Abstract
Salmonella genomic island 1 (SGI1) and variants (SGI1-I and the new variant SGI1-O) were mapped in five strains of Proteus mirabilis isolated from humans and food in China. Sequencing showed that SGI1 and variants were integrated at the 3′ end of the chromosomal thdF gene as previously described for Salmonella strains.
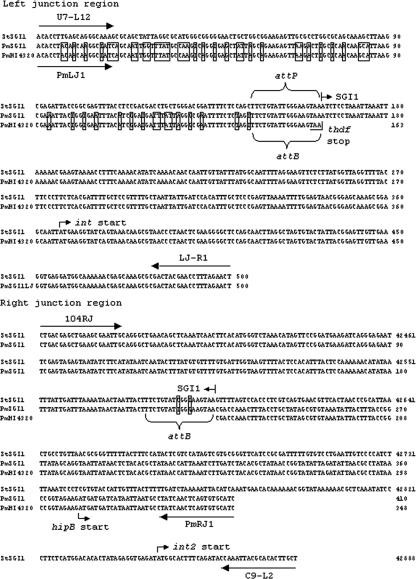
Salmonella genomic island 1 (SGI1) is a 43-kb integrative mobilizable element initially characterized in Salmonella enterica serovar Typhimurium phage type DT104 strains (3, 7). SGI1 contains a multidrug resistance region (MDR), which is a complex class 1 integron containing all the genes necessary for resistance located between a resolvase-like gene (res) and a gene of unknown function (S044) (Fig. 1). SGI1 and variants of it named SGI1-A to SGI1-N have been described for other Salmonella enterica serovars (5, 11, 12, 14, 17, 18). In Salmonella SGI1 is integrated in the last 18 bp of the thdF gene (specific attB site) (7, 14). SGI1 has been shown to be mobilized using a conjugative helper plasmid and thus transferred to Salmonella or Escherichia coli recipient strains (7). Typically SGI1 integration can be confirmed by PCR targeting the left junction region with primers U7-L12 (thdF specific) and LJ-R1 (SGI1 int specific) and/or the right junction region with primer 104RJ (SGI1 S044 specific) and either primer C9-L2 (serovar Typhimurium retron int specific) or primer 104D (yidY specific) (Fig. 1) (2, 3). Recently the presence of an SGI1 variant (SGI1-L) was described in clinical strain Proteus mirabilis 18306, the first bacterium other than Salmonella shown to harbor this element (1). In P. mirabilis 18306 the left junction region could not be detected by PCR, and it was concluded that SGI1 may be integrated elsewhere in the genome (1). In a comment on that report it was pointed out that the thdF gene of P. mirabilis HI430 is only 70% identical to the serovar Typhimurium gene and that in fact the primer U7-L12 binding site contains seven nucleotide differences and, thus, this may explain the failure of the left junction PCR (Fig. 1) (8).
FIG. 1.
Sequence alignment of the left (top) and right (bottom) junction regions of SGI1 from S. enterica serovar Typhimurium DT104 (StSGI1) and P. mirabilis B02012 (PmSGI1) and the genomic region from P. mirabilis HI4320 containing thdF and the downstream region. The nucleotide differences between the Proteus and Salmonella thdF genes are boxed (top) as are the ones in the attB site (bottom). The nucleotide differences between the Salmonella and Proteus sequences after the right junction (bottom) are not indicated, as these sequences are not homologous. Base pair coordinates for StSGI1 are from the sequence with accession no. AF261825. Primers used to detect the junction regions are indicated by arrows.
Strains characterized in this study are listed in Table 1. Primers used in this study are listed in Table 2. PCR was carried out with Amplitaq Gold (Applied Biosystems, Foster City, CA) or the FailSafe PCR System (Epicentre Biotechnologies, Madison, WI). Sequencing was by automated dideoxy cycle sequencing.
TABLE 1.
P. mirabilis strains characterized in this study
| Strain | Integron cassette(s)
|
SGI1 type | Antibiogramc | Isolate source | |
|---|---|---|---|---|---|
| Gene(s)a | Size(s) (kb)b | ||||
| B02012 | aadA2, blaPSE-1 | 1, 1.2 | SGI1 | AmChCiKaNaStSuTeTm | Patient |
| C02035 | aadA2, dfrA1-orfC | 1, 1.2 | SGI1-I | ChStSuTeTmd | Food |
| C04014 | dfrA1-orfC, blaPSE-1-aadA2 | 1.2, 2.0 | SGI1-O | AmChCiKaNaStSuTeTme | Food |
| C05022 | dfrA1-orfC | 1.2 | SGI1-O | AmChKaStSuTeTm | Patient |
| C05023 | dfrA1-orfC | 1.2 | SGI1-O | AmChKaStSuTeTm | Patient |
| P06020 | dfrA7 | 0.75 | None | (Am)ChKaStSuTeTm | Patient |
| P06021 | dfrA7 | 0.75 | None | AmChKaStSuTeTm | Patient |
As determined by PCR mapping and/or DNA sequencing.
