ovalbumin-conjugated synthetic peptide comprising aa 81 to 106 of GP5 for the detection of anti-EAV antibodies in equine sera has been described previously (49). That ELISA had slightly higher sensitivity and specificity (96.75% and 95.6%, respectively) than the MIA based on GP555-98 used in this study. Similarly, an ELISA based on a cocktail of three major structural proteins (GP5, M, and N) expressed by baculovirus was able to detect antibodies in most equine sera that were positive by the VNT following natural or experimental infection (92.3% sensitivity and 100% specificity) (33). It should be stressed that the latter two studies (33, 49) were undertaken using a limited number of EAV VNT antibody-positive (400 and 73, respectively) and antibody-negative (400 and 111, respectively) equine sera. By comparison, in the present study the sensitivity (92.6%) and specificity (92.9%) of the GP555-98 MIA were evaluated based on the testing of 2,500 diagnostic equine sera.
The MIA based on GP555-98 has several advantages over the VNT, as well as over various ELISAs. ELISAs that utilize whole-virus antigen preparations can give rise to a large number of false-positive reactions when sera are derived from horses vaccinated with tissue culture-derived vaccines. Such animals can develop antibodies to cell culture proteins remaining in the EAV whole-virus antigen preparation (19). Furthermore, these cell protein-specific antibodies cause cytotoxicity at the lower serum dilutions that in turn can interfere with the interpretation of the VNT results (29). The GP555-98 MIA circumvents both of these problems, since it utilizes a recombinant protein that represents the immunodominant GP5 protein of EAV. Furthermore, the GP555-98 MIA is not subjective in interpretation and is less time-consuming, is less expensive, and uses a smaller sample volume than the VNT. Accordingly, the GP555-98 MIA can be considered a more convenient and rapid serologic testing procedure for EAV infection, especially for screening large numbers of sera. The MIA can be completed within a few hours; in comparison, the VNT takes several days. The MIA does not require the use of live virus and so does not require virus containment facilities.
In summary, we have developed and validated several MIAs that use recombinant EAV structural proteins to detect antibodies to EAV in equine serum. Among eight recombinant proteins, the highest concordance with the VNT results using selected equine sera was obtained with the partial GP5 protein GP555-98. The GP555-98 MIA and VNT outcomes correlated significantly (r = 0.84; P < 0.0001). Although the GP555-98 MIA is less sensitive than the VNT, it has the potential to provide an accurate, rapid, convenient, and more economical test for screening equine sera for the presence of antibodies to EAV, with the VNT used as a confirmatory assay.
Acknowledgments
We thank James MacLachlan, Department of Veterinary Pathology, Microbiology and Immunology, School of Veterinary Medicine, University of California at Davis, for providing EAV strain-specific polyclonal antisera from experimentally infected horses. We are grateful to Adriana Verschoor, Wadsworth Center, New York State Department of Health, for editorial assistance.
This work was supported by the Grayson Jockey Club Research Foundation and the Frederick Van Lennep Endowment. Yun Young Go is the recipient of a Geoffrey C. Hughes Foundation graduate fellowship.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Balasuriya, U. B., J. C. Dobbe, H. W. Heidner, V. L. Smalley, A. Navarrette, E. J. Snijder, and N. J. MacLachlan. 2004. Characterization of the neutralization determinants of equine arteritis virus using recombinant chimeric viruses and site-specific mutagenesis of an infectious cDNA clone. Virology 321:235-246. [DOI] [PubMed] [Google Scholar]
- 2.Balasuriya, U. B., H. W. Heidner, N. L. Davis, H. M. Wagner, P. J. Hullinger, J. F. Hedges, J. C. Williams, R. E. Johnston, W. David Wilson, I. K. Liu, and N. James MacLachlan. 2002. Alphavirus replicon particles expressing the two major envelope proteins of equine arteritis virus induce high level protection against challenge with virulent virus in vaccinated horses. Vaccine 20:1609-1617. [DOI] [PubMed] [Google Scholar]
