The marC gene of Escherichia coli is divergently transcribed from the marRAB operon involved in resistance to multiple antibiotics (5, 8), oxidative stress agents (2), and organic solvents (3, 16). Previous data from our laboratory had suggested a role for marC in intrinsic multiple antibiotic resistance (4, 9, 16), and the gene has been so annotated in most databases.
Because that earlier work had suggested that marC was regulated by the repressor MarR and induced by tetracycline, we sought the transcriptional start site for marC to see if the marC promoter might overlap the MarR binding sites within the marRAB promoter (5′ rapid amplification of cDNA end [RACE] system of Gibco/BRL Life Technologies, cells grown with 2 μg/ml tetracycline to increase the amount of mRNA). Transcription of marC started 30 nucleotides upstream from the putative ATG initiation codon of MarC (bp 1266 of Cohen et al. [4]). Therefore, the marC promoter does not contain the MarR binding sites. Moreover, Northern blot analysis of AG100 and its isogenic marR mutant AG112 (in which MarR is inactive [8, 12]) showed no differences in levels of marC mRNA between the two strains (data not shown), nor was expression of marC induced by salicylate, which inactivates the repressor MarR (1) (Fig. 1). We conclude that marC is not regulated by MarR. Since chloramphenicol does not bind to MarR (1), the apparent up-regulation by tetracycline and chloramphenicol (Fig. 1) (4) likely reflects stabilization of mRNA rather than true induction (11, 13, 14).
FIG. 1.
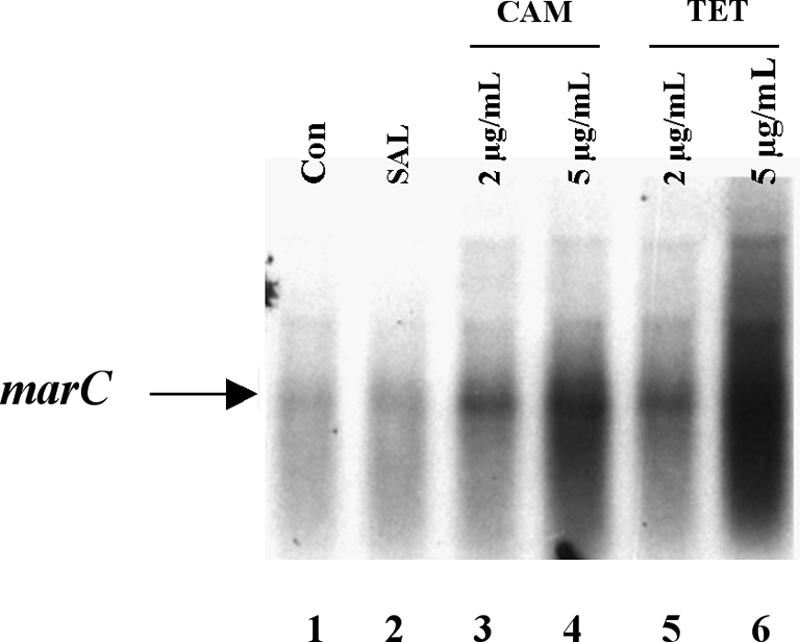
Northern blot analysis of marC from E. coli. RNA was isolated from mid-exponential-phase cells grown in LB broth at 30°C and treated for 1 h with the specified compounds. Separate cultures of E. coli AG100 following exposure to 5 mM salicylate (SAL), tetracycline (TET), or chloramphenicol (CAM) at the indicated concentrations were probed with marC. Controls (Con) represent RNA from nontreated cultures. Total cellular RNA was prepared by cesium chloride gradient as previously described (15). RNA (20 μg) was separated by electrophoresis in a 1.5% agarose gel containing 20 mM guanidinium thiocyanate, transferred to Hybond N+ nylon membranes (Amersham), and probed using 32P-radiolabeled marC. Hybridization signals were visualized using a phosphorimager (Molecular Dynamics, Sunnyvale, CA).
