Abstract
The yeast Candida albicans possesses a gene family that encodes secreted aspartic proteases (Saps), which are important for the virulence of this human fungal pathogen. Inhibitors of the Saps could therefore be used as novel antimycotic agents for the treatment of C. albicans infections. In the present study, we established a bioassay which allows testing of the activity of potential protease inhibitors against specific Sap isoenzymes by their ability to inhibit protease-dependent growth of C. albicans. In a medium containing bovine serum albumin (BSA) as the sole source of nitrogen, C. albicans specifically expresses the Sap2p isoenzyme, which degrades the BSA and thereby enables the fungus to grow. As the other SAP genes are not significantly expressed under these conditions, mutants lacking SAP2 are unable to utilize BSA as a nitrogen source and cannot grow in such a medium. To investigate whether forced expression of SAP genes other than SAP2 would also allow growth on BSA, we constructed a set of strains expressing each of the 10 SAP genes from a tetracycline-inducible promoter in a sap2Δ mutant background. Expression of Sap1p, Sap2p, Sap3p, Sap4p, Sap5p, Sap6p, Sap8p, and a C-terminally truncated, secreted Sap9p restored the growth of the sap2Δ mutant with different efficiencies. This set of strains was then used to test the activities of various aspartic protease inhibitors against specific Sap isoenzymes by monitoring growth on BSA in the presence of the inhibitors. While pepstatin blocked the activity of all of the Saps tested, the human immunodeficiency virus protease inhibitors ritonavir and saquinavir inhibited growth of the strains expressing Sap1p to Sap3p and Sap1p, respectively, but not that of strains expressing other Saps. Therefore, the strain set can be used to test the activity of new protease inhibitors against individual C. albicans Sap isoenzymes by their ability to block the growth of the pathogen.
Candida albicans is a major human fungal pathogen which can cause superficial, as well as life-threatening systemic, mycoses in immunocompromised patients (35). Potent drugs for the treatment of C. albicans infections are available; however, problems with the toxicity of amphotericin B and the development of resistance to the other drugs have stimulated the search for new pharmaceuticals with different drug targets (8). An alternative approach to cure C. albicans infections could be the inhibition of specific pathogenicity-related factors of the fungal cells, which should decrease their virulence and help the remaining host defense mechanisms to successfully combat the pathogen (38). Secreted aspartic proteases (Saps) are known virulence factors of C. albicans, and many investigations performed in the last decades have revealed that these enzymes contribute to the pathogenicity of C. albicans in different ways. The Saps can provide nutrients by degrading host proteins but also support adherence to host surfaces and invasion of tissue barriers (12, 32, 46, 52). They are encoded by a family of 10 homologous genes which are differentially regulated during infection, indicating that the individual isoenzymes fulfill specific functions (33, 34, 43, 47). The hypothesis that C. albicans infections can be attenuated by inhibition of the Saps was supported to some extent in animal models by treatment with the aspartic protease inhibitor pepstatin. Whereas a protective role in mucosal and peritoneal infections was demonstrated (13, 27), results obtained in systemic-infection models were contradictory, a finding which was partly attributed to the inappropriate pharmacokinetics of this compound (16, 18, 42, 56). Nevertheless, the idea of using protease inhibitors in the treatment of candidiasis has received new attention in recent years. It was observed that highly active antiretroviral therapy, which includes human immunodeficiency virus (HIV) aspartic protease inhibitors, coincided with decreasing numbers of C. albicans infections in HIV and AIDS patients (10, 20, 21, 36, 55). A direct inhibitory effect of HIV protease inhibitors on C. albicans was supported by experimental in vitro and in vivo infection models. Using concentrations which are nontoxic for the fungal cells, some of the HIV protease inhibitors decreased C. albicans adherence and also attenuated mucosal infection (3, 7, 9, 26). However, the limited specificity of these inhibitors for the Saps and the finding that they act on only some of the different isoenzymes are expected to prevent their application against disseminated disease (7). Since different Sap isoenzymes contribute to the progression of C. albicans infections, new Sap inhibitors should block the action of as many of the Saps as possible in order to paralyze the fungus most efficiently. Analysis of the inhibitory effect of protease inhibitors on individual Saps requires the expression of these enzymes under in vitro conditions. Some of the Saps have been expressed as recombinant proteins in the heterologous hosts Escherichia coli (24), Saccharomyces cerevisiae (45), and Pichia pastoris (6), but most of the Saps cannot easily be expressed in the native host under laboratory conditions. It has long been known that C. albicans secretes protease during growth in a medium containing a protein, e.g., bovine serum albumin (BSA), as the sole source of nitrogen, and growth in such media can be blocked by the addition of pepstatin (41, 46). It was later found that of all of the members of the Sap family, only the Sap2p isoenzyme is significantly expressed under these conditions and inactivation of the SAP2 gene rendered the mutants unable to grow on BSA (22, 23, 48). Therefore, it seemed possible that forced expression of other members of the SAP gene family in a sap2Δ mutant background might bypass the requirement for SAP2 and enable the cells to grow under these conditions. This, in turn, would allow testing of the activity of protease inhibitors against specific C. albicans Sap isoenzymes by assessment of their ability to block the growth of strains expressing the corresponding SAP gene. In the present work, we generated a set of C. albicans reporter strains expressing individual SAP genes from a tetracycline-inducible promoter and demonstrated the feasibility of this approach.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans strains used in this study are listed in Table 1. All strains were stored as frozen stocks with 15% glycerol at −80°C. The strains were routinely grown in YPD medium (10 g yeast extract, 20 g peptone [BBL Trypticase Peptone; Becton Dickinson, Sparks, MD], and 20 g glucose per liter) at 30°C. For solid medium, 1.5% agar was added before autoclaving. To select nourseothricin-resistant (Nour) transformants, 200 μg ml−1 of nourseothricin (Werner Bioagents, Jena, Germany) was added to YPD agar. To obtain nourseothricin-sensitive (Nous) derivatives in which the SAT1 flipper was excised by FLP-mediated recombination, transformants were cultivated for 6 h in YPM medium (10 g yeast extract, 20 g peptone, and 20 g maltose per liter) without selective pressure to induce the MAL2 promoter. One hundred to 200 cells were spread on YPD plates containing 20 μg ml−1 nourseothricin and grown for 2 days at 30°C. Nous clones were identified by their small colony size and confirmed by restreaking on YPD plates containing 100 μg ml−1 nourseothricin as described previously (40).
TABLE 1.
C. albicans strains used in this study
| Strain(s) | Parent | Relevant genotype or characteristicsa | Reference |
|---|---|---|---|
| SC5314 | Wild type | 19 | |
| SAP2MS1A | SC5314 | sap2-1::SAT1-FLIP/SAP2-2 | This study |
| SAP2MS1B | SC5314 | SAP2-1/sap2-2::SAT1-FLIP | This study |
| SAP2MS2A | SAP2MS1A | sap2-1::FRT/SAP2-2 | This study |
| SAP2MS2B | SAP2MS1B | SAP2-1/sap2-2::FRT | This study |
| SAP2MS3A | SAP2MS2A | sap2-1::FRT/sap2-2::SAT1-FLIP | This study |
| SAP2MS3B | SAP2MS2B | sap2-1::SAT1-FLIP/sap2-2::FRT | This study |
| SAP2MS4A | SAP2MS3A | sap2-1::FRT/sap2-2::FRT | This study |
| SAP2MS4B | SAP2MS3B | sap2-1::FRT/sap2-2::FRT | This study |
| SAP2KS3A | SAP2MS4A | sap2-1::FRT/sap2-2::PSAP2-2-SAP2-2-TACT1-SAT1-FLIP | This study |
| SAP2KS3B | SAP2MS4B | sap2-1::FRT/sap2-2::PSAP2-2-SAP2-2-TACT1-SAT1-FLIP | This study |
| SAP2KS4A | SAP2KS3A | sap2-1::FRT/sap2-2::PSAP2-2-SAP2-2-TACT1-FRT | This study |
| SAP2KS4B | SAP2KS3B | sap2-1::FRT/sap2-2::PSAP2-2-SAP2-2-TACT1-FRT | This study |
| SAP1ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP1 | This study |
| SAP2ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP2-1 | This study |
| SAP3ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP3 | This study |
| SAP4ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP4 | This study |
| SAP5ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP5 | This study |
| SAP6ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP6-2 | This study |
| SAP7ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP7 | This study |
| SAP8ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP8 | This study |
| SAP9ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP9 | This study |
| SAP10ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP10 | This study |
| SAP9ΔC1ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP9ΔC520 | This study |
| SAP9ΔC2ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP9ΔC493 | This study |
| SAP10ΔC1ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP10ΔC427 | This study |
| SAP10ΔC2ex4A and -B | SAP2MS4B | ADH1/adh1::Ptet-SAP10ΔC387 | This study |
SAT1-FLIP denotes the SAT1 flipper cassette.