- 3.Balasuriya, U. B., H. W. Heidner, J. F. Hedges, J. C. Williams, N. L. Davis, R. E. Johnston, and N. J. MacLachlan. 2000. Expression of the two major envelope proteins of equine arteritis virus as a heterodimer is necessary for induction of neutralizing antibodies in mice immunized with recombinant Venezuelan equine encephalitis virus replicon particles. J. Virol. 74:10623-10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasuriya, U. B., and N. J. MacLachlan. 2004. The immune response to equine arteritis virus: potential lessons for other arteriviruses. Vet. Immunol. Immunopathol. 102:107-129. [DOI] [PubMed] [Google Scholar]
- 5.Balasuriya, U. B., N. J. Maclachlan, A. A. De Vries, P. V. Rossitto, and P. J. Rottier. 1995. Identification of a neutralization site in the major envelope glycoprotein (GL) of equine arteritis virus. Virology 207:518-527. [DOI] [PubMed] [Google Scholar]
- 6.Balasuriya, U. B., J. F. Patton, P. V. Rossitto, P. J. Timoney, W. H. McCollum, and N. J. MacLachlan. 1997. Neutralization determinants of laboratory strains and field isolates of equine arteritis virus: identification of four neutralization sites in the amino-terminal ectodomain of the G(L) envelope glycoprotein. Virology 232:114-128. [DOI] [PubMed] [Google Scholar]
- 7.Balasuriya, U. B., P. V. Rossitto, C. D. DeMaula, and N. J. MacLachlan. 1993. A 29K envelope glycoprotein of equine arteritis virus expresses neutralization determinants recognized by murine monoclonal antibodies. J. Gen. Virol. 74:2525-2529. [DOI] [PubMed] [Google Scholar]
- 8.Balasuriya, U. B., P. Y. Shi, S. J. Wong, V. L. Demarest, I. A. Gardner, P. J. Hullinger, G. L. Ferraro, J. D. Boone, C. L. De-Cino, A. L. Glaser, R. W. Renshaw, M. Ledizet, R. A. Koski, and N. J. MacLachlan. 2006. Detection of antibodies to West Nile virus in equine sera using microsphere immunoassay. J. Vet. Diagn. Investig. 18:392-395. [DOI] [PubMed] [Google Scholar]
- 9.Balasuriya, U. B., E. J. Snijder, H. W. Heidner, J. Zhang, J. C. Zevenhoven-Dobbe, J. D. Boone, W. H. McCollum, P. J. Timoney, and N. J. Maclachlan. 2007. Development and characterization of an infectious cDNA clone of the virulent Bucyrus strain of equine arteritis virus. J. Gen. Virol. 88:918-924. [DOI] [PubMed] [Google Scholar]
- 10.Balasuriya, U. B., E. J. Snijder, L. C. van Dinten, H. W. Heidner, W. D. Wilson, J. F. Hedges, P. J. Hullinger, and N. J. MacLachlan. 1999. Equine arteritis virus derived from an infectious cDNA clone is attenuated and genetically stable in infected stallions. Virology 260:201-208. [DOI] [PubMed] [Google Scholar]
- 11.Balasuriya, U. B., P. J. Timoney, W. H. McCollum, and N. J. MacLachlan. 1995. Phylogenetic analysis of open reading frame 5 of field isolates of equine arteritis virus and identification of conserved and nonconserved regions in the GL envelope glycoprotein. Virology 214:690-697. [DOI] [PubMed] [Google Scholar]
- 12.Barajas-Rojas, J. A., H. P. Riemann, and C. E. Franti. 1993. Notes about determining the cut-off value in enzyme-linked immunosorbent assay (ELISA). Prev. Vet. Med. 15:231-233. [Google Scholar]
- 13.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 14.Chirnside, E. D., A. A. de Vries, J. A. Mumford, and P. J. Rottier. 1995. Equine arteritis virus-neutralizing antibody in the horse is induced by a determinant on the large envelope glycoprotein GL. J. Gen. Virol. 76:1989-1998. [DOI] [PubMed] [Google Scholar]
- 15.Chirnside, E. D., P. M. Francis, A. A. de Vries, R. Sinclair, and J. A. Mumford. 1995. Development and evaluation of an ELISA using recombinant fusion protein to detect the presence of host antibody to equine arteritis virus. J. Virol. Methods 54:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chirnside, E. D., P. M. Francis, and J. A. Mumford. 1995. Expression cloning and antigenic analysis of the nucleocapsid protein of equine arteritis virus. Virus Res. 39:277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho, H. J., S. C. Entz, D. Deregt, L. T. Jordan, P. J. Timoney, and W. H. McCollum. 2000. Detection of antibodies to equine arteritis virus by a monoclonal antibody-based blocking ELISA. Can. J. Vet. Res. 64:38-43. [PMC free article] [PubMed] [Google Scholar]