We replaced the marC locus in E. coli AG100 with a kanamycin cassette as described previously (10) and looked for any increase in susceptibilities to antimicrobials. MICs were determined on LB agar by use of Etest strips (AB Biodisk, Sölna, Sweden) with about three dozen different agents, including beta lactams, tetracyclines, fluoroquinolones, cephalosporins, imipenem, macrolides, aminoglycosides, chloramphenicol, fusidic acid, trimethoprim, and rifampin. Gradient plates (6) were used for oxidative stress agents (plumbagin, paraquat, phenylmethylsulfonate, dinitrophenol, and menadione) and for ethidium bromide. No differences in susceptibilities were seen for the marC::kan deletion mutant relative to the wild type. We then replaced the genes for the E. coli MarC paralogs YchE and YhgN in the marC::kan strain by use of spectinomycin and gentamicin cassettes, respectively, to create a triple knockout mutant, but again no differences in susceptibilities were seen.
We also used three plasmid constructs designed to overexpress marC via the araBAD, T7, or native marC promoter. Plasmid pHA-1::marC (obtained from D. Daley) specifies membrane-bound MarC-PhoA regulated by Salmonella enterica serovar Typhimurium pBAD/AraC (7). We constructed pETmarC11 (T7 promoter/lac operator with lac repressor; specifying MarC-6H) and pACmarC1 (marC promoter starting 58 bp upstream of the transcriptional start site; specifying native MarC) by cloning PCR-amplified DNA into vectors pET21b (Novagen) and pACYC184, respectively. None of these three plasmids led to a change in susceptibility of cells to a variety of antibiotics and oxidative stress agents.
We suggest that MarC no longer be classified as a multiple antibiotic resistance protein. However, we do not advise a name change until a function is found.
Acknowledgments
We thank Bonnie Marshall and Sonya Bodeis for help with some of the susceptibility tests and Dan Daley for strain CC118 and plasmid pHA-1::marC.
This work was supported by United States Public Health Service grant AI56021 from the National Institutes of Health.
Footnotes
Published ahead of print on 22 October 2007.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181:4669-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ariza, R. R., S. P. Cohen, N. Bachhawat, S. B. Levy, and B. Demple. 1994. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 176:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asako, H., H. Nakajima, K. Kobayashi, M. Kobayashi, and R. Aono. 1997. Organic solvent tolerance and antibiotic resistance increased by overexpression of marA in Escherichia coli. Appl. Environ. Microbiol. 63:1428-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, S. P., H. Hachler, and S. B. Levy. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, S. P., L. M. McMurry, D. C. Hooper, J. S. Wolfson, and S. B. Levy. 1989. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob. Agents Chemother. 33:1318-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curiale, M. S., and S. B. Levy. 1982. Two complementation groups mediate tetracycline resistance determined by Tn10. J. Bacteriol. 151:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321-1323. [DOI] [PubMed] [Google Scholar]
- 8.George, A. M., and S. B. Levy. 1983. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J. Bacteriol. 155:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hachler, H., S. P. Cohen, and S. B. Levy. 1991. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 173:5532-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez, P. J., I. Marchand, O. Yarchuk, and M. Dreyfus. 1998. Translation inhibitors stabilize Escherichia coli mRNAs independently of ribosome protection. Proc. Natl. Acad. Sci. USA 95:6067-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oethinger, M., W. V. Kern, A. S. Jellen-Ritter, L. M. McMurry, and S. B. Levy. 2000. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob. Agents Chemother. 44:10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pato, M. L., P. M. Bennett, and K. von Meyenburg. 1973. Messenger ribonucleic acid synthesis and degradation in Escherichia coli during inhibition of translation. J. Bacteriol. 116:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pepe, C. M., C. Suzuki, C. Laurie, and R. W. Simons. 1997. Regulation of the “tetCD” genes of transposon Tn10. J. Mol. Biol. 270:14-25. [DOI] [PubMed] [Google Scholar]
- 15.Reddy, K. J., R. Webb, and L. A. Sherman. 1990. Bacterial RNA isolation with one hour centrifugation in a table-top ultracentrifuge. BioTechniques 8:250-251. [PubMed] [Google Scholar]
- 16.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]


