Abstract
Quorum sensing (QS) is a key regulator of virulence and biofilm formation in Pseudomonas aeruginosa and other medically relevant bacteria. Aqueous extracts of six plants, Conocarpus erectus, Chamaesyce hypericifolia, Callistemon viminalis, Bucida buceras, Tetrazygia bicolor, and Quercus virginiana, were examined in this study for their effects on P. aeruginosa virulence factors and the QS system. C. erectus, B. buceras, and C. viminalis caused a significant inhibition of LasA protease, LasB elastase, pyoverdin production, and biofilm formation. Additionally, each plant presented a distinct effect profile on the las and rhl QS genes and their respective signaling molecules, suggesting that different mechanisms are responsible for efficacy. Extracts of all plants caused the inhibition of QS genes and QS-controlled factors, with marginal effects on bacterial growth, suggesting that the quorum-quenching mechanisms are unrelated to static or cidal effects.
Pneumonia due to microbial infections is a major cause of morbidity and mortality in immunocompromised patients. Pseudomonas aeruginosa hails as the leading pathogen among patients with cystic fibrosis, diffused panbronchitis, and chronic obstructive pulmonary disease (16, 29, 37). In addition, P. aeruginosa remains one of the major causes of nosocomial infections (10). The success of this organism is largely due to the production of a myriad of virulence factors (including LasA protease, LasB elastase, pyoverdin, pyocyanin, and alginate) and its ability to form intractable biofilms (38).
Expression of many of the virulence factors in P. aeruginosa is controlled by a quorum-sensing (QS) system (59), an intercellular communication scheme in which bacteria are able to detect the population density (via signaling molecules and receptors) and control gene expression accordingly (55). P. aeruginosa elaborates two main sets of QS systems: lasI-lasR and rhlI-rhlR (55). LasI and RhlI are synthetases that manufacture the autoinducer signaling molecules N-(3-oxododecanoyl)-l-homoserine lactone (OdDHL) and N-butanoyl-l-homoserine lactone (BHL), respectively. These molecules diffuse out into the environment, and when they reach a putative threshold concentration, they activate the receptors lasR and rhlR. These receptors, in turn, coordinate the regulation of pathogenicity. A third signal, the Pseudomonas quinolone signal, also plays an integral role in the QS system (50). This secondary metabolite of P. aeruginosa is incorporated into the QS hierarchy in times of cell stress (43). This intricate communication system of P. aeruginosa is mirrored in many gram-negative pathogenic bacteria, where it coordinates the regulation of virulence, including motility, biofilm formation, and toxin production (18, 19, 21, 48, 54).
The misuse and abuse of antibiotics in pharmacotherapy have led to the development of widespread resistance in the target organism. The failure of existing antibiotics to control infection makes it crucial to find alternatives to currently available drugs. Since pathogenicity in many bacteria is regulated by QS, inhibition of this system may cause the attenuation of virulence and protect against infection (25, 32, 56). In fact, an anti-QS approach has already shown promise in the battle against P. aeruginosa infections (27, 62).
Anti-QS agents were first characterized in the red marine alga (Delisea pulchra) (40, 41) and, more recently, in a south Florida alga (15) and a few higher plants (6, 23, 57). It has been shown that terrestrial plants not only produce autoinducer mimics to confound the bacterial QS system but also receive and respond to microbial signals (1, 3). Given the promise of anti-QS compounds, efficient screening for these agents becomes imperative. In a previous study we utilized an ethnobotanically directed search for anti-QS activity (1). We confirmed that six south Florida medicinal plants, Conocarpus erectus, Chamaesyce hypericifolia, Callistemon viminalis, Bucida buceras, Tetrazygia bicolor, and Quercus virginiana, have anti-QS properties using Chromobacterium violaceum and Agrobacterium tumefaciens NTL4 strains as biomonitors (1). These plants were chosen on the basis of their traditional use against respiratory and skin infections, conditions potentially caused or complicated by bacteria such as P. aeruginosa.
In the study described here we have taken this work a step further by exploring the effects of these six plants on the production of virulence factors and biofilms, acylated homoserine lactone (AHL) levels, and QS gene transcription in this organism. We demonstrate a significant decrease in the production of LasA protease, LasB elastase, pyoverdin, and biofilms in the presence of the extracts. Furthermore, each plant has a unique pattern of effect on the QS genes lasI-lasR and rhlI-rhlR and their respective signaling molecules, OdDHL and BHL.
MATERIALS AND METHODS
Plant extraction.
Conocarpus erectus (Combretaceae), Chamaesyce hypericifolia (Euphorbiaceae), Callistemon viminalis (Myrtaceae) (inflorescence), Bucida buceras (Combretaceae), Tetrazygia bicolor (Melastomataceae), and Quercus virginiana (Fagaceae) were collected and processed by the methods described previously (1). Briefly, pulverized plant material was extracted into boiling water, freeze-dried with a lyophilizer, and stored at −20°C until it was needed. Leaf extracts were tested unless otherwise noted. To complement the six active plants from the previous study, an additional plant with no anti-QS activity (Schefflera actinophylla, Apiaceae) was chosen as a negative control.