- 18.Choi, Y. K., W. O. Johnson, M. T. Collins, and I. A. Gardner. 2006. Bayesian estimation of ROC curves in the absence of a gold standard. J. Agric. Biol. Environ. Stat. 11:210-229. [Google Scholar]
- 19.Cook, R. F., S. J. Gann, and J. A. Mumford. 1989. The effects of vaccination with tissue culture-derived viral vaccines on detection of antibodies to equine arteritis virus by enzyme-linked immunosorbent assay (ELISA). Vet. Microbiol. 20:181-189. [DOI] [PubMed] [Google Scholar]
- 20.Cullinane, A. A. 2004. Testing for equine arteritis virus. Vet. Rec. 155:647-648. [PubMed] [Google Scholar]
- 21.Denac, H., C. Moser, J. D. Tratschin, and M. A. Hofmann. 1997. An indirect ELISA for the detection of antibodies against porcine reproductive and respiratory syndrome virus using recombinant nucleocapsid protein as antigen. J. Virol. Methods 65:169-181. [DOI] [PubMed] [Google Scholar]
- 22.Deregt, D., A. A. de Vries, M. J. Raamsman, L. D. Elmgren, and P. J. Rottier. 1994. Monoclonal antibodies to equine arteritis virus proteins identify the GL protein as a target for virus neutralization. J. Gen. Virol. 75:2439-2444. [DOI] [PubMed] [Google Scholar]
- 23.de Vries, A. A., E. D. Chirnside, M. C. Horzinek, and P. J. Rottier. 1992. Structural proteins of equine arteritis virus. J. Virol. 66:6294-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Vries, A. A., S. M. Post, M. J. Raamsman, M. C. Horzinek, and P. J. Rottier. 1995. The two major envelope proteins of equine arteritis virus associate into disulfide-linked heterodimers. J. Virol. 69:4668-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Vries, A. A. F., M. C. Horzinek, P. J. M. Rottier, and R. J. de Groot. 1997. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dias, D., J. Van Doren, S. Schlottmann, S. Kelly, D. Puchalski, W. Ruiz, P. Boerckel, J. Kessler, J. M. Antonello, T. Green, M. Brown, J. Smith, N. Chirmule, E. Barr, K. U. Jansen, and M. T. Esser. 2005. Optimization and validation of a multiplexed luminex assay to quantify antibodies to neutralizing epitopes on human papillomaviruses 6, 11, 16, and 18. Clin. Diagn. Lab. Immunol. 12:959-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doll, E. R., J. T. Bryans, J. C. Wilson, and W. H. McCollum. 1968. Immunization against equine viral arteritis using modified live virus propagated in cell cultures of rabbit kidney. Cornell Vet. 48:497-524. [PubMed] [Google Scholar]
- 28.Fukunaga, Y., H. Imagawa, E. Tabuchi, and Y. Akiyama. 1981. Clinical and virological findings on experimental equine viral arteritis in horses. Bull. Equine Res. Inst. 18:110-118. [Google Scholar]
- 29.Geraghty, R. J., J. R. Newton, J. Castillo Olivares, J. M. Cardwell, and J. A. Mumford. 2003. Testing for equine arteritis virus. Vet. Rec. 152:478-479. [PubMed] [Google Scholar]
- 30.Glaser, A. L., A. A. de Vries, and E. J. Dubovi. 1995. Comparison of equine arteritis virus isolates using neutralizing monoclonal antibodies and identification of sequence changes in GL associated with neutralization resistance. J. Gen. Virol. 76:2223-2233. [DOI] [PubMed] [Google Scholar]
- 31.Greiner, M. 1996. Two-graph receiver operating characteristic (TG-ROC): update version supports optimisation of cut-off values that minimise overall misclassification costs. J. Immunol. Methods 191:93-94. [DOI] [PubMed] [Google Scholar]
- 32.Greiner, M., D. Sohr, and P. Gobel. 1995. A modified ROC analysis for the selection of cut-off values and the definition of intermediate results of serodiagnostic tests. J. Immunol. Methods 185:123-132. [DOI] [PubMed] [Google Scholar]
- 33.Hedges, J. F., U. B. Balasuriya, S. Ahmad, P. J. Timoney, W. H. McCollum, T. Yilma, and N. J. MacLachlan. 1998. Detection of antibodies to equine arteritis virus by enzyme linked immunosorbant assays utilizing G(L), M and N proteins expressed from recombinant baculoviruses. J. Virol. Methods 76:127-137. [DOI] [PubMed] [Google Scholar]
