Abstract
Exposure-response analyses were performed to test the microbiological and clinical efficacies of tigecycline in complicated intra-abdominal infections where Escherichia coli and Bacteroides fragilis are the predominant pathogens. Data from evaluable patients enrolled in three clinical trials were pooled. Patients received intravenous tigecycline (100-mg loading dose followed by 50 mg every 12 h or 50-mg loading dose followed by 25 mg every 12 h). At the test-of-cure visit, microbiological and clinical responses were evaluated. Patients were prospectively classified into cohorts based on infection with a baseline pathogen(s): E. coli only (cohort 1), other mono- or polymicrobial Enterobacteriaceae (cohort 2), at least one Enterobacteriaceae pathogen plus an anaerobe(s) (cohort 3), at least one Enterobacteriaceae pathogen plus a gram-positive pathogen(s) (cohort 4), and all other pathogens (cohort 5). The cohorts were prospectively combined to increase sample size. Logistic regression was used to evaluate ratio of steady-state 24-hour area under the concentration-time curve (AUC) to MIC as a response predictor, and classification-and-regression-tree (CART) analyses were utilized to determine AUC/MIC breakpoints. Analysis began with cohorts 1, 2, and 3 pooled, which included 71 patients, with 106 pathogens. The small sample size precluded evaluation of cohorts 1 (34 patients, 35 E. coli pathogens) and 2 (16 patients, 24 Enterobacteriaceae). CART analyses identified a significant AUC/MIC breakpoint of 6.96 for microbiological and clinical responses (P values of 0.0004 and 0.399, respectively). The continuous AUC/MIC ratio was also borderline predictive of microbiological response (P = 0.0568). Cohort 4 (21 patients, 50 pathogens) was evaluated separately; however, an exposure-response relationship was not detected; cohort 5 (31 patients, 60 pathogens) was not evaluated. The prospective approach of creating homogenous populations of pathogens was critical for identifying exposure-response relationships in complicated intra-abdominal infections.
Evaluating exposure-response relationships by use of clinical trial data is an essential component of optimizing antimicrobial treatment, yet it is often quite challenging. A single dosing regimen is often used, thus limiting the range of observed drug exposure, and it is difficult to collect on an individual-patient basis the three integral pieces of information required to perform such analyses: pharmacokinetic (PK), clinical, and microbiological outcome data. The value of such analyses, however, has become increasingly important in quantifying drug efficacy and in contributing to the establishment of appropriate in vitro MIC susceptibility breakpoints by regulatory and clinical agencies (e.g., the Clinical and Laboratory Standards Institute [CLSI] and the European Committee on Antimicrobial Susceptibility Testing [EUCAST]) (5). Utilizing results from PK-pharmacodynamic (PK-PD) analyses may allow a better understanding of the causes of variability in responses among subgroups of patients.
Complicated intra-abdominal infection (cIAI) encompasses a variety of forms, but all require similar approaches to treatment: adequate source control (e.g., surgical intervention) and targeted antimicrobial therapy. Analyses of data from trials of cIAI are especially challenging due to the polymicrobial nature of the infection and the intrinsic heterogeneity in the patient and pathogen populations. To further complicate the issue, the virulence levels of the bacteria that cause these infections can be enhanced when certain microorganisms are combined. Generally, three anaerobic and two facultative anaerobic organisms are isolated from each individual patient (7, 11, 13). Anaerobes are present in 80 to 90% of intra-abdominal infections, with Bacteroides spp. accounting for two-thirds of these organisms. Although Bacteroides fragilis makes up only a small fraction of the normal oral and colonic flora, it is the organism most commonly isolated from both intra-abdominal abscesses and peritonitis (6). This may be attributed to several virulence factors identified for B. fragilis, including a critical capsular polysaccharide complex found on the bacterial cell surface. Animal models of infection have suggested a key role for B. fragilis in the formation of abdominal abscesses (18, 23). Escherichia coli is the most commonly isolated facultative anaerobe (6). The prevalences of E. coli and B. fragilis are understandable given that members of the Enterobacteriaceae family are common colonizers of the human gastrointestinal tract and Bacteroides species are the predominant organisms in the colon. An animal model of intra-abdominal sepsis has demonstrated the necessity of treating both the facultative enteric gram-negative bacilli (E. coli) to prevent acute mortality and the anaerobic gram-negative bacilli (B. fragilis) to avert abscess formation (2, 23). Clinically, antimicrobial agents or combinations of agents are selected for their activities against the more virulent pathogens in the infective mixture.