Strains and media.
Prototypic P. aeruginosa strain PAO1 (30) and its derivatives (35) were used throughout this study. In addition, Staphylococcus aureus (ATCC 12600) was used in the LasA assay. Cells were maintained on Luria-Bertani (LB) plates and in LB liquid for overnight cultures. For quantitative assays, either LB (LasA assay only) or Agrobacterium (AB) minimum medium (to which glucose and casein amino acids [20%, wt/vol] was added) was used (11).
Culture conditions.
For all assays except those for biofilm formation, the culture conditions were as follows. Overnight cultures of strain PAO1 were grown in LB medium at 37°C with shaking. The cultures were then diluted 100-fold in AB or LB medium and allowed to grow to an optical density at 600 nm (OD600) of 1.7 (early stationary phase). At this point, the culture was divided into 10-ml aliquots and an additional 1 ml of fresh medium containing crude plant extract (or media control) was added to a final concentration of 1 mg/ml extract. Cultures were recovered at late stationary phase (approximately 12 h after addition). The cells were separated from the growth medium by centrifugation at 10,000 × g for 10 min.
LasA staphylolytic assay.
LasA protease activity was determined by measuring the ability of culture supernatants to lyse boiled S. aureus cells (35). A 100-μl aliquot of P. aeruginosa LB medium culture supernatant with or without plant extracts was added to 900 μl of a boiled S. aureus suspension. The OD600 was determined after 0, 5, 10, 20, 30, 45, and 60 min. Activity was expressed as the change in the OD600/hour per μg protein.
LasB elastolytic assay.
The elastolytic activity of the AB medium culture supernatants was determined by using elastin Congo red (ECR; Sigma, St. Louis, MO) (46). A 100-μl aliquot was added to 900 μl of ECR buffer (100 mM Tris, 1 mM CaCl2, pH 7.5) containing 20 mg ECR. This mixture was then incubated with shaking for 3 h at 37°C. Insoluble ECR was removed by centrifugation, and the absorption of the supernatant was measured at 495 nm. Cell-free AB medium alone and AB medium with plant extracts were used as negative controls. Activity was expressed as the change in the OD495 per μg protein.
Pyoverdin assay.
The pyoverdin assay was adapted from the methods of Cox and Adams (14). The AB medium culture supernatant was diluted 10-fold in Tris-HCl buffer (pH 7.4), and 100-μl aliquots were added to 96-well microtiter plates on ice. The relative concentration of pyoverdin was based on the fluorescence of the supernatant at an excitation wavelength of 405 nm and an emission wavelength of 465 nm (on a Tecan GENios FluorSpec instrument). Activity was expressed in relative fluorescence units. Although we consider pyoverdin to be a marker of QS, a drop in production may be due to an indirect effect via pH or iron concentration changes (31). To eliminate the chance of false-positive results, the iron concentration was tested by using 1,10-phenanthroline and desferal (63), and the solution pH was checked throughout the experiment.
Polyvinyl chloride biofilm formation assay.
The effect of plant extracts on the attachment phase of biofilm formation was measured by using the polyvinyl chloride biofilm formation assay (47). Briefly, overnight cultures of PAO1 were resuspended in fresh AB medium in the presence and the absence of plant extracts. After a 10-h incubation at 30°C, the biofilms in the microtiter plates were visualized by staining with a crystal violet solution. The plates were rinsed to remove planktonic cells, and the surface-attached cells were then quantified by solubilizing the dye in ethanol and measuring the absorbance at OD650.
AHL assay.
AHLs were extracted from the AB culture supernatants with acidified ethyl acetate, dried under nitrogen, and quantified by electrospray mass spectroscopy after the methods of Makemson et al. (39). The peak intensities for BHL (m/z = 172), OdDHL (m/z = 298), and their sodium adducts (m/z = 194 and 230, respectively) were combined and converted to concentrations by using a standard curve generated from the pure compounds. Background readings from samples extracted with alkaline ethyl acetate were subtracted from those of the acid-extracted bacterial cultures before conversion, as the lactone ring is broken by alkaline hydrolysis, making AHLs too polar to be fully extracted into ethyl acetate.
Growth curves.
The effect of plant extracts on cell proliferation was determined by monitoring the strain PAO1 growth curve. Briefly, an overnight culture (in LB medium) of PAO1 was diluted 100-fold into 1 liter AB or LB medium. The OD600 was monitored at 45-min intervals until an OD600 of ∼1.7 was obtained (approximately 8 h). The culture was then divided into 28-ml aliquots, to which 2 ml of AB or LB medium (controls) or 2 ml concentrated extract was added. The final extract concentration was 1 mg/ml. The of OD600s of cultures with added extracts were normalized to the control OD600 of 1.7 at this time to account for plant pigmentation. The OD was monitored at 1.5-h intervals until a final time point of 24 h. All OD600 measurements were verified at a 1/10 dilution for greater accuracy.