Size of amplicon produced with primers 5′-CS and 3′-CS.
Resistant or intermediate resistance (parentheses) to drugs shown as determined by broth microdilution. Drugs tested: amikacin, ampicillin (Am), amoxicillin-clavulanic acid, cefoxitin, ceftiofur, ceftriaxone, chloramphenicol (Ch), ciprofloxacin (Ci), gentamicin, kanamycin (Ka), nalidixic acid (Na), streptomycin (St), sulfizoxazole (Su), tetracycline (Te), and trimethoprim-sulfamethoxazole (Tm).
By streptomycin Etest exhibited a MIC of 8 μg/ml with colonies in the zone. A single colony from the zone exhibited a MIC of 96 μg/ml.
By streptomycin Etest exhibited a MIC of 32 μg/ml with colonies in the zone. A single colony from the zone exhibited a MIC of 96 μg/ml.
TABLE 2.
Primers used in this study
| Primera | Sequence (5′-3′) | Target regionb | PCR
|
Reference | |
|---|---|---|---|---|---|
| No.c | Size (bp) | ||||
| U9-L1 (+) | TACTACAAGCAGATAACGCC | 2771-3679 | St1 | 909 | 13 |
| P1-R1 (−) | TAGAAACGACAAAGCGCGTG | 13 | |||
| P134-L2 (+) | TGACCCAATTCCAAAGCCAC | 16784-17839 | St4 | 1,490 | 13 |
| P134-R1 (−) | GTGTTTGGGCAAGATCCCAG | 13 | |||
| StGP21 (+) | ATAACGGCAGGTTCCGGTTC | 20173-21108 | St5 | 936 | 13 |
| StGP6 (−) | CGATGAAGCGCACAAATTTG | 13 | |||
| StGP24 (+) | TCAAGATTCCTATCTGCAGG | 24363-25201 | St6 | 838 | 13 |
| StGP28 (−) | AGAGTTACTAGACCAAGCGC | 13 | |||
| LJ-R1 (−) | AGTTCTAAAGGTTCGTAGTCG | int | 1 | 500 | 3 |
| PmLJ1 (+) | ACACCTACAACAAGGCTATC | thdF of P. mirabilis | 1 | 500 | This study |
| 23 | 348 | ||||
| 104RJ (+) | CTGACGAGCTGAAGCGAATTG | S044 | 2 | 476 | 3 |
| PmRJ1 (−) | GATGCACACTGAGTTGATAG | hipB of P. mirabilis | 2 | 476 | This study |
| 23 | 348 | ||||
| 5′-CS (+) | GGCATCCAAGCAGCAAGC | 5′-CS region | 3 | Variabled | 10 |
| 3′-CS (−) | AAGCAGACTTGACCTGAT | 3′-CS region | 3 | Variabled | 10 |
| tetG-1s (+) | GCTCGGTGGTATCTCTGCTC | tet(G) | 4 | 468 | 15 |
| tetG-2s (−) | AGCAACAGAATCGGGAACAC | tet(G) | 4 | 468 | 15 |
| StCm-L (+) | CACGTTGAGCCTCTATATGG | floR | 5 | 888 | 2 |
| StCm-R (−) | ATGCAGAAGTAGAACGCGAC | floR | 5 | 888 | 2 |
| F4 (+) | TTCCTCACCTTCATCCTACC | floR | 6 | 599 | 6 |
| F6 (−) | TTGGAACAGACGGCATGG | tetR | 6 | 599 | 6 |
| MDR-13 (+) | TCCCGATTCTGTTGCTGCTTG | tet(G) | 7 | 1,078 | This study |
| MDR-B3 (−) | AAGCATGGCTGCTGACAAC | orf1 | 7 | 1,078 | This study |
| floR-R1 (−) | TCAACGTGAGTTGGATCATAG | floR | 8 | 1,430 | This study |