To test C. albicans transformants expressing the different SAP genes for growth on BSA as the sole source of nitrogen, strains were pregrown overnight in SD medium (6.7 g yeast nitrogen base without amino acids [YNB; Bio 101, Vista, CA] and 20 g glucose per liter) at 30°C and then diluted 1:100 in YCB-BSA medium (23.4 g yeast carbon base and 4 g BSA per liter). Depending on the expressed Sap, the pH of YCB-BSA medium was adjusted to 4.0 or 5.0 and the culture was incubated at 30°C or 37°C (pH 4.0 and 30°C for expression of SAP1, SAP2, SAP3, and SAP8 and pH 5.0 and 37°C for expression of SAP4, SAP5, SAP6, and SAP9). To induce the expression of SAP genes from the Tet promoter, 50 μg ml−1 doxycycline was added to cultures incubated at pH 4.0 and 30°C and 25 μg ml−1 was added to cultures incubated at pH 5.0 and 37°C because 50 μg ml−1 doxycycline had a slight adverse effect on growth of the cells under the latter conditions. The aspartic protease inhibitor pepstatin A (Sigma, Deisenhofen, Germany) was added at a concentration of 7.3 μM, and the HIV protease inhibitors ritonavir (Abbott, Chicago, IL) and saquinavir (Roche, Welwyn Garden City, United Kingdom) were used at a concentration of 100 μM (0.125% dimethyl sulfoxide end concentration).
Plasmid constructions.
To generate a SAP2 deletion construct, an ApaI-SalI fragment from plasmid pSFL213 containing 1.1 kb of SAP2 upstream sequences (47) was used to replace the OPT1 upstream region in ApaI/XhoI-digested plasmid pOPT1M3 (40), resulting in plasmid pSAP2MS1. A SAP2 downstream fragment was obtained by PCR with primers SAP2M and SAP2N (Table 2 contains the sequences of the primers used in this study), digested at the introduced SacII and SacI sites, and substituted for the OPT1 downstream region in pSAP2MS1, yielding plasmid pSAP2MS2, in which the SAT1 flipper cassette is flanked by SAP2 upstream and downstream sequences (Fig. 1A). For reinsertion of a functional SAP2 copy at its original locus in sap2Δ mutants, we first ligated a KpnI-SalI fragment from plasmid pMEP1M4 (4), which contained the transcription termination sequence of the ACT1 gene, together with an XhoI-PstI fragment from pSFS2 (40) containing part of the SAT1 flipper cassette into KpnI/PstI-digested pSAP2MS2 to generate pSAP2KS1. An ApaI-BamHI fragment containing the coding region and 0.3 kb of upstream sequences of the SAP2-1 allele was then amplified with primers SAP2P5 and SAP2ex2 and cloned into ApaI/BglII-digested pSAP2KS1 to result in pSAP2KS2. Finally, the complete coding region and 2 kb of upstream sequences of the SAP2-2 allele were PCR amplified with primers SAP2P1 and SAP2E by using genomic DNA from the heterozygous sap2-1Δ/SAP2-2 mutant SAP2MS2A as a template. The PCR product was digested with ApaI/XbaI and with XbaI/BglII, and the two fragments were ligated together into ApaI/BglII-digested pSAP2KS2 to result in pSAP2KS3 (Fig. 1B).
TABLE 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| SAP2M | 5′-CTGCCTTGACCGCGGATGAAGGGGTG-3′ |
| SAP2N | 5′-GGGCCCATGACGAGCTCTAGTTTGAGC-3′ |
| SAP2P1 | 5′-TTGTTGGGCCCGTTGTCAATTTATGGGCCGATCTG-3′ |
| SAP2P5 | 5′-ATATAGGGCCCGCATTTGAATAAACGGCATC-3′ |
| SAP2E | 5′-CCACCCCTTCATCTGCAGTCAAGGCAGAAATAC-3′ |
| SAP1ex1 | 5′-TCTAGTCGACAATGTTTTTAAAGAATATTTTCAT-3′ |
| SAP1ex2 | 5′-AGTAGGGATCCCTAGGTAAGAGCAGCAATGTTTGAAGC-3′ |
| SAP2ex1 | 5′-ACCAGTCGACAATGTTTTTAAAGAATATTTTCAT-3′ |
| SAP2ex2 | 5′-ACCCCGGATCCTTAGGTCAAGGCAGAAATACTGGAAGC-3′ |
| SAP3ex1 | 5′-ATATGTCGACAATGTTTTTAAAAAATATCTTTAT-3′ |
| SAP3ex2 | 5′-TGTACGGATCCCTAAGTAAGAGCAGCAATGTTAGAAGC-3′ |
| SAP4ex1 | 5′-CTCACTCGAGAATGTTCTTACAAAATATCTTGAG-3′ |
| SAP4ex2 | 5′-ACCAAGGATCCCTAATTAATAGCAACAATGTTAGACTG-3′ |
| SAP56ex1 | 5′-CTCAGTCGACAATGTTCTTGAAAAATATCTTGAG-3′ |
| SAP5ex2 | 5′-GTCAAGGATCCTTAATTAATAGCAACAATGTCAGACTCGG-3′ |
| SAP6ex2 | 5′-ACCAAGGATCCCTAATTAATAGCACCAATGTTAGACTC-3′ |
| SAP7ex1 | 5′-GAAGGTCGACAATGCAAAGAGTATTAGAGTTATT-3′ |
| SAP7ex2 | 5′-TTCTAGGATCCTTACTCGATAGGAACAACGGCATGGTT-3′ |
| SAP8ex1 | 5′-AACCGTCGACAATGGTCTCCATTATTACTTTTAC-3′ |
| SAP8ex2 | 5′-GAACAGGATCCCTATAAAGTAGAAATACTTGAAGAAGT-3′ |
| SAP9ex1 | 5′-TTTCGTCGACAATGAGACTCAATTCTGTTGCGTT-3′ |
| SAP9ex2 | 5′-TCTCTGGATCCTTAAACCAAAACATAGTAGGATATCAA-3′ |
| SAP9ex3 | 5′-TATAGGATCCTTATGAATGACGTGTGCTGGTAC-3′ |
| SAP9ex4 | 5′-TATAGGATCCTTAACCAATGACTTCAATCGATTCC-3′ |
| SAP10ex1 | 5′-TATAGTCGACAATGGACCTAGTAATAATGAATTT-3′ |
| SAP10ex2 | 5′-ATTCTGGATCCCTATATAATATGTATATACACGAGGAA-3′ |
| SAP10ex3 | 5′-TATAGGATCCTTAGCTAGTAGTGTTCTTGTCCATGTTC-3′ |
| SAP10ex4 | 5′-TATAGGATCCTTAGTTTAAAATCTCTTCAATATCTTCGTCTTC-3′ |
| SAP5CF | 5′-CGGGGATCCATGGTTAACTGTCGGTTTAAG-3′ |
| SAP5CR | 5′-CGCTCGAGATTAATAGCAACAATGT-3′ |
Restriction sites introduced into the primers are underlined; the start and stop (reverse sequence) codons of the SAP genes are in bold.