β-Galactosidase assay.
The transcriptional activity of the QS gene promoters was assayed by using strain PAO1-derived strains harboring the promoter-lacZ fusions PlasI-lacZ (pPCS223), PlasR-lacZ (pPCS1001), PrhlI-lacZ (pLPR1), and PrhlR-lacZ (pPCS1002) and, as a control, a promoterless-lacZ fusion strain (pLP170) (35). The cultures were grown in AB medium and monitored under the same conditions used for strain PAO1, with the extract added once the cultures reached an OD600 of 1.7. Assays for β-galactosidase activity in P. aeruginosa were performed with o-nitrophenyl-β-d-galactopyranoside, as described previously (42).
Bradford assay.
In addition to growth curve monitoring, the Bradford assay (7) was performed to confirm that the reduction in virulence factors was not due to a decrease in cell density. Raw data from all assays were normalized to the total protein concentration; however, there was no significant difference between sample sets.
Statistical analysis.
All experiments were performed independently in triplicate with pooled samples of biological replicates, as described by Adonizio et al. (1). Data were analyzed by one-way analysis of variance, with a P value of 0.05 being significant, by using the SPSS (Chicago, IL) statistical software package.
RESULTS
LasA protease activity in the presence of plant extracts.
LasA staphylolytic protease is a 20-kDa zinc metalloendopeptidase belonging to the β-lytic endopeptidase family of proteases (33). There was a significant decrease in LasA activity compared to that of the control when strain PAO1 was grown in the presence of B. buceras (96% decrease), C. erectus (94% decrease), T. bicolor (89% decrease), C. viminalis (71% decrease), or C. hypericifolia (49% decrease) (Table 1). Addition of Q. virginiana had no significant effect on LasA protease production. As expected, the negative control, S. actinophylla, also showed no significant change in LasA activity (Table 1).
TABLE 1.
Effect of plant aqueous extracts on P. aeruginosa virulence factors
| Culture condition | LasA activitya | Elastase activityb | Pyoverdin productionc | Biofilm formationd |
|---|---|---|---|---|
| Medium only | 0.274 ± 0.016 | 145.5 ± 6.9 | 4,918 ± 281 | 0.64 ± 0.01 |
| C. erectus | 0.017 ± 0.005e | 48.2 ± 11.8e | 453 ± 85e | 0.38 ± 0.19 |
| B. buceras | 0.010 ± 0.011e | 130.8 ± 31.5 | 800 ± 275e | 0.14 ± 0.01e |
| C. viminalis | 0.080 ± 0.004e | 53.9 ± 4.2e | 1,997 ± 271e | 0.07 ± 0.01e |
| T. bicolor | 0.030 ± 0.015e | 105.6 ± 14.3 | 2,134 ± 304e | 0.13 ± 0.02e |
| Q. virginiana | 0.197 ± 0.012 | 499.1 ± 36.0 | 4423 ± 422 | 0.08 ± 0.03e |
| C. hypericifolia | 0.139 ± 0.012e | 553.6 ± 19.4 | 3,068 ± 295e | 0.70 ± 0.07 |
| S. actinophylla | 0.224 ± 0.014 | 172.4 ± 9.2 | 4,069 ± 611 | 0.53 ± 0.16 |
LasA activity is expressed as the reduction in the OD600 per hour per microgram of total protein.
Elastase activity is expressed as the absorbance at OD495 per microgram of protein·1,000.
Pyoverdin production is expressed as the fluorescence at 465 nm (excitation λ = 405 nm) per microgram of protein.
Biofilm production is expressed as the OD650 after incubation with crystal violet.
Significance at P = 0.05.
LasB elastase activity in the presence of plant extracts.
LasB elastase is a zinc metalloprotease capable of destroying or inactivating a wide range of biological tissues and immunological agents (5). There was a significant decrease in LasB activity compared to that of the control when strain PAO1 was grown in the presence of C. erectus (65% decrease) or C. viminalis (63% decrease). The growth of PAO1 with B. buceras, T. bicolor, or S. actinophylla (negative control) caused no significant effect, whereas the growth of PAO1 with C. hypericifolia and Q. virginiana caused an increase in elastase activity.
Extracts alter pyoverdin production.
Pyoverdins are virulence factors, in that they compete with mammalian transferrin for iron, the successful sequestration of which essentially starves the host tissues (44). They also promote pathogenicity by stimulating bacterial growth (14). One of the pyoverdins is suggested to be a QS-like molecule, regulating both itself and the production of other toxins (4, 36). All plant extracts with the exception of those of Q. virginiana and S. actinophylla (negative control) showed a significant reduction of pyoverdin production; however, the results for C. hypericifolia were only marginally significant (Table 1). The most active extracts were C. erectus and B. buceras, with a substantial decrease in pyoverdin activity (91% and 84% decreases, respectively) compared to that of the control (Table 1). All of the culture supernatants retained a pH of ∼7.0, regardless of the amount or the type of extract added. Although statistically significant differences between the iron concentrations in the extracts were found, there was no trend which could be correlated with activity against pyoverdin (data not shown).