| MDR-5 (+) | TAGGTATGGGGCTCATAATTG | qacEΔ1 | 8 | 1,430 | This study |
| 104-D4 (+) | ATGCCTAGCATTCACCTTCC | sulI | 9 | 1,183e | This study |
| DB-B4 (−) | ATCACATCACCCTGGAAATGG | IS6100 | 9 | 1,183e | This study |
| 21 | 2,954e | ||||
| 22 | 2,642e | ||||
| DB-T1 (+) | TGCCACGCTCAATACCGAC | IS6100 | 10 | 930 | 2 |
| DB-B6 (−) | CTGTGCCTTCTTGCGAGC | S044 | 10 | 930 | This study |
| StGP-23 (+) | CCTTGGTACGTTCGCTAATC | res | 11 | 1,417 | This study |
| 16 | 1,848 | ||||
| 20 | 2,262 | ||||
| int-R (−) | GCCTTGCTGTTCTTCTACGG | intI1 | 11 | 1,417 | This study |
| AadA2-L (+) | TGTTGGTTACTGTGGCCG | aadA2 | 12 | 538 | 15 |
| AadA2-R2 (−) | TGCTTAGCTTCAAGTAAGACG | aadA2 | 12 | 538 | 3 |
| 15 | 1,316, 2,360f | ||||
| MDR-20 (+) | TAGTTCAAAGTTTCAGCAAG | blaPSE-1 | 13 | 765 | This study |
| 17 | 1,751, 2,608g | ||||
| PSE-R2 (−) | ACAATCGCATCATTTCGCTC | blaPSE-1 | 13 | 765 | 3 |
| PSE-R3 (−) | CTGAAACTTTGAACTACTTGC | blaPSE-1 | 14 | 935 | This study |
| MDR-19 (−) | AACCGGATCAGAAATCCATGC | groEL/int | 14 | 935 | This study |
| 19 | 1,254 | ||||
| MDR-1 (+) | TGATCGAAATCCAGATCCTG | intI1 | 15 | 1,316, 2,360f | This study |
| AadA2-R3 (−) | GGTTCGAAATTTCGATGGTC | aadA2 | 16 | 1,848 | This study |
| QS-2 (−) | TGAGTGCATAACCACCAGCC | sul1 | 17 | 1,751, 2,608g | 3 |
| dfrA1-F (+) | CGAAGAATGGAGTTATCGG | dfrA1 | 18 | 919 | 11 |
| orfC-R (−) | TCTCGAATCAAGCAGGAACC | orfC | 18 | 919 | 11 |
| dfrA1-R (−) | TTAGAGGCGAAGTCTTGG | dfrA1 | 19 | 1,254 | 11 |
| 20 | 2,262 | ||||
| orfC-F (+) | CATTACGAAGCGAA GCACC | orfC | 21 | 2,954e | 11 |
| PSE-D1 (+) | AGAGCGAAATGATGCGATTG | blaPSE-1 | 22 | 2,642e | This study |
| Ag5 (−) | ACAAACGACAAGCCACGT | orf513 | Negh | 750 | This study |
| Ag6 (+) | TATCGTCTATCGTACACTCTC | orf513 | Negh | 750 | This study |
Coordinates are from SGI1 (accession no. AF261825), and genes are as shown in Fig. 2.
PCR numbers as shown in Fig. 2.
Size varies depending on number of cassettes in this region.
Size would be ∼2.9 kb larger if this region contained common region 1 (CR1) and dfrA10 as found in some SGI1 variants (2, 14).
Size was 1,316 bp in SGI1 but 2,360 bp in P. mirabilis C04014.
Size was 1,751 bp in SGI1 but 2,608 bp in P. mirabilis C04014.
PCR with Ag5/Ag6 primers was negative for all strains tested but would be expected to give a product of 750 bp if CR1 was present.