Tigecycline is a glycylcycline antimicrobial agent approved in 2004 by the FDA, and subsequently in more than 50 other countries, for the treatment of complicated skin and skin structure infections and cIAI (24). This agent has demonstrated an expanded spectrum of in vitro activity against gram-positive and gram-negative aerobic and anaerobic bacteria, including sensitive and multiple-drug-resistant strains of methicillin-resistant Staphylococcus aureus, streptococci, and vancomycin-resistant enterococcal species. In addition, the susceptibilities of most Enterobacteriaceae, including extended-spectrum-beta-lactamase-positive and -negative Escherichia coli, Klebsiella pneumoniae, and Klebsiella oxytoca and most strains of Bacteroides fragilis, is noteworthy in the setting of cIAI.
The objectives of these analyses were to assess the relationships between tigecycline exposure and microbiological and clinical responses and to determine patient demographic characteristics, drug exposure measurements, and other covariates predictive of clinical and microbiological outcome in the treatment of patients with cIAI. It was postulated that a more homogenous pathogen population would enhance the ability to establish an association between microbiological response and tigecycline exposure measures. Thus, a methodology for patient and pathogen classification similar to that used in previous exposure-response analyses for the efficacy of tigecycline in the treatment of patients with complicated skin and skin structure infections was developed (14). Based on data from animal models (20), in conjunction with the prolonged in vivo postantibiotic effect observed with tigecycline and the relatively long half-life (approximately 40 h) in humans, the ratio of area under the concentration-time curve (AUC) to MIC (AUC/MIC) is the PK-PD index most likely to be predictive of efficacy (15).
MATERIALS AND METHODS
Clinical and PK data acquisition.
Data from three clinical trials of tigecycline were pooled for analysis: one randomized, open-label phase 2 study and two randomized, double-blind phase 3 comparison trials of the safety and efficacy of intravenous (i.v.) tigecycline in the treatment of hospitalized patients with cIAI (1). The appropriateness of pooling data was assessed prior to analyses by reviewing protocols for trial design and inclusion and exclusion criteria to determine if the patients represented in these trials were homogenous in nature. Aside from the obvious open-label and comparator-controlled designs for the phase 2 and 3 programs, respectively, the clinical and microbiological endpoints for the trials were comparable. Inclusion and exclusion criteria were slightly modified from phase 2 to phase 3 to further refine the characterization of cIAI. All changes noted, however, were deemed unlikely to affect the analyses. Additional information regarding the extent of the original infection (presence of abscesses, fecal contamination, and presence of peritonitis) and surgical interventions (extent of residual contamination and likelihood of surgical control of infection) were collected in the phase 3 program. A surgical review board was also established to assess the adequacy of surgical source control with regard to the initial surgical or interventional radiology procedure in patients determined to be clinical failures.
Following pooling of phase 2 and 3 data and identification of a microbiologically and clinically evaluable population, patients were reviewed from a clinical standpoint by using recorded medical and diagnostic histories to ensure appropriate inclusion for analysis. All patients had clinical signs and symptoms of cIAI requiring hospitalization. Clinical entities included complicated appendicitis or cholecystitis, perforation of the small or large intestines, and gastric or duodenal ulcer perforation. All patients underwent laparotomy, laparoscopy, or percutaneous drainage of an intra-abdominal abscess, in addition to requiring antimicrobial therapy. Patients suspected of having spontaneous bacterial peritonitis, simple cholecystitis, gangrenous cholecystitis without rupture, simple appendicitis, acute