Plant extracts have an inhibitory effect on biofilm formation.
P. aeruginosa has the ability to form biofilms, a partially QS-controlled phenomenon (17) in which cells are organized into layers and enmeshed in a matrix of mucoid polysaccharides (12). A switch to the biofilm mode of growth confers increased antibacterial resistance and creates a considerably more severe infection in the lungs of patients with cystic fibrosis (38). There was a significant decrease in biofilm formation compared to that of the control when strain PAO1 was grown in the presence of C. viminalis (89% decrease), Q. virginiana (88% decrease), T. bicolor (80% decrease), or B. buceras (78% decrease) (Table 1). C. erectus caused a 41% decrease in biofilm formation; however, this result was marginally significant.
Plant extracts affect QS gene expression.
With the exception of Q. virginiana, C. hypericifolia, and the negative plant control (S. actinophylla), most extracts had a significant effect on QS gene expression (Table 2). The most significant decreases in lasI expression were found with C. viminalis (80% decrease), B. buceras (49% decrease), T. bicolor (41% decrease), and C. erectus (38% decrease). C. erectus and C. viminalis showed the greatest reduction of lasR expression (56% and 48% decreases, respectively), whereas T. bicolor (42% decrease), B. buceras (55% decrease), and C. erectus (40% decrease) showed the greatest reductions in rhlI expression. The same three plants also effected the greatest reductions in rhlR expression (66%, 62%, and 66% decreases, respectively).
TABLE 2.
Effect of plant aqueous extracts on P. aeruginosa QS genes and AHL production
| Culture condition | AHL production (μM)
|
Gene expressiona
|
||||
|---|---|---|---|---|---|---|
| C12-AHL | C4-AHL | lasI | lasR | rhlI | rhlR | |
| Medium only | 1.216 ± 0.19 | 0.789 ± 0.10 | 3,363 ± 311 | 5,008 ± 256 | 4855 ± 459 | 8271 ± 655 |
| C. erectus | 0.981 ± 0.14 | 0.597 ± 0.01 | 2,101 ± 270c | 1,489 ± 102c | 2,917 ± 265c | 3,004 ± 406c |
| B. buceras | 0.751 ± 0.16b | 0.468 ± 0.12c | 1,718 ± 147c | 2,377 ± 179c | 2,200 ± 249c | 3,339 ± 440c |
| C. viminalis | 0.659 ± 0.15c | 0.533 ± 0.02c | 662 ± 86c | 1,745 ± 182c | 3,370 ± 300c | 3,751 ± 120c |
| T. bicolor | 1.501 ± 0.29 | 0.483 ± 0.03c | 2,000 ± 183c | 2,038 ± 202c | 2,810 ± 449c | 2,930 ± 282c |
| Q. virginiana | 1.041 ± 0.19 | 0.586 ± 0.09 | 2,806 ± 204 | 3,374 ± 460c | 3,128 ± 299c | 5,324 ± 306c |
| C. hypericifolia | 0.892 ± 0.02 | 0.511 ± 0.08c | 2,765 ± 249 | 3,324 ± 97c | 3,433 ± 412c | 3,635 ± 319c |
| S. actinophylla | 1.146 ± 0.17 | 0.761 ± 0.10 | 3,715 ± 363 | 4,692 ± 200 | 4,748 ± 181 | 7,347 ± 628 |
Gene expression was measured as the β-galactosidase activity of the lacZ gene fusion products and is expressed in Miller units.
Significance at P = 0.10.
Significance at P = 0.05.
Plant extracts affect the production of AHL molecules.
Notable decreases in OdDHL levels were seen with C. viminalis (46% reduction from that for the control) and B. buceras (38% reduction). Significant decreases in BHL levels corresponded to the addition of B. buceras, T. bicolor, and C. hypericifolia extracts, with reductions of 41%, 39%, and 35%, respectively.
Extracts have a minimal effect on PAO1 growth after log phase.