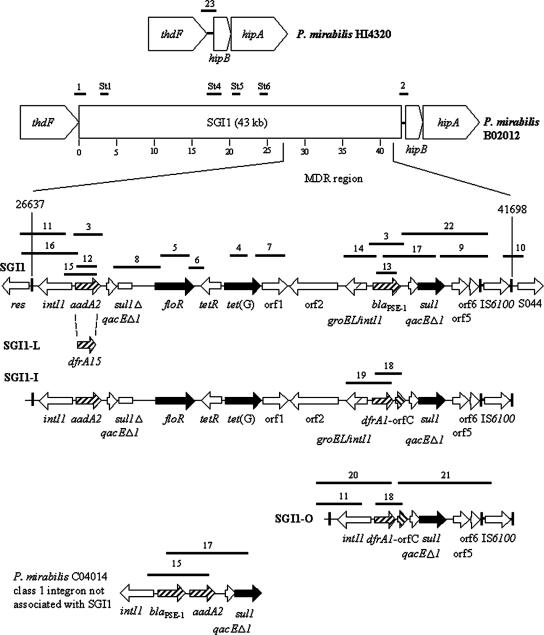
We recently screened 30 clinical P. mirabilis strains isolated from the environment, food, and stool of diarrheic patients in China and P. mirabilis ATCC 29906 and ATCC 29245 for the cassettes of class 1 integrons (5′-CS/3′-CS primers) and detected amplicons in seven strains. (Table 1). All seven strains were multidrug resistant, with the number of drugs ranging from four up to nine (Table 1). PCR mapping and/or DNA sequencing of the cassettes revealed that one strain harbored aadA2 and blaPSE-1, one strain harbored aadA2 and dfrA1-orfC, three strains harbored dfrA1-orfC, and two strains harbored dfrA7 (Table 1; also see below). We used the thdF gene of serovar Typhimurium as a query sequence in a BLAST search of the completely sequenced genome of P. mirabilis HI4320 (http://www.sanger.ac.uk/Projects/P_mirabilis) to find the thdF homologue and flanking sequence and designed primers PmLJ1 and PmRJ1 (Table 2; Fig. 1). We note that the P. mirabilis HI4320 thdF gene is located 145 bp upstream from the start of hipB/hipA toxin/antitoxin homologues (Fig. 2). We conducted PCR with primers PmLJ1 and LJ-R1 (left junction) on all 30 P. mirabilis clinical strains and the two ATCC strains and obtained an amplicon of the predicted size (500 bp if SGI1 was present) only with DNA from the five strains that harbored 1- and/or 1.2-kb cassettes. The seven strains that harbored cassettes were then analyzed by PCR with primers 104RJ and PmRJ1 (right junction), and only the five strains positive for the left junction region produced an amplicon of the expected size (410 bp). The two strains harboring the dfrA7 gene (accession number X58425; 0.75-kb cassette) were not further studied, as it was concluded that although they contained integrons and were multidrug resistant, they were not associated with SGI1 (Table 1). Sequence analysis of both the left and right junction amplicons from P. mirabilis B02012 confirmed that the last 18 bp of the thdF gene corresponding to the left direct repeat (DR-L) is 100% identical to the right 18-bp direct repeat (DR-R) (Fig. 1). This result is in accordance with the sequenced genome of P. mirabilis HI4320, in which the last 18 bp of the putative thdF gene (the attB site) is identical to the previously described attP site of SGI1 (7, 8). Thus, five P. mirabilis strains appeared to have SGI1 or a variant integrated at the end of the thdF gene as in Salmonella. Further, all five strains were positive in PCRs St1, St4, St5, and St6 designed to amplify various regions of SGI1 outside of the MDR (Fig. 2) (13). We used various combinations of the primers listed in Table 2 to map the SGI1s in the five strains (Fig. 2). One strain contained SGI1 (3), one strain contained SGI1-I (11), and three strains contained a new variant named SGI1-O, which contained only the drfA1-orfC cassette. The orf513 gene from common region 1 (CR1) found in some SGI1 variants was not detected (2). Interestingly, one strain with SGI1-O, P. mirabilis C04014 isolated from chicken, was positive by PCR for blaPSE-1 and aadA2. However, though mapping showed these to be adjacent to one another as the only cassettes in a class 1 integron, they were not linked to regions upstream (res) or downstream (IS6100) of the MDR of SGI1 (Fig. 2).
FIG. 2.
Schematic view of the SGI1 and SGI1 variants as integrated in P. mirabilis. The P. mirabilis HI4320 thdF gene and downstream region are shown at the top, and the MDRs of SGI1, SGI1-I, SGI1-L, and SGI1-O (not to scale) are shown at the bottom. Base pair coordinates are from the sequence with accession no. AF261825. PCRs carried out to map the SGI1s are indicated by thick black bars and are numbered (Table 1). SGI1 was mapped as shown, and SGI1-I was mapped as for SGI1 and also with PCRs 18, 19, and 21. The blaPSE-1/aadA2 integron from P. mirabilis C04014 was negative in PCRs 9, 10, 11, and 16.
Thus, when present SGI1 is integrated via the specific attB site defined by the last 18 bp of the thdF gene in P. mirabilis as predicted (Fig. 1) (8).
The identification of SGI1 and its variants in P. mirabilis isolated from food and human clinical isolates from China highlights the increased dissemination of the SGI1 element both geographically and biologically. The documented potential for increased virulence in strains harboring this element in conjunction with the multidrug-resistant nature of the SGI1 element raises concerns especially since these new strains harboring SGI1 have been identified from food sources (4, 9, 16, 19).
Nucleotide sequence accession numbers.