- 34.Hedges, J. F., U. B. Balasuriya, P. J. Timoney, W. H. McCollum, and N. J. MacLachlan. 1999. Genetic divergence with emergence of novel phenotypic variants of equine arteritis virus during persistent infection of stallions. J. Virol. 73:3672-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huntington, P. J., A. J. Forman, and P. M. Ellis. 1990. The occurrence of equine arteritis virus in Australia. Aust. Vet. J. 67:432-435. [DOI] [PubMed] [Google Scholar]
- 36.Jacobson, R. H. 1998. Validation of serological assays for diagnosis of infectious diseases. Rev. Sci. Tech. Off. Int. Epizoot. 17:469-526. [DOI] [PubMed] [Google Scholar]
- 37.Jeronimo, C., and D. Archambault. 2002. Importance of M-protein C terminus as substrate antigen for serodetection of equine arteritis virus infection. Clin. Diagn. Lab. Immunol. 9:698-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson, A. J., R. C. Cheshier, G. Cosentino, H. P. Masri, V. Mock, R. Oesterle, R. S. Lanciotti, D. A. Martin, A. J. Panella, O. Kosoy, and B. J. Biggerstaff. 2007. Validation of a microsphere-based immunoassay for detection of anti-West Nile virus and anti-St. Louis encephalitis virus immunoglobulin M antibodies. Clin. Vaccine Immunol. 14:1084-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondo, T., Y. Fukunaga, K. Sekiguchi, T. Sugiura, and H. Imagawa. 1998. Enzyme-linked immunosorbent assay for serological survey of equine arteritis virus in racehorses. J. Vet. Med. Sci. 60:1043-1045. [DOI] [PubMed] [Google Scholar]
- 40.Lenihan, P. 2004. Use of the virus microneutralisation (VN) test for equine arteritis virus (EAV) serology, p. 52-53. Int. Workshop Diagn. Equine Arteritis Virus Infect., Lexington, KY.
- 41.MacLachlan, N. J., U. B. Balasuriya, J. F. Hedges, T. M. Schweidler, W. H. McCollum, P. J. Timoney, P. J. Hullinger, and J. F. Patton. 1998. Serologic response of horses to the structural proteins of equine arteritis virus. J. Vet. Diagn. Investig. 10:229-236. [DOI] [PubMed] [Google Scholar]
- 42.MacLachlan, N. J., U. B. Balasuriya, P. V. Rossitto, P. A. Hullinger, J. F. Patton, and W. D. Wilson. 1996. Fatal experimental equine arteritis virus infection of a pregnant mare: immunohistochemical staining of viral antigens. J. Vet. Diagn. Investig. 8:367-374. [DOI] [PubMed] [Google Scholar]
- 43.McCollum, W. H. 1969. Development of a modified virus strain and vaccine for equine viral arteritis. J. Am. Vet. Med. Assoc. 155:318-322. [PubMed] [Google Scholar]
- 44.McCollum, W. H. 1986. Responses of horses vaccinated with avirulent modified-live equine arteritis virus propagated in the E. Derm (NBL-6) cell line to nasal inoculation with virulent virus. Am. J. Vet. Res. 47:1931-1934. [PubMed] [Google Scholar]
- 45.McCollum, W. H. 1970. Vaccination for equine viral arteritis, p. 143-151. In Proceedings of the Second International Conference on Equine Infectious Diseases, Paris, France. S. Karger, Basel, Switzerland.
- 46.McCollum, W. H., and J. T. Bryans. 1972. Serological identification of infection by equine arteritis virus in horses of several countries, p. 256-263. In Proceedings of the Third International Conference on Equine Infectious Diseases, Paris, France. S. Karger, Basel, Switzerland.
- 47.McCollum, W. H., K. Shuck, J. Zhang, and P. J. Timoney. 2004. Factors important to the isolation of equine arteritis virus in cell culture, p. 20. Int. Workshop Diagn. Equine Arteritis Virus Infect., Lexington, KY.
- 48.Moraillon, A., and R. Moraillon. 1978. Results of an epidemiological investigation on viral arteritis in France and some other European and African countries. Ann. Rech. Vet. 9:43-54. [PubMed] [Google Scholar]
- 49.Nugent, J., R. Sinclair, A. A. deVries, R. Y. Eberhardt, J. Castillo Olivares, N. Davis Poynter, P. J. Rottier, and J. A. Mumford. 2000. Development and evaluation of ELISA procedures to detect antibodies against the major envelope protein (G(L)) of equine arteritis virus. J. Virol. Methods 90:167-183. [DOI] [PubMed] [Google Scholar]
- 50.Organisation Mondiale de la Santé Animale. 2004. OIE manual of diagnostic tests and vaccines for terrestrial animals, 5th ed., vol. 2. Organisation Mondiale de la Santé Animale, Paris, France.