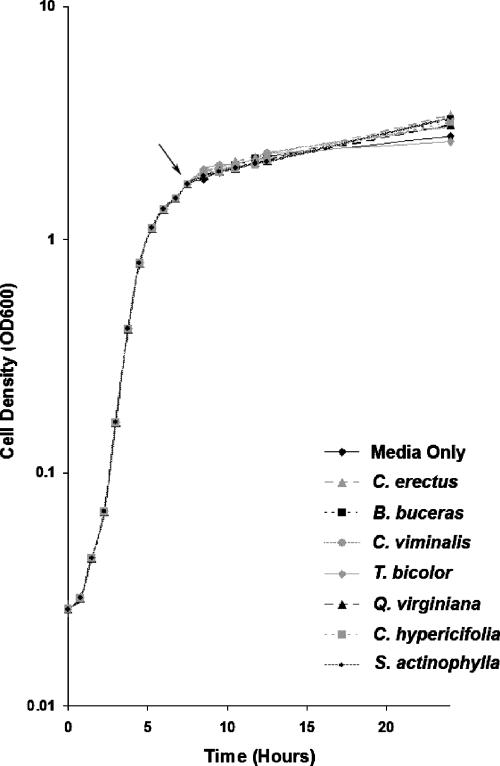
Addition of extracts at early stationary phase was chosen to limit any confounding effects on growth. When the extracts were added at the beginning of the cell cycle (time zero), there were changes (a slight delay or acceleration) in the logarithmic phase (data not shown). This did not, however, affect the endpoint cell density in most cases. To confirm an anti-QS mode of action rather than logarithmic changes, a growth curve was taken by controlling for the latter (Fig. 1). Cultures of strain PAO1 were grown to early stationary phase before addition of the compound (Fig. 1, arrow). This ensures that all samples would have the same opportunity (length of time and cell density) to reach the point of QS-controlled production of virulence factors. Stationary phase was reached in all samples (including the control) approximately 8 h after extract addition. Addition of the extracts did not significantly affect the cell density or the total protein concentration.
FIG. 1.
Influence of medicinal plant extracts on growth of P. aeruginosa (semilogarithmic graph). Extracts were added during early stationary phase (approximately 8 h), as indicated by the arrow. The data represent the mean values of experiments performed in triplicate.
The majority of the samples had little effect on the growth curve of strain PAO1 after log phase (Fig. 1). The growth curve for cultures to which T. bicolor was added closely followed that of the medium-only control, whereas the growth curves for Q. virginiana, B. buceras, C. hypericifolia, C. erectus, and S. actinophylla all exhibited slight increases in cell density (Fig. 1). This increase was not significant and was possibly due to the added nutrients. However, it verifies that the anti-QS effects of these plants are not due to cell death.
Incidentally, addition of T. bicolor and Q. virginiana at time zero resulted in cell densities below the cell density for the control (data not shown). This suggests an additional antibacterial effect of these plant extracts. However, the total protein concentration (measured by the Bradford assay) of these samples did not differ significantly from that of the control.
DISCUSSION
Six south Florida medicinal plants, C. erectus (Combretaceae), C. hypericifolia (Euphorbiaceae), C. viminalis (Myrtaceae), B. buceras (Combretaceae), T. bicolor (Melastomataceae), and Q. virginiana (Fagaceae), were examined for their anti-QS activities against P. aeruginosa PAO1. The virulence of P. aeruginosa is owed to its capacity to degrade host tissue with proteases and toxins and to evade antibiotic attack by the formation of biofilms. Biofilm formation and the virulence factors examined in this study are under QS control (17, 34, 44, 45). Thus, the plant extracts were examined for their ability to interfere with the QS-dependent production of the P. aeruginosa virulence factors LasA, LasB, and pyoverdin. In addition, we examined the ability of the extracts to inhibit biofilm formation, QS gene expression, and AHL synthesis.
Plant extracts differentially affect biofilm formation.
Since QS is involved in biofilm formation (17), we expected the plants possessing anti-QS activity to have a significant effect. Indeed, all extracts tested, with the exception of the extract of C. hypericifolia, effected a decrease in biofilm formation. Interestingly, Q. virginiana exhibited an effect on biofilm production, but not on any other virulence factors, suggesting either a physical inhibition of biofilm growth or the repression of biofilm genes and components outside the QS system. Disruption of the QS system with furanones has also been shown to inhibit biofilm growth (26). Previous work with garlic and D. pulchra furanones showed a qualitative change in biofilm morphology and a reduction in thickness; however, these analyses were not quantified (26, 27, 51).
Plant extracts differentially affect production of virulence factors.
P. aeruginosa proteases LasA and LasB are believed to play a major role in pathogenesis via host tissue degradation (34, 45). With the exception of growth with Q. virginiana extracts, growth with the extracts of all other plant species tested resulted in a significant decrease in the LasA activity of P. aeruginosa, with the most drastic reductions seen in B. buceras, C. erectus, and T. bicolor extract cultures. No prior studies have considered LasA activity in the presence of anti-QS compounds.
The LasB elastase activity was significantly reduced in the presence of C. erectus and C. viminalis extracts. Extracts from B. buceras and T. bicolor had no significant effect, whereas those from C. hypericifolia and Q. virginiana caused an increase in elastase activity. It is likely that the compounds in the last two plants may upregulate the production of LasB and/or enhance the elastase activity. The compounds are not likely to be elastase-like proteins in the plants, since the extraction process would have denatured most proteins. In comparison, recent studies with garlic (at 2%, vol/vol) showed a 50% decrease in LasB activity (51), whereas studies with purified halogenated furanone from D. pulchra (10 μM) resulted in an approximately 90% decrease (27).