The dfrA1-orfC cassette region from SGI1-O and the blaPSE-1-aadA2 cassette region from P. mirabilis C04014 have been assigned accession numbers EU006710 and EU006711, respectively, and the left and right junction regions of SGI1 in P. mirabilis B02012 have been assigned accession numbers EU180707 and EU180708, respectively, in the GenBank database.
Acknowledgments
We gratefully acknowledge the expert technical assistance of Dennis Le and Romeo Hizon. The sequence data for P. mirabilis were produced by the Proteus mirabilis Sequencing Group at the Sanger Institute and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/pm.
Footnotes
Published ahead of print on 19 November 2007.
REFERENCES
- 1.Ahmed, A. M., A. I. Hussein, and T. Shimamoto. 2007. Proteus mirabilis clinical isolate harbouring a new variant of Salmonella genomic island 1 containing the multiple antibiotic resistance region. J. Antimicrob. Chemother. 59:184-190. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, D. A., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D. A., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson, S. A., Z. P. McCuddin, and M. T. Wu. 2005. SlyA regulates the collagenase-mediated cytopathic phenotype in multiresistant Salmonella. Microb. Pathog. 38:181-187. [DOI] [PubMed] [Google Scholar]
- 5.Cloeckaert, A., K. Praud, B. Doublet, M. Demartin, and F.-X. Weill. 2006. Variant Salmonella genomic island 1-L antibiotic resistance cluster in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 50:3944-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloeckaert, A., K. Sidi Boumedine, G. Flaujac, H. Imberechts, I. D'Hooghe, and E. Chaslus-Dancla. 2000. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. enterica serovar Agona. Antimicrob. Agents Chemother. 44:1359-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doublet, B., D. A. Boyd, M. R. Mulvey, and A. Cloeckaert. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911-1924. [DOI] [PubMed] [Google Scholar]
- 8.Doublet, B., G. R. Golding, M. R. Mulvey, and A. Cloeckaert. 2007. Potential integration sites of the Salmonella genomic island 1 in Proteus mirabilis and other bacteria. J. Antimicrob. Chemother. 59:801-803. [DOI] [PubMed] [Google Scholar]
- 9.Golding, G. R., A. B. Olsen, B. Doublet, A. Cloeckaert, S. Christianson, M. R. Graham, and M. R. Mulvey. 2007. The effect of the Salmonella genomic island 1 on in vitro global gene expression in Salmonella enterica serovar Typhimurium LT2. Microbes Infect. 9:21-27. [DOI] [PubMed] [Google Scholar]
- 10.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levings, R. S., D. Lightfoot, S. R. Partridge, R. M. Hall, and S. P. Djordjevic. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levings, R. S., S. R. Partridge, S. P. Djordjevic, and R. M. Hall. 2007. SGI1-K, a variant of the SGI1 genomic island carrying a mercury resistance region, in Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 51:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulvey, M. R., D. A. Boyd, A. Cloeckaert, R. Ahmed, and L.-K. Ng. 2004. Emergence of multidrug resistant Salmonella Paratyphi B dT+, Canada. Emerg. Infect. Dis. 10:1307-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulvey, M. R., D. A. Boyd, A. B. Olson, B. Doublet, and A. Cloeckaert. 2006. The genetics of Salmonella genomic island 1. Microbes Infect. 8:1915-1922. [DOI] [PubMed] [Google Scholar]
- 15.Ng, L.-K., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen, M. A., S. A. Carlson, S. K. Franklin, Z. P. McCuddin, M. T. Wu, and V. K. Sharma. 2005. Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect. Immun. 73:4668-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vo, A. T. T., E. van Duijkeren, A. C. Fluit, and W. Gaastra. 2007. A novel Salmonella genomic island 1 and rare types in Salmonella Typhimurium isolates from horses in The Netherlands. J. Antimicrob. Chemother. 59:594-599. [DOI] [PubMed] [Google Scholar]
- 18.Vo, A. T. T., E. van Duijkeren, A. C. Fluit, and W. Gaastra. 2 April 2007. Antimicrobial resistance, class 1 integrons, and a novel variant of genomic island 1 in Salmonella isolates from Vietnam. Antimicrob. Agents Chemother. doi: 10.1128/AAC.01093-06. [DOI] [PubMed]
- 19.Wu, M. T., S. A. Carlson, and D. K. Meyerholz. 2002. Cytopathic effects observed upon expression of a repressed collagenase gene present in Salmonella and related pathogens: mimicry of a cytotoxin from multiple antibiotic-resistant Salmonella enterica serotype Typhimurium phagetype DT104. Microb. Pathog. 33:279-287. [DOI] [PubMed] [Google Scholar]