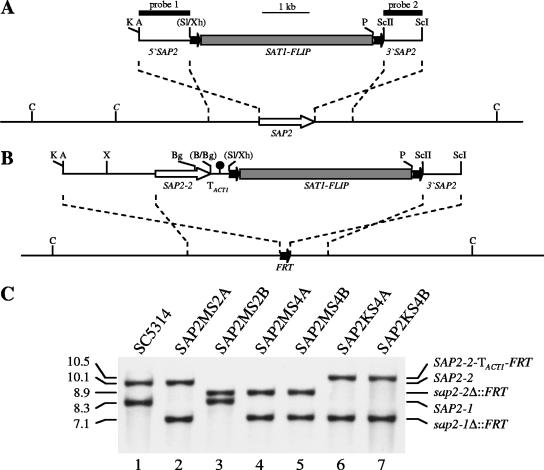
FIG. 1.
Construction of sap2Δ mutants and complemented strains. (A) Structure of the deletion cassette from plasmid pSAP2MS2 (top), which was used to delete both SAP2 alleles, and genomic structure of the SAP2 locus in strain SC5314 (bottom). The SAP2 coding region is represented by the white arrow, and the upstream and downstream regions are represented by the solid lines. Details of the SAT1 flipper cassette (gray rectangle bordered by FRT sites [black arrows]) have been presented elsewhere (40). The 34-bp FRT sites are not drawn to scale. The probes used for Southern hybridization analysis of the mutants are indicated by the black bars. (B) Structure of the DNA fragment from pSAP2KS3 (top), which was used for reinsertion of the SAP2-2 allele into its original site in the sap2Δ mutants (bottom). The transcription termination sequence of the ACT1 gene (TACT1) is indicated by the filled circle. Only the following relevant restriction sites are given in panels A and B: A, ApaI; B, BamHI; Bg, BglII; C, ClaI; K, KpnI; P, PstI; ScI, SacI; ScII, SacII; Sl, SalI; X, XbaI; Xh, XhoI. Sites in parentheses were destroyed by the cloning procedure. The ClaI site in italics is present only in the SAP2-1 allele. (C) Southern hybridization of ClaI-digested genomic DNA of parental strain SC5314 (lane 1), heterozygous sap2 mutants SAP2MS2A (lane 2) and SAP2MS2B (lane 3), homozygous sap2Δ mutants SAP2MS4A (lane 4) and SAP2MS4B (lane 5), and reconstituted strains SAP2KS4A (lane 6) and SAP2KS4B (lane 7) with SAP2-specific probe 1. The sizes of the hybridizing fragments (in kilobases) are on the left side of the blot, and their identities are on the right.
To express the individual SAP genes from a Tet-inducible promoter (Ptet) (37), the coding region of each of the SAP genes was amplified from genomic DNA of strain SC5314 by PCR with the following primer pairs: SAP1ex1 and SAP1ex2 for SAP1, SAP2ex1 and SAP2ex2 for SAP2 (allele 1), SAP3ex1 and SAP3ex2 for SAP3, SAP4ex1 and SAP4ex2 for SAP4, SAP56ex1 and SAP5ex2 for SAP5, SAP56ex1 and SAP6ex2 for SAP6 (allele 2), SAP7ex1 and SAP7ex2 for SAP7, SAP8ex1 and SAP8ex2 for SAP8, SAP9ex1 and SAP9ex2 for SAP9, and SAP10ex1 and SAP10ex2 for SAP10. Truncated SAP9 and SAP10 alleles were generated by the introduction of a stop codon within the coding region, in front of the putative cleavage sites for addition of the glycosylphosphatidylinositol (GPI) anchor. Two different truncations (ΔC1 and ΔC2) were produced for each of the SAP9 (SAP9ΔC520 and SAP9ΔC493) and SAP10 (SAP10ΔC427 and SAP10ΔC387) genes by PCR with the primer pairs SAP9ex1/SAP9ex3, SAP9ex1/SAP9ex4, SAP10ex1/SAP10ex3, and SAP10ex1/SAP10ex4, respectively. The PCR products were digested at the restriction sites introduced in front of the start codon and behind the stop codon and cloned between Ptet and TACT1 in a SalI/BglII-digested derivative of plasmid pNIM1 (37), resulting in plasmids pSAP1ex4, pSAP2ex4, pSAP3ex4, pSAP4ex4, pSAP5ex4, pSAP6ex4, pSAP7ex4, pSAP8ex4, pSAP9ex4, pSAP10ex4, pSAP9ΔC1ex4, pSAP9ΔC2ex4, pSAP10ΔC1ex4, and pSAP10ΔC2ex4.
C. albicans transformation.
C. albicans strains were transformed by electroporation (25) with the following gel-purified, linear DNA fragments: the ApaI-SacI fragment from pSAP2MS2 to inactivate the SAP2 gene in strain SC5314, the ApaI-SacI fragment from pSAP2KS3 to reinsert the SAP2-2 allele in the sap2Δ deletion mutants, and the ApaI-SacII fragments from plasmids pSAP1ex4, pSAP2ex4, pSAP3ex4, pSAP4ex4, pSAP5ex4, pSAP6ex4, pSAP7ex4, pSAP8ex4, pSAP9ex4, pSAP10ex4, pSAP9ΔC1ex4, pSAP9ΔC2ex4, pSAP10ΔC1ex4, and pSAP10ΔC2ex4 to integrate the individual Ptet-SAP fusions into the ADH1 locus of the sap2Δ mutant SAP2MS4B. Single-copy integration of all constructs was confirmed by Southern hybridization.
Isolation of genomic DNA and Southern hybridization.
Genomic DNA from C. albicans strains was isolated as described previously (28). DNA (15 μg) was digested with appropriate restriction enzymes, separated in 1% (wt/vol) agarose gels and, after ethidium bromide staining, transferred by vacuum blotting onto nylon membranes and fixed by UV cross-linking. Southern hybridization with enhanced-chemiluminescence (ECL)-labeled probes was performed with the Amersham ECL Direct Nucleic Acid Labeling and Detection System (GE Healthcare, Braunschweig, Germany) according to the instructions of the manufacturer.
Generation of antibodies for Western blot experiments.
A fusion protein consisting of glutathione S-transferase (GST) and the C-terminal 148 amino acids of Sap5p (GST-Sap5F) was prepared by using the GST gene fusion system from Amersham. A SAP5 DNA fragment that encodes the C-terminal part of the Sap5 protein from Leu271 to Asn418 was obtained by PCR with primers SAP5CF and SAP5CR. Plasmid pCA5 (31) was used as the DNA template for the PCR. The amplified fragment was cloned via the introduced BamHI and XhoI sites into BamHI/XhoI-digested vector pGEX-4T (Pharmacia, Uppsala, Sweden) to obtain an in-frame fusion of GST and the C-terminal part of Sap5p. The fusion protein was produced and purified by affinity chromatography according to the recommendations of the manufacturer. Rabbit antisera were made by Eurogentec (Liège, Belgium). Antiserum to Sap2p was produced by using a 15-amino-acid peptide (H2N-DPSGSSASQDLNTPF-CONH2) as an antigen. Antiserum to Sap5p was produced by using GST-Sap5F as an antigen.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot experiments.
Fifteen microliters of the culture supernatant of C. albicans strains grown in YCB-BSA was analyzed on an SDS-12% polyacrylamide gel. Protein bands were visualized by staining with colloidal Coomassie dye. The molecular weights of the proteins were determined by use of the Precision Plus protein standards all blue size marker (Bio-Rad, Munich, Germany). Proteins were transferred onto nitrocellulose membranes with a Semi-Dry-Trans-Blot SD blot apparatus (Bio-Rad). Sap bands were detected with the antibodies raised against Sap2p and Sap5p; as a second antibody, an anti-rabbit antibody was used. Signals were detected with ECL detection solutions on ECL films (Amersham, Braunschweig, Germany).
RESULTS
Construction of sap2Δ mutants of C. albicans wild-type strain SC5314.
As we wanted to use wild-type C. albicans model strain SC5314 for our experiments, we first created a sap2Δ mutant of this strain by using the SAT1-flipping strategy (40) for sequential deletion of both SAP2 alleles. Strain SC5314 was transformed with a DNA fragment in which the SAT1 flipper cassette was flanked by upstream and downstream sequences of the SAP2 gene (Fig. 1A). Two different heterozygous mutants in which the deletion cassette had been inserted into the SAP2-1 or the SAP2-2 allele, followed by FLP-mediated excision of the cassette (strains SAP2MS2A and SAP2MS2B, Fig. 1C, lanes 2 and 3), were selected. These strains were transformed again with the same deletion cassette to inactivate the remaining wild-type SAP2 allele, generating homozygous sap2Δ mutants SAP2MS4A and SAP2MS4B (Fig. 1C, lanes 4 and 5). Reinsertion of the SAP2-2 allele at its original locus in both independently generated sap2Δ mutants with the help of the SAT1 flipper cassette (Fig. 1B), which was subsequently excised again, resulted in complemented strains SAP2KS4A and SAP2KS4B (Fig. 1C, lanes 6 and 7).
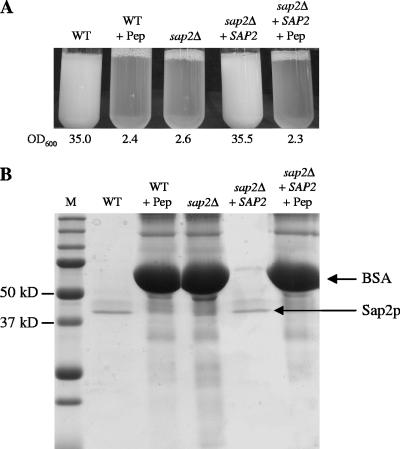
The heterozygous and homozygous sap2Δ mutants were tested for the ability to grow in YCB-BSA medium. In contrast to wild-type strain SC5314, homozygous sap2Δ mutants SAP2MS4A and SAP2MS4B were unable to grow under these conditions, as expected from previous work (23, 48). After 48 h of growth, the optical densities of the corresponding cultures were similar to that of the wild type incubated in the presence of the protease inhibitor pepstatin (Fig. 2A). We had previously demonstrated that the two SAP2 alleles in strain CAI4, a derivative of strain SC5314, are differentially regulated and that inactivation of the SAP2-2 allele abolished growth in YCB-BSA, while inactivation of the SAP2-1 allele had no effect (48). The same result was obtained in the present study with the heterozygous sap2 mutants of wild-type strain SC5314. Strain SAP2MS2A, in which the SAP2-1 allele was deleted, grew like the wild type in YCB-BSA medium, while strain SAP2MS2B, which lacked the SAP2-2 allele, was unable to grow under these conditions, like the homozygous sap2Δ mutants (data not shown). Reinsertion of the SAP2-2 allele at its native locus in homozygous sap2Δ mutants SAP2MS4A and SAP2MS4B restored the growth of resulting strains SAP2KS4A and SAP2KS4B to wild-type levels (Fig. 2A). Accordingly, the BSA in the culture medium was almost completely degraded during growth and the Sap2p band could be detected in the culture supernatants of the wild-type and complemented strains (Fig. 2B).
FIG. 2.
The protease Sap2p is required for growth of C. albicans strain SC5314 in a medium with BSA as the sole source of nitrogen. (A) Growth of wild-type strain SC5314 (WT), sap2Δ mutant SAP2MS4B (sap2Δ), and complemented strain SAP2KS4B (sap2Δ + SAP2) in YCB-BSA. Overnight cultures of each strain in SD medium were diluted 1:100 in YCB-BSA, incubated for 48 h at 30°C, and photographed. Strain SC5314 and complemented strain SAP2KS4B were also incubated in the presence the protease inhibitor pepstatin (+ Pep). The optical densities at 600 nm (OD600) of the cultures are given below each tube. The two independently constructed sap2Δ mutants and complemented strains behaved identically, and only one is shown. (B) Expression of Sap2p in wild-type strain SC5314, the sap2Δ mutants, and the complemented strains. Supernatants of the cultures shown in panel A were analyzed by SDS-PAGE. The bands corresponding to BSA and Sap2p are indicated. Lane M, molecular size markers.
Tetracycline-inducible expression of individual SAP genes in a sap2Δ mutant.
The inability of C. albicans sap2Δ mutants to grow in YCB-BSA medium allowed us to test whether inducible expression of individual members of the SAP gene family would restore the growth of the mutants under these conditions. Therefore, we cloned the coding regions of the SAP1 to SAP10 genes and placed them under the control of a tetracycline-inducible promoter (37). The resulting expression cassettes (Fig. 3) were used to transform the sap2Δ mutant SAP2MS4B, and correct integration into the ADH1 locus was confirmed by Southern hybridization with ADH1 upstream and downstream probes, as well as SAP-specific probes (data not shown). In each case, two independent transformants were kept for further analysis (Table 1).
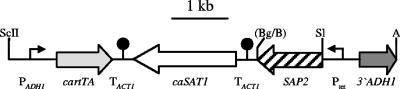
FIG. 3.
Structure of the DNA cassettes used to integrate the Tet-inducible SAP genes into the ADH1 locus of C. albicans strain SAP2MS4B (in this example shown for SAP2 expression). Bent arrows symbolize promoters (P); filled circles represent TACT1, which serves for proper transcription termination of the Candida-adapted, reverse tetracycline-dependent transactivator (cartTA) and the target genes in this cassette (37). Only the following relevant restriction sites used to construct the plasmids or to excise the whole cassette from the vector backbone are shown: A, ApaI; B, BamHI; Bg, BglII; S, SalI; ScII, SacII.
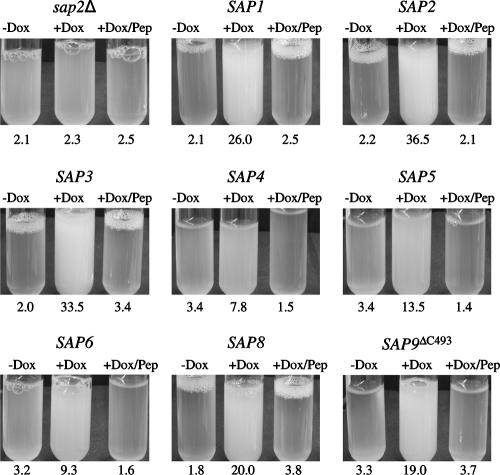
In order to test whether expression of individual SAP genes would result in BSA degradation and growth of the sap2Δ mutants, precultures of strains SAP1ex4A/B to SAP10ex4A/B grown overnight in liquid SD medium were diluted in YCB-BSA medium with and without doxycycline and incubated under different conditions, as the parameters that allow optimal proteolytic activity vary for the different Saps (33). Doxycycline-induced expression of most of the SAP genes allowed the growth of the sap2Δ mutants. Dense growth of transformants expressing SAP1, SAP2, SAP3, and SAP8 was reached after 48 h at pH 4.0 and 30°C, whereas growth of strains expressing SAP4, SAP5, and SAP6 was strongest after 90 to 120 h at pH 5.0 and 37°C (Fig. 4), although the latter, especially the strains expressing SAP4, grew more slowly in YCB-BSA and the corresponding cultures did not reach as high densities as those expressing SAP1, SAP2, SAP3, or SAP8. Therefore, the various Sap isoenzymes differ in the ability to mediate growth on BSA as a nitrogen source. After prolonged incubation at pH 5.0 and 37°C, a slight increase in the optical density of the sap2Δ mutant (and transformants grown in the absence of doxycycline) was also observed. However, this growth was significantly weaker than the growth of the cells in which expression of the analyzed SAP genes was induced by doxycycline.
FIG. 4.
Tet-inducible expression of SAP1, SAP2, SAP3, SAP4, SAP5, SAP6, SAP8, and SAP9ΔC493 allows growth of a sap2Δ mutant in YCB-BSA. Overnight cultures of strains SAP1ex4A, SAP2ex4A, SAP3ex4A, SAP4ex4A, SAP5ex4A, SAP6ex4A, SAP8ex4A, and SAP9ΔC2ex4A were diluted 1:100 in YCB-BSA without (−Dox) or with (+Dox) doxycycline. In addition, the strains were also incubated in the presence of both doxycycline and pepstatin (+Dox/Pep). Untransformed sap2Δ mutant SAP2MS4B served as a control. The tubes were photographed, and the optical densities at 600 nm of the cultures were measured after 48 h (for SAP2MS4B and strains expressing SAP1, SAP2, SAP3, and SAP8) or 96 h (for strains expressing SAP4, SAP5, SAP6, and SAP9ΔC493). The OD600 values are given below each tube. The two independently constructed transformants for Tet-inducible expression of the individual SAP genes behaved identically, and only one of them is shown in each case.
The transformants expressing SAP7, SAP9, or SAP10 did not grow in YCB-BSA medium under any of the conditions tested (data not shown). The inability of the strains expressing SAP9 and SAP10 from the Tet-inducible promoter to grow in YCB-BSA might be due to the fact that Sap9p and Sap10p are located in the cell membrane and/or the cell wall and, unlike the other Sap isoenzymes, are not released into the culture medium (1). Therefore, we expressed truncated SAP9 and SAP10 genes lacking the sequences required for GPI anchor addition. Because different putative GPI anchorage sites have been proposed for Sap9p and Sap10p (1, 14, 17, 30), two different C-terminally truncated alleles were generated for each of these two SAP genes (see Materials and Methods and Table 1). Strains expressing SAP9 alleles lacking the region that encodes the last 24 (SAP9ΔC520) or 51 (SAP9ΔC493) amino acids were able to grow in YCB-BSA medium at a pH of 5.0 and 37°C, indicating that Sap9p, when secreted into the culture medium, could mediate the degradation of BSA for use as a nitrogen source (Fig. 4 and data not shown). In contrast, doxycycline-induced expression of neither of the truncated SAP10 alleles restored the growth of the sap2Δ mutant (data not shown).
The culture supernatants of strains expressing SAP1, SAP2, SAP3, SAP4, SAP5, SAP6, SAP8, and SAP9ΔC493 were analyzed by SDS-PAGE. In all cases, the BSA was almost completely degraded, and specific protein bands in the size range of approximately 35 kDa to 45 kDa could be detected for Sap1p, Sap2p, Sap3p, Sap4p, Sap5p, Sap6p, and Sap8p (Fig. 5A). In contrast, several bands of different sizes were observed in the culture supernatant of the strain expressing SAP9ΔC493. Sap9p has been reported to be cleaved into two subfragments when heterologously expressed and secreted in P. pastoris (1, 11), but we were unable to decide which of the proteins present in our culture supernatants corresponded to mature Sap9p. To confirm the identities of the different Sap isoenzymes in the culture supernatants, we performed Western immunoblotting experiments with antibodies raised against Sap2p and Sap5p. The anti-Sap2p antibody specifically recognized only Sap2p (Fig. 5B), whereas the anti-Sap5p antibody showed cross-reactivity with all of the Sap1p-Sap6p proteins (Fig. 5C). Neither of the two antibodies reacted with the proteins secreted by the strains expressing SAP8 or SAP9ΔC493. The proteins recognized by the antibodies in the Western blot experiments exhibited the same sizes as the protease bands in the corresponding Coomassie-stained gel (compare Fig. 5A and C), confirming that no other Sap isoenzyme than the one expressed from the Tet promoter was secreted at detectable levels by these strains.
FIG. 5.
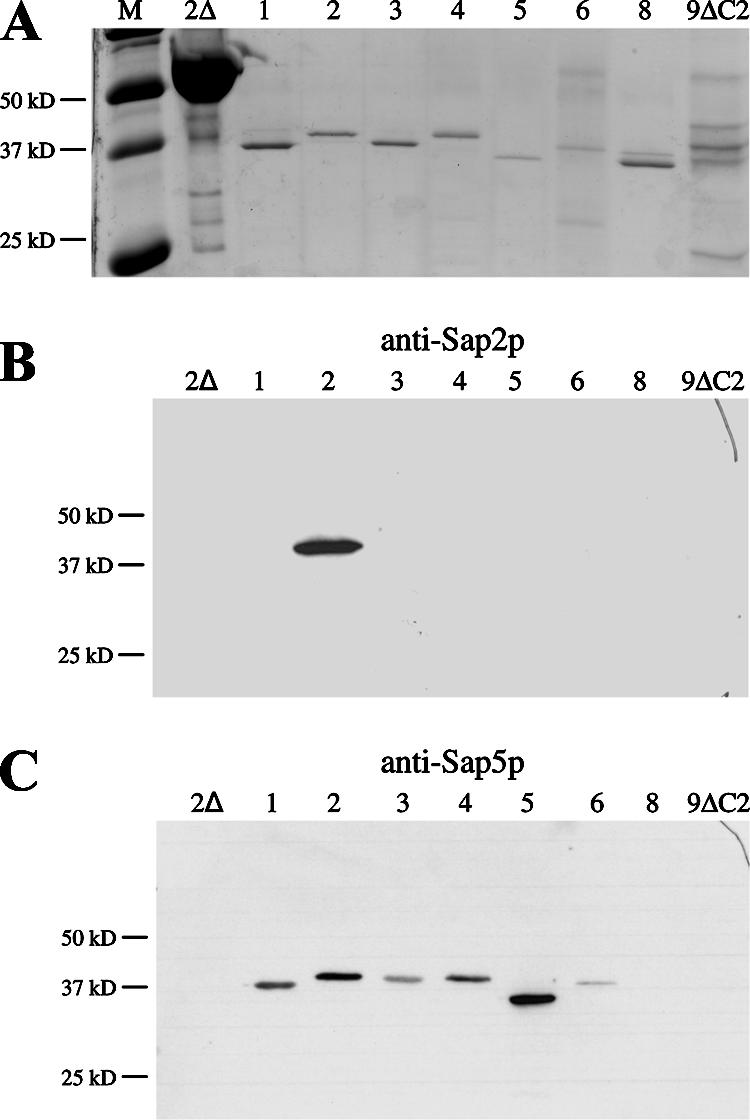
Detection of individual Saps in the culture supernatants of SAP-expressing transformants. sap2Δ mutant SAP2MS4B and strains expressing specific SAP genes, as indicated by the numbers above the lanes, were cultivated in YCB-BSA medium with doxycycline (for details, see Materials and Methods). (A) Culture supernatants of the strains were analyzed on an SDS-polyacrylamide gel stained with colloidal Coomassie dye. Lane M, molecular size markers. (B and C) Detection of the Saps in the supernatants of the analyzed strains by Western immunoblotting with antibodies raised against Sap2p (B) and Sap5p (C).
In summary, doxycycline-inducible expression of 8 of the 10 members of the SAP gene family enabled C. albicans sap2Δ mutants to grow in a medium containing BSA as the sole nitrogen source. Therefore, this phenotype could be used in a simple bioassay to test the activities of various protease inhibitors against individual Sap isoenzymes.
Inhibition of Sap-dependent growth by different protease inhibitors.
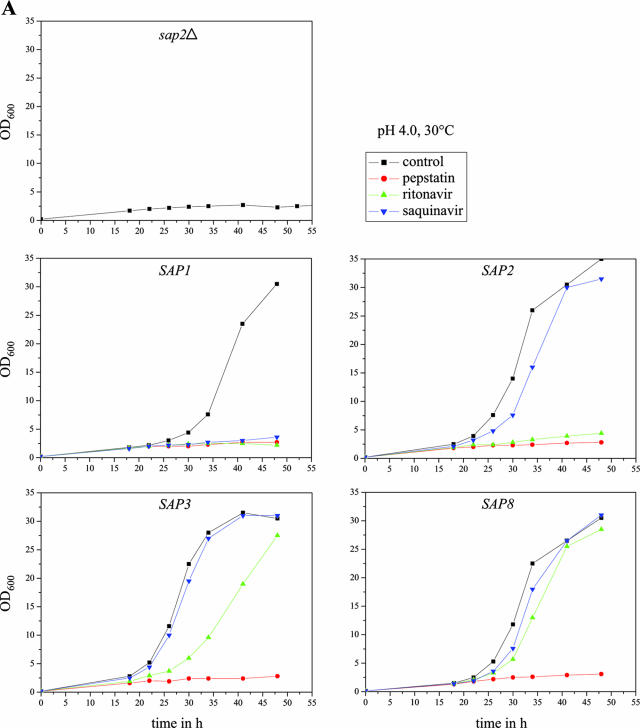
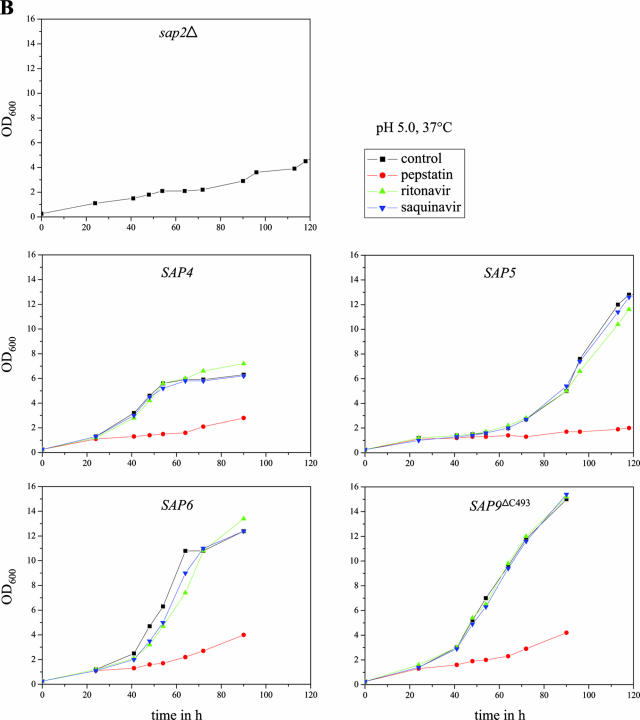
We then tested the abilities of various aspartic protease inhibitors to block the growth of strains expressing specific Sap isoenzymes under control of the Tet promoter in YCB-BSA medium. The strains were incubated under conditions allowing optimal activity of the corresponding Sap isoenzyme, and growth was monitored by measuring the optical densities of the cultures over time in the absence or presence of the inhibitors. All experiments were performed in duplicate with two independent transformants, which yielded essentially identical results. As demonstrated in Fig. 6A and B, the aspartic protease inhibitor pepstatin was active against all of the eight Saps under the conditions tested; i.e., none of the SAP-expressing strains was able to grow on BSA in the presence of this inhibitor. In contrast, the HIV protease inhibitors ritonavir and saquinavir displayed a differential activity against the individual Saps at the tested concentration of 100 μM. Ritonavir blocked the growth of the strains expressing SAP1 and SAP2 and slightly delayed the growth of the strains expressing SAP3. In contrast, saquinavir only blocked the growth of the strains expressing SAP1. Neither of the two HIV protease inhibitors was able to inhibit the growth of the strains expressing SAP4, SAP5, SAP6, SAP8, or SAP9ΔC493 under the conditions tested, in agreement with previously reported activities of the same protease inhibitors on recombinantly expressed, purified Sap isoenzymes (7, 29), demonstrating that protease-dependent growth of strains expressing specific SAP genes can be used as a simple assay to test the activities of novel protease inhibitors against almost all of the members of the secreted aspartic protease family of C. albicans.
FIG. 6.
Inhibition of Sap-dependent growth of C. albicans by different protease inhibitors. Overnight cultures of sap2Δ mutant SAP2MS4B and strains SAP1ex4A, SAP2ex4A, SAP3ex4A, SAP4ex4A, SAP5ex4A, SAP6ex4A, SAP8ex4A, and SAP9ΔC2ex4A were diluted 1:100 in YCB-BSA medium with doxycycline in the absence (control, containing dimethyl sulfoxide only) or presence of the protease inhibitor pepstatin, ritonavir, or saquinavir. The optical densities at 600 nm (OD600) of the cultures were measured at the indicated times. (A) Cultures incubated in YCB-BSA, pH 4.0, at 30°C. (B) Cultures incubated in YCB-BSA, pH 5.0, at 37°C. The two independently constructed transformants for Tet-inducible expression of the individual SAP genes yielded essentially identical results, and for better clarity, only one of them is shown in each case.
DISCUSSION
Since proteolytic activity was discovered in C. albicans, the secreted aspartic proteases have been intensively studied virulence attributes of this human-pathogenic yeast. Because of their importance for the virulence of C. albicans, the Saps have also been considered as potential drug targets for a long time (49). As C. albicans possesses 10 SAP genes, which are differentially expressed during infection and have different roles in colonization, invasion, and proliferation of the fungus in host tissues, potent inhibitors should block the activities of as many of the Sap isoenzymes as possible. In this work, we established a bioassay which allows the screening of potential protease inhibitors for activity against 8 of the 10 Saps by their ability to inhibit protease-dependent growth of C. albicans. In contrast to approaches that are based on the expression of Saps in a heterologous host and in vitro testing of inhibitors against the purified proteases, this assay enables for the first time inhibitor testing against Saps other than Sap2p in their native host. The assay relies on the fact that C. albicans can grow in YCB-BSA medium by secreting Sap2p, which is the only Sap isoenzyme expressed by most C. albicans strains under these conditions (22, 53). The expression of SAP genes under the control of an inducible promoter in a sap2Δ mutant therefore rendered the growth of the so-generated strains dependent on the activity of the corresponding Sap isoenzyme. As a first step, we had to evaluate whether the expression of the various SAP genes from the Tet promoter would indeed allow the growth of sap2Δ mutants in YCB-BSA. Expression of all of the SAP genes except SAP7 and SAP10 resulted in BSA degradation and growth of the strains, albeit with various efficiencies even after optimization of the culture conditions to allow maximum growth of the different strains. Under our standard conditions for culturing C. albicans in YCB-BSA (30°C, pH 4.0), expression of SAP1, SAP2, SAP3, and SAP8 resulted in dense growth of the strains. In contrast, significant growth of strains expressing SAP4, SAP5, SAP6, or SAP9ΔC493 was only observed at an elevated temperature (37°C) and pH (5.0). After prolonged incubation at 37°C, the sap2Δ mutant also exhibited some residual growth, which may have been caused by some self-degradation of BSA over time under these conditions. However, the growth of the strains expressing the various SAP genes from the Tet promoter was significantly higher in all cases, and without doxycycline induction the strains behaved like the sap2Δ mutant, confirming that growth was indeed mediated by the protease expressed from the Tet promoter.
SAP9 could mediate growth in YCB-BSA only after removal of the GPI-anchoring sequence, supporting the view that release of the secreted proteases from the cells is necessary for efficient extracellular proteolysis. When Sap9p was expressed in the heterologous host P. pastoris, secretion also occurred only after deletion of the GPI anchor sequence (1). However, recombinantly expressed Sap9p was not able to hydrolyze serum albumin in that study. Cell surface-anchored Sap9p and Sap10p, which display sequence homology to the S. cerevisiae yapsins, have functions other than the degradation of extracellular proteins and are involved in cell surface integrity, cell separation, and adherence of the fungal cells to host tissue (1, 30). Nevertheless, our results indicate that forced secretion of Sap9p can also mediate the growth of C. albicans on BSA and that this phenotype can be used to search for Sap9p inhibitors.
Unlike the other SAP genes, Tet-induced expression of SAP7 and SAP10 did not allow growth of the sap2Δ mutant. These two genes, together with SAP9, are the most divergent genes within the SAP gene family. It is possible that BSA is not a good substrate for Sap7p and Sap10p or that the enzymes are not active or not functionally expressed under the growth conditions used in our experiments.
Most of the studies on C. albicans Sap inhibition have been carried out with the aspartic protease inhibitor pepstatin (51). Its high potential to block Sap activity, which was demonstrated before for heterologously expressed enzymes Sap1p to Sap6p (7), was confirmed in our experiments, as none of the SAP-expressing strains could grow in BSA medium in the presence of pepstatin. In contrast, the HIV protease inhibitors ritonavir and saquinavir were active only against some of the Saps. The potential of ritonavir, but not saquinavir, to inhibit the growth of a C. albicans wild-type strain in YCB-BSA has been demonstrated before (5). As C. albicans usually expresses only SAP2 under these conditions, the growth inhibition presumably reflected the ability of ritonavir to inhibit Sap2p. Using our set of test strains, we could demonstrate that ritonavir also inhibits Sap1p-dependent and (partially) Sap3p-dependent growth and that saquinavir could inhibit Sap1p-dependent, but not Sap2p- or Sap3p-dependent, growth of C. albicans. These findings are consistent with recent observations that ritonavir had a stronger inhibitory effect on these three Saps than did saquinavir (2). Until now, HIV protease inhibitors have only once been tested against Sap4p-Sap6p expressed in P. pastoris (7). The data of this former study were confirmed by our results. Ritonavir and saquinavir had no detectable inhibitory effect on Sap4p to Sap6p. In addition to Sap1p to Sap6p, we could now also test protease inhibitors against Sap8p and Sap9p. Neither ritonavir nor saquinavir could inhibit the growth of C. albicans mediated by these enzymes, at least at the concentration tested.
The Tet-inducible expression of individual Sap enzymes in their native host, C. albicans, not only offers the possibility to test the abilities of potential Sap inhibitors to block the growth of C. albicans but also allows the purification of Saps from culture supernatant for further detailed characterization. Expression of Saps in their native host should circumvent possible caveats related to their expression in a heterologous host, like missing or inappropriate posttranslational modifications. Except for Sap9p, the individual Saps in the culture supernatants of the Sap-expressing strains were easily detectable in Coomassie-stained gels. Their presence and size were also confirmed in Western blotting experiments using Sap-specific antibodies. As demonstrated before, antibodies raised against protease Sap4p, Sap5p, or Sap6p displayed cross-reactivity with other Saps (6). However, Sap8p and Sap9p were not recognized by the antibodies we used.
Until now, none of the few Sap inhibitors tested was able to reproducibly inhibit or cure an experimental systemic infection (18, 49). Efforts have thus been made to identify new Sap inhibitors by different approaches; however, promising candidates for a therapeutic application have not been identified so far. As Sap2p is the only isoenzyme that can be easily expressed in C. albicans under laboratory conditions in large amounts, the applied screening assays were mostly based on this isoenzyme (15, 39, 44, 50, 54) or on Sap1p to Sap3p (2). However, because deep C. albicans mycoses are associated with the expression of many different Sap isoenzymes, including Sap4p to Sap6p, new inhibitors are needed which act against as many of the Saps as possible and at the same time are applicable in vivo. The set of reporter strains created in the present work should be highly useful for the identification and analysis of newly synthesized protease inhibitors. Compounds which specifically act on the C. albicans Saps would be promising candidates in the treatment of C. albicans infections in the future.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (DFG grants MO 846/1 and SFB630) and the Austrian Science Fund (FWF P17043-B13). Peter Staib is the recipient of a fellowship from the Deutsche Akademie der Naturforscher Leopoldina (Förderkennzeichen BMBF-LPD 9901/8-146).
Footnotes
Published ahead of print on 22 October 2007.
REFERENCES
- 1.Albrecht, A., A. Felk, I. Pichova, J. R. Naglik, M. Schaller, P. de Groot, D. Maccallum, F. C. Odds, W. Schafer, F. Klis, M. Monod, and B. Hube. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J. Biol. Chem. 281:688-694. [DOI] [PubMed] [Google Scholar]
- 2.Backman, D., M. Monod, and U. H. Danielson. 2006. Biosensor-based screening and characterization of HIV-1 inhibitor interactions with Sap 1, Sap 2, and Sap 3 from Candida albicans. J. Biomol. Screen. 11:165-175. [DOI] [PubMed] [Google Scholar]
- 3.Bektić, J., C. P. Lell, A. Fuchs, H. Stoiber, C. Speth, C. Lass-Florl, M. Borg-von Zepelin, M. P. Dierich, and R. Wurzner. 2001. HIV protease inhibitors attenuate adherence of Candida albicans to epithelial cells in vitro. FEMS Immunol. Med. Microbiol. 31:65-71. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, K., and J. Morschhäuser. 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56:649-669. [DOI] [PubMed] [Google Scholar]
- 5.Blanco, M. T., C. Hurtado, C. Perez-Giraldo, F. J. Moran, C. Gonzalez-Velasco, and A. C. Gomez-Garcia. 2003. Effect of ritonavir and saquinavir on Candida albicans growth rate and in vitro activity of aspartyl proteinases. Med. Mycol. 41:167-170. [DOI] [PubMed] [Google Scholar]
- 6.Borg-von Zepelin, M., S. Beggah, K. Boggian, D. Sanglard, and M. Monod. 1998. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol. Microbiol. 28:543-554. [DOI] [PubMed] [Google Scholar]
- 7.Borg-von Zepelin, M., I. Meyer, R. Thomssen, R. Wurzner, D. Sanglard, A. Telenti, and M. Monod. 1999. HIV-protease inhibitors reduce cell adherence of Candida albicans strains by inhibition of yeast secreted aspartic proteases. J. Investig. Dermatol. 113:747-751. [DOI] [PubMed] [Google Scholar]
- 8.Boucher, H. W., A. H. Groll, C. C. Chiou, and T. J. Walsh. 2004. Newer systemic antifungal agents: pharmacokinetics, safety and efficacy. Drugs 64:1997-2020. [DOI] [PubMed] [Google Scholar]
- 9.Cassone, A., F. De Bernardis, A. Torosantucci, E. Tacconelli, M. Tumbarello, and R. Cauda. 1999. In vitro and in vivo anticandidal activity of human immunodeficiency virus protease inhibitors. J. Infect. Dis. 180:448-453. [DOI] [PubMed] [Google Scholar]
- 10.Cauda, R., E. Tacconelli, M. Tumbarello, G. Morace, F. De Bernardis, A. Torosantucci, and A. Cassone. 1999. Role of protease inhibitors in preventing recurrent oral candidosis in patients with HIV infection: a prospective case-control study. J. Acquir. Immune Defic. Syndr. 21:20-25. [DOI] [PubMed] [Google Scholar]
- 11.Cawley, N. X., M. Chino, A. Maldonado, Y. M. Rodriguez, Y. P. Loh, and J. A. Ellman. 2003. Synthesis and characterization of the first potent inhibitor of yapsin 1. Implications for the study of yapsin-like enzymes. J. Biol. Chem. 278:5523-5530. [DOI] [PubMed] [Google Scholar]
- 12.Colina, A. R., F. Aumont, N. Deslauriers, P. Belhumeur, and L. de Repentigny. 1996. Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infect. Immun. 64:4514-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Bernardis, F., M. Boccanera, D. Adriani, E. Spreghini, G. Santoni, and A. Cassone. 1997. Protective role of antimannan and anti-aspartyl proteinase antibodies in an experimental model of Candida albicans vaginitis in rats. Infect. Immun. 65:3399-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Groot, P. W., K. J. Hellingwerf, and F. M. Klis. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20:781-796. [DOI] [PubMed] [Google Scholar]
- 15.Dostál, J., P. Hamal, L. Pavlickova, M. Soucek, T. Ruml, I. Pichova, and O. Hruskova-Heidingsfeldova. 2003. Simple method for screening Candida species isolates for the presence of secreted proteinases: a tool for the prediction of successful inhibitory treatment. J. Clin. Microbiol. 41:712-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edison, A. M., and M. Manning-Zweerink. 1988. Comparison of the extracellular proteinase activity produced by a low-virulence mutant of Candida albicans and its wild-type parent. Infect. Immun. 56:1388-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenhaber, B., G. Schneider, M. Wildpaner, and F. Eisenhaber. 2004. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 337:243-253. [DOI] [PubMed] [Google Scholar]
- 18.Fallon, K., K. Bausch, J. Noonan, E. Huguenel, and P. Tamburini. 1997. Role of aspartic proteases in disseminated Candida albicans infection in mice. Infect. Immun. 65:551-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 20.Gruber, A., C. Speth, E. Lukasser-Vogl, R. Zangerle, M. Borg-von Zepelin, M. P. Dierich, and R. Wurzner. 1999. Human immunodeficiency virus type 1 protease inhibitor attenuates Candida albicans virulence properties in vitro. Immunopharmacology 41:227-234. [DOI] [PubMed] [Google Scholar]
- 21.Hoegl, L., E. Thoma-Greber, M. Rocken, and H. C. Korting. 1998. HIV protease inhibitors influence the prevalence of oral candidosis in HIV-infected patients: a 2-year study. Mycoses 41:321-325. [DOI] [PubMed] [Google Scholar]
- 22.Hube, B., M. Monod, D. A. Schofield, A. J. Brown, and N. A. Gow. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 23.Hube, B., D. Sanglard, F. C. Odds, D. Hess, M. Monod, W. Schafer, A. J. Brown, and N. A. Gow. 1997. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect. Immun. 65:3529-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koelsch, G., J. Tang, J. A. Loy, M. Monod, K. Jackson, S. I. Foundling, and X. Lin. 2000. Enzymic characteristics of secreted aspartic proteases of Candida albicans. Biochim. Biophys. Acta 1480:117-131. [DOI] [PubMed] [Google Scholar]
- 25.Köhler, G. A., T. C. White, and N. Agabian. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korting, H. C., M. Schaller, G. Eder, G. Hamm, U. Bohmer, and B. Hube. 1999. Effects of the human immunodeficiency virus (HIV) proteinase inhibitors saquinavir and indinavir on in vitro activities of secreted aspartyl proteinases of Candida albicans isolates from HIV-infected patients. Antimicrob. Agents Chemother. 43:2038-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kretschmar, M., B. Hube, T. Bertsch, D. Sanglard, R. Merker, M. Schroder, H. Hof, and T. Nichterlein. 1999. Germ tubes and proteinase activity contribute to virulence of Candida albicans in murine peritonitis. Infect. Immun. 67:6637-6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millon, L., A. Manteaux, G. Reboux, C. Drobacheff, M. Monod, T. Barale, and Y. Michel-Briand. 1994. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J. Clin. Microbiol. 32:1115-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monod, M., M. Borg-von Zepelin, A. Telenti, and D. Sanglard. 1999. The inhibition of Candida-albicans-secreted aspartic proteases by three different HIV protease inhibitors. Dermatology 198:412-414. [PubMed] [Google Scholar]
- 30.Monod, M., B. Hube, D. Hess, and D. Sanglard. 1998. Differential regulation of SAP8 and SAP9, which encode two new members of the secreted aspartic proteinase family in Candida albicans. Microbiology 144(Pt. 10):2731-2737. [DOI] [PubMed] [Google Scholar]
- 31.Monod, M., G. Togni, B. Hube, and D. Sanglard. 1994. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol. Microbiol. 13:357-368. [DOI] [PubMed] [Google Scholar]
- 32.Morschhäuser, J., R. Virkola, T. K. Korhonen, and J. Hacker. 1997. Degradation of human subendothelial extracellular matrix by proteinase-secreting Candida albicans. FEMS Microbiol. Lett. 153:349-355. [DOI] [PubMed] [Google Scholar]
- 33.Naglik, J. R., S. J. Challacombe, and B. Hube. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67:400-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naglik, J. R., G. Newport, T. C. White, L. L. Fernandes-Naglik, J. S. Greenspan, D. Greenspan, S. P. Sweet, S. J. Challacombe, and N. Agabian. 1999. In vivo analysis of secreted aspartyl proteinase expression in human oral candidiasis. Infect. Immun. 67:2482-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Odds, F. C. 1988. Candida and candidosis: a review and bibliography. Bailliere Tindall, London, United Kingdom.
- 36.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 37.Park, Y. N., and J. Morschhäuser. 2005. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell 4:1328-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perfect, J. R. 1996. Fungal virulence genes as targets for antifungal chemotherapy. Antimicrob. Agents Chemother. 40:1577-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pichova, I., L. Pavlickova, J. Dostal, E. Dolejsi, O. Hruskova-Heidingsfeldova, J. Weber, T. Ruml, and M. Soucek. 2001. Secreted aspartic proteases of Candida albicans, Candida tropicalis, Candida parapsilosis and Candida lusitaniae. Inhibition with peptidomimetic inhibitors. Eur. J. Biochem. 268:2669-2677. [DOI] [PubMed] [Google Scholar]
- 40.Reuβ, O., Å. Vik, R. Kolter, and J. Morschhäuser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 41.Rüchel, R. 1981. Properties of a purified proteinase from the yeast Candida albicans. Biochim. Biophys. Acta 659:99-113. [DOI] [PubMed] [Google Scholar]
- 42.Rüchel, R., B. Ritter, and M. Schaffrinski. 1990. Modulation of experimental systemic murine candidosis by intravenous pepstatin. Zentralbl. Bakteriol. 273:391-403. [DOI] [PubMed] [Google Scholar]
- 43.Schaller, M., B. Hube, M. W. Ollert, W. Schafer, M. Borg-von Zepelin, E. Thoma-Greber, and H. C. Korting. 1999. In vivo expression and localization of Candida albicans secreted aspartyl proteinases during oral candidiasis in HIV-infected patients. J. Investig. Dermatol. 112:383-386. [DOI] [PubMed] [Google Scholar]
- 44.Skrbec, D., and D. Romeo. 2002. Inhibition of Candida albicans secreted aspartic protease by a novel series of peptidomimetics, also active on the HIV-1 protease. Biochem. Biophys. Res. Commun. 297:1350-1353. [DOI] [PubMed] [Google Scholar]
- 45.Smolenski, G., P. A. Sullivan, S. M. Cutfield, and J. F. Cutfield. 1997. Analysis of secreted aspartic proteinases from Candida albicans: purification and characterization of individual Sap1, Sap2 and Sap3 isoenzymes. Microbiology 143(Pt. 2):349-356. [DOI] [PubMed] [Google Scholar]
- 46.Staib, F. 1965. Serum-proteins as nitrogen source for yeastlike fungi. Sabouraudia 4:187-193. [DOI] [PubMed] [Google Scholar]
- 47.Staib, P., M. Kretschmar, T. Nichterlein, H. Hof, and J. Morschhäuser. 2000. Differential activation of a Candida albicans virulence gene family during infection. Proc. Natl. Acad. Sci. USA 97:6102-6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staib, P., M. Kretschmar, T. Nichterlein, H. Hof, and J. Morschhäuser. 2002. Host versus in vitro signals and intrastrain allelic differences in the expression of a Candida albicans virulence gene. Mol. Microbiol. 44:1351-1366. [DOI] [PubMed] [Google Scholar]
- 49.Stewart, K., and C. Abad-Zapatero. 2001. Candida proteases and their inhibition: prospects for antifungal therapy. Curr. Med. Chem. 8:941-948. [DOI] [PubMed] [Google Scholar]
- 50.Tossi, A., F. Benedetti, S. Norbedo, D. Skrbec, F. Berti, and D. Romeo. 2003. Small hydroxyethylene-based peptidomimetics inhibiting both HIV-1 and C. albicans aspartic proteases. Bioorg. Med. Chem. 11:4719-4727. [DOI] [PubMed] [Google Scholar]
- 51.Umezawa, H., T. Aoyagi, H. Morishima, M. Matsuzaki, and M. Hamada. 1970. Pepstatin, a new pepsin inhibitor produced by Actinomycetes. J. Antibiot. (Tokyo) 23:259-262. [DOI] [PubMed] [Google Scholar]
- 52.Watts, H. J., F. S. Cheah, B. Hube, D. Sanglard, and N. A. Gow. 1998. Altered adherence in strains of Candida albicans harbouring null mutations in secreted aspartic proteinase genes. FEMS Microbiol. Lett. 159:129-135. [DOI] [PubMed] [Google Scholar]
- 53.White, T. C., and N. Agabian. 1995. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J. Bacteriol. 177:5215-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, Z., H. N. ElSohly, M. R. Jacob, D. S. Pasco, L. A. Walker, and A. M. Clark. 2002. Natural products inhibiting Candida albicans secreted aspartic proteases from Tovomita krukovii. Planta Med. 68:49-54. [DOI] [PubMed] [Google Scholar]
- 55.Zingman, B. S. 1996. Resolution of refractory AIDS-related mucosal candidiasis after initiation of didanosine plus saquinavir. N. Engl. J. Med. 334:1674-1675. [DOI] [PubMed] [Google Scholar]
- 56.Zotter, C., U. F. Haustein, C. Schonborn, H. D. Grimmecke, and H. Wand. 1990. Effect of pepstatin A on Candida albicans infection in the mouse. Dermatol. Monatsschr. 176:189-198. [PubMed] [Google Scholar]