All plant extracts, with the exception of the Q. virginiana extract, caused a significant reduction in pyoverdin production. Mixed results on pyoverdin production have previously been observed with furanones. A naturally occurring furanone from D. pulchra [(5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone] actually increased the production of pyoverdin (53), whereas furanone C-30, a synthetic derivative of a compound from D. pulchra, conferred a 90% reduction in pyoverdin levels (27). Neither iron limitation nor pH is sufficient to explain the effect on virulence factor production. There were also no significant changes in cell growth corresponding to pyoverdin production, leaving an anti-QS effect as the most plausible hypothesis.
Mechanistic musings: multiple targets or global effect.
QS inhibition occurs via a number of different mechanisms, with the most well known being signal mimicry, such as in the case of the furanones (40, 41). Other methods include signal degradation by proteins such as lactonase and acylase (20), signal binding, the inhibition of genetic regulation systems, or the interruption of downstream virulence and biofilm genes (for a review, see reference 60).
The current view of the P. aeruginosa QS hierarchy suggests that las controls rhl with virulence proteins at the bottom of the ladder (55). The virulence factors LasA (staphylolytic protease) and LasB (elastase) are generally thought to be under the control of the lasI-lasR system (22, 55); however, rhlI-rhlR also controls activity to a lesser extent (8, 49). Pyoverdin is believed to be under the control of rhlI-rhlR (8, 61), whereas biofilm production is only partially under the control of the QS system (17). The redundant and autoregulatory nature of the QS system is quite convoluted (58). This fact, compounded with the complex phytochemistry of plant extracts, prevents us from precisely linking QS gene expression, the AHL level, and virulence factor production.
Although the mechanism of action of these plant extracts is a complex problem, there was an overall inhibition of the QS system with each of the plant extracts tested (with the exception of the control S. actinophylla extract). This somewhat general effect points to one of two explanations. The first one suggests that multiple chemicals in the plants may be causing distinct effects on different aspects of the QS system. The second explanation is that the effect is not directly on the las-rhl system but, rather, on a more global QS regulator, such as Vfr (2) or GacA (52).
There is also a trend in which the plant species that have less of an effect on AHL production and las-rhl expression also have less activity against P. aeruginosa virulence factors. This was most clearly seen with Q. virginiana, C. hypericifolia, and the control, S. actinophylla (Tables 1 and 2). Although it is not absolute, the converse is generally true. Overall, the most significant effects on the QS system were found with C. viminalis, B. buceras, and C. erectus.
The distinct patterns of thin-layer chromatography migration of these three extracts (data not shown) suggest that they contain multiple active compounds and that they perhaps function with separate mechanisms. At this point we do not have sufficient data to pinpoint the method of quorum quenching. We will therefore withhold further speculation on the mechanism of action until these compounds are purified from the plants.
In summary, this work describes six plant species, representing five distinct families, which have a differential but significant effects on virulence factors, biofilms, QS gene expression, and signal production at a concentration of 1 mg/ml. This concentration, although high, is relevant to traditional medicinal use in teas and poultices. More significantly, this concentration represents that of a crude aqueous extract, and therefore, the concentration should be much lower when the purified agent is used as a putative anti-QS compound. Concentrations down to 0.25 mg/ml were tested; and some extracts, notably, that of C. erectus, still had an effect on virulence factor production.
None of the plant extracts tested had a significant effect on the growth of strain PAO1 when they were added at early stationary phase (Fig. 1, arrow). However, addition of T. bicolor and Q. virginiana at time zero resulted in cell densities below the density for the control (data not shown), which suggests that these plant extracts have additional antibacterial effects. All other plant extracts significantly reduced one to four QS-controlled virulence factors, genes, and AHL levels without a reduction in growth. This would strongly suggest an anti-QS effect rather than an antibacterial effect.
The top candidates for further anti-QS investigation are C. erectus, B. buceras, and C. viminalis. C. erectus significantly decreases LasA, LasB, and pyoverdin production but not biofilm formation, whereas B. buceras affects all but elastase activity. B. buceras also has a significant effect on the expression of lasI, lasR, rhlI, and rhlR and the concentrations of OdDHL and BHL. C. erectus exhibits the same pattern of effect on QS genes and signaling molecules, although to a lesser extent than B. buceras. C. viminalis decreases all three virulence factors and biofilm formation, all QS genes tested, and OdDHL concentrations. Research is under way in our laboratory to isolate the active chemicals from these species.
The plant kingdom has long been a source of medicines, and as such, there have been many ethnobotanically directed searches for agents that can be used to treat infections (e.g., (9, 13, 24, 28). However, most studies focus solely on bactericidal effects. Since the plants in this study showed little, if any, cidal activity (1), quorum inhibition remains a potential mode of action. A shift of our focus to anti-QS and antivirulence properties may reveal new quorum-quenching compounds from medicinal plants and provide a novel method for the treatment of infections.
In conclusion, the effects of the plant extracts studied on P. aeruginosa are quite complicated and perhaps extend beyond the domain of the QS control hypothesis. However, the reduction of QS gene expression and signaling molecule levels and the end effect on virulence factor production provide some insight into why these plants were used in the past and how they can be used in the future to combat P. aeruginosa and other bacterial infections.
Acknowledgments
We gratefully acknowledge the support of the National Institutes of Health National Center for Alternative and Complementary Medicine (grant NRSA 1-T32-AT01060-01 to A.A. and 1-R15-AT002626-01 to K.M.), Cystic Fibrosis Foundation Traineeship Grant ADONIZ06H0 (to A.A.), NIGMS Research Initiative Award R25 GM61347 (to A.A.), Biomedical Research Initiative Award R25 GM61347, and Florida International University Teaching Assistantship (to K.-F.K.).
We thank John Makemson, members of the K. Mathee lab for assistance and support (especially Melissa Doud, Deepak Balasubramanian, and Lance Umansky), and members of the D. Roy lab for gracious use of the fluorescence spectrophotometer. We also thank the reviewers for their critical reading of the manuscript and their invaluable suggestions and comments.
Footnotes
Published ahead of print on 15 October 2007.
REFERENCES
- 1.Adonizio, A. L., K. Downum, B. C. Bennett, and K. Mathee. 2006. Anti-quorum sensing activity of medicinal plants in southern Florida. J. Ethnopharmacol. 103:427-435. [DOI] [PubMed] [Google Scholar]
- 2.Albus, A., E. Pesci, L. Runyen-Janecky, S. West, and B. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, W. D., and U. Mathesius. 2004. Plant responses to bacterial quorum sensing signals. Curr. Opin. Plant Biol. 7:429-433. [DOI] [PubMed] [Google Scholar]
- 4.Beare, P. A., R. J. For, L. W. Martin, and I. L. Lamont. 2003. Siderophore-mediated cell signaling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 47:195-207. [DOI] [PubMed] [Google Scholar]
- 5.Bever, R. A., and B. H. Iglewski. 1988. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J. Bacteriol. 170:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjarnsholt, T., P. O. Jensen, T. B. Rasmussen, L. Christophersen, H. Calum, M. Hentzer, H.-P. Hougen, J. Rygaard, C. Moser, L. Eberl, N. Hoiby, and M. Givskov. 2005. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 151:3873-3880. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Brint, J., and D. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camporese, A., M. J. Balick, R. Arvigo, R. G. Esposito, N. Morsellino, F. D. Simone, and A. Tubaro. 2003. Screening of anti-bacterial activity of medicinal plants from Belize (Central America). J. Ethnopharmacol. 87:103-107. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention, Health Inspections Program. 2004. National Nosocomial Infections Surveillance (NNIS) report, data summary from January 1992-June 2004, issued October 2004: a report from the NNIS System. Am. J. Infect. Control 32:470-485. [DOI] [PubMed] [Google Scholar]
- 11.Clark, D. J., and O. Maaloe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 12.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 13.Cowan, M. M. 1999. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 12:564-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox, C. D., and P. Adams. 1985. Siderophore activity of pyoverdin for Pseudomonas aeruginosa. Infect. Immun. 48:130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cumberbatch, A. 2002. Characterization of the anti-quorum sensing activity exhibited by marine macroalgae of south Florida. Undergraduate honors thesis. Florida International University, Miami.
- 16.Cystic Fibrosis Foundation. 2005. Patient registry 2004 annual report. Cystic Fibrosis Foundation, Bethesda, MD.
- 17.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 18.De Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donabedian, H. 2003. Quorum sensing and its relevance to infectious diseases. J. Infect. 46:207-214. [DOI] [PubMed] [Google Scholar]
- 20.Dong, Y. H., L. H. Wang, J. L. Xu, H. B. Zhang, X. F. Zhang, and L. H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 21.Fuqua, C., and E. P. Greenberg. 1998. Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1:183-189. [DOI] [PubMed] [Google Scholar]
- 22.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 179:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao, M., M. Teplitski, J. B. Robinson, and W. D. Bauer. 2003. Production of substances by Medicago truncatula that affect bacterial quorum sensing. Mol. Plant Microbe Interact. 16:827-834. [DOI] [PubMed] [Google Scholar]
- 24.Gnanamani, A., K. Shanmuga Priya, N. Radhakrishnan, and M. Babu. 2003. Antibacterial activity of two plant extracts on eight burn pathogens. J. Ethnopharmacol. 86:59-61. [DOI] [PubMed] [Google Scholar]
- 25.Hentzer, M., and M. Givskov. 2003. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Investig. 112:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hentzer, M., K. Reidel, T. B. Rasmussen, A. Heydorn, J. B. Anderson, M. R. Parsek, S. A. Rice, L. Eberl, S. Molin, N. Hoiby, S. Kjelleberg, and M. Givskov. 2002. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87-102. [DOI] [PubMed] [Google Scholar]
- 27.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez, T., M. Canales, J. G. Avila, A. Duran, J. Caballero, A. Romo de Vivar, and R. Lira. 2003. Ethnobotany and antibacterial activity of some plants used in traditional medicine of Zapotitlan de las Salinas, Puebla (Mexico). J. Ethnopharmacol. 88:181-188. [DOI] [PubMed] [Google Scholar]
- 29.Hoiby, N. 1994. Diffuse panbronchiolitis and cystic fibrosis: East meets West. Thorax 49:531-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holloway, B. W., and A. F. Morgan. 1986. Genome organization in Pseudomonas. Annu. Rev. Microbiol. 40:79-105. [DOI] [PubMed] [Google Scholar]
- 31.Jacques, P., M. Ongena, F. Bernard, R. Fuchs, H. Budzikiewicz, and P. Thonart. 2003. Fluorescent Pseudomonas mainly produce the dihydroform of pyoverdine at low specific growth rate. Lett. Appl. Microbiol. 36:259-262. [DOI] [PubMed] [Google Scholar]
- 32.Juhas, M., L. Eberl, and B. Tummler. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ. Microbiol. 7:459-471. [DOI] [PubMed] [Google Scholar]
- 33.Kessler, E. 1995. Beta-lytic endopeptidases. Methods Enzymol. 248:740-756. [DOI] [PubMed] [Google Scholar]
- 34.Kharazmi, A. 1989. Interactions of Pseudomonas aeruginosa proteases with the cells of the immune system. Antibiot. Chemother. 42:42-49. [DOI] [PubMed] [Google Scholar]
- 35.Kong, K.-F., S. R. Jayawardena, S. D. Indulkar, A. del Puerto, C.-L. Koh, N. Hoiby, and K. Mathee. 2005. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB β-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 49:4567-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamont, I. L., P. A. Beare, U. Ochsner, and M. L. Vasil. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 99:7072-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieberman, D. 2003. Pseudomonal infections in patients with COPD: epidemiology and management. Am. J. Respir. Med. 2:459-468. [DOI] [PubMed] [Google Scholar]
- 38.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makemson, J., A. Eberhard, and K. Mathee. 2006. Simple electrospray mass spectrometry detection of acylhomoserine lactones. Luminescence 21:1-6. [DOI] [PubMed] [Google Scholar]
- 40.Manefield, M., R. de Nys, N. Kumar, R. Read, M. Givskov, P. Steinberg, and S. Kjelleberg. 1999. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 145:283-291. [DOI] [PubMed] [Google Scholar]
- 41.Manefield, M., T. B. Rasmussen, M. Henzter, J. B. Andersen, P. Steinberg, S. Kjelleberg, and M. Givskov. 2002. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148:1119-1127. [DOI] [PubMed] [Google Scholar]
- 42.Mathee, K., C. McPherson, and D. Ohman. 1997. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J. Bacteriol. 179:3711-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer, J., A. Neely, A. Stintzi, C. Georges, and I. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morihara, K., and J. Y. Homma. 1985. Pseudomonas proteases, p. 41-79. In I. Holder (ed.), Bacterial enzymes and virulence. CRC Press, Inc., Boca Raton, FL.
- 46.Ohman, D. E., S. J. Cryz, and B. H. Iglewski. 1980. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 48.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearson, J., E. Pesci, and B. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pesci, E. C., J. B. J. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rasmussen, T. B., T. Bjarnsholt, M. E. Skindersoe, M. Hentzer, P. Kristoffersen, M. Kote, J. Nielsen, L. Eberl, and M. Givskov. 2005. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 187:1799-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 53.Ren, D., R. Zuo, and T. K. Wood. 2005. Quorum-sensing antagonist (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone influences siderophore biosynthesis in Pseudomonas putida and Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 66:689-695. [DOI] [PubMed] [Google Scholar]
- 54.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Gene Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 55.Schuster, M., and E. P. Greenberg. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:73-81. [DOI] [PubMed] [Google Scholar]
- 56.Smith, R. S., and B. H. Iglewski. 2003. Pseudomonas aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 57.Teplitski, M., J. B. Robinson, and W. D. Bauer. 2000. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant Microbe. Interact. 13:637-648. [DOI] [PubMed] [Google Scholar]
- 58.Van Delden, C., and B. H. Iglewski. 1998. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venturi, V. 2006. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30:274-291. [DOI] [PubMed] [Google Scholar]
- 60.Whitehead, N. A., M. Welch, and G. P. C. Salmond. 2001. Silencing the majority. Nat. Biotechnol. 19:735-736. [DOI] [PubMed] [Google Scholar]
- 61.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, H., Z. Song, M. Hentzer, J. B. Andersen, S. Molin, M. Givskov, and N. Hoiby. 2004. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J. Antimicrob. Chemother. 53:1054-1061. [DOI] [PubMed] [Google Scholar]
- 63.Yegorov, D. Y., A. V. Kozlov, O. A. Azizova, and Y. A. Vladimirov. 1993. Simultaneous determination of Fe(III) and Fe(II) in water solutions and tissue homogenates using desferal and 1,10-phenanthroline. Free Radical Biol. Med. 15:565-574. [DOI] [PubMed] [Google Scholar]