- 51.Patton, J. F., U. B. Balasuriya, J. F. Hedges, T. M. Schweidler, P. J. Hullinger, and N. J. MacLachlan. 1999. Phylogenetic characterization of a highly attenuated strain of equine arteritis virus from the semen of a persistently infected Standardbred stallion. Arch. Virol. 144:817-827. [DOI] [PubMed] [Google Scholar]
- 52.Pepe, M. S. 2003. Oxford statistical science series, vol. 31. The statistical evaluation of medical tests for classification and prediction. Oxford University Press, New York, NY.
- 53.Senne, D. A., J. E. Pearson, and E. A. Carbrey. 1985. Equine viral arteritis: a standard procedure for the virus neutralization test and comparison of results of a proficiency test performed at five laboratories, p. 29-34. In Proceedings of the 89th Annual Meeting of the United States Animal Health Association, Milwaukee, WI. Carter Printing, Richmond, VA.
- 54.Shuck, K. M., P. J. Timoney, J. Zhang, and W. H. McCollum. 2004. Reliability of the virus neutralization test for determining the serologic status of horses to equine arteritis, virus, p. 55. Int. Workshop Diagn. Equine Arteritis Virus Infect., Lexington, KY.
- 55.Snijder, E. J. 2001. Arteriviruses, p. 1205-1220. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, PA.
- 56.Snijder, E. J., J. C. Dobbe, and W. J. Spaan. 2003. Heterodimerization of the two major envelope proteins is essential for arterivirus infectivity. J. Virol. 77:97-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 58.Snijder, E. J., H. van Tol, K. W. Pedersen, M. J. Raamsman, and A. A. de Vries. 1999. Identification of a novel structural protein of arteriviruses. J. Virol. 73:6335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spiegelhalter, D., A. Thomas, N. Best, and D. Lunn. 2003. WinBUGS (Bayesian inference using Gibbs sampling) user manual, version 1.4. MRC Biostatistics Unit, Cambridge, United Kingdom.
- 60.Starik, E., A. Ginter, and P. Coppe. 2001. ELISA and direct immunofluorescence test to detect equine arteritis virus (EAV) using a monoclonal antibody directed to the EAV-N protein. J. Vet. Med. B 48:1-9. [DOI] [PubMed] [Google Scholar]
- 61.Timoney, P. J., and W. H. McCollum. 1993. Equine viral arteritis. Vet. Clin. N. Am. Equine Pract. 9:295-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vickers, M. L. 2004. Virus neutralization testing for equine arteritis virus antibodies at the LDDC-Kentucky: progress along the way from 7,000 to 10,000 tests per year, p. 56. Int. Workshop Diagn. Equine Arteritis Virus Infect., Lexington, KY.
- 63.Wagner, H. M., U. B. Balasuriya, and N. James MacLachlan. 2003. The serologic response of horses to equine arteritis virus as determined by competitive enzyme-linked immunosorbent assays (c-ELISAs) to structural and non-structural viral proteins. Comp. Immunol. Microbiol. Infect. Dis. 26:251-260. [DOI] [PubMed] [Google Scholar]
- 64.Wieringa, R., A. A. de Vries, M. J. Raamsman, and P. J. Rottier. 2002. Characterization of two new structural glycoproteins, GP3 and GP4, of equine arteritis virus. J. Virol. 76:10829-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong, S. J., R. H. Boyle, V. L. Demarest, A. N. Woodmansee, L. D. Kramer, H. Li, M. Drebot, R. A. Koski, E. Fikrig, D. A. Martin, and P. Y. Shi. 2003. Immunoassay targeting nonstructural protein 5 to differentiate West Nile virus infection from dengue and St. Louis encephalitis virus infections and from flavivirus vaccination. J. Clin. Microbiol. 41:4217-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong, S. J., V. L. Demarest, R. H. Boyle, T. Wang, M. Ledizet, K. Kar, L. D. Kramer, E. Fikrig, and R. A. Koski. 2004. Detection of human anti-flavivirus antibodies with a West Nile virus recombinant antigen microsphere immunoassay. J. Clin. Microbiol. 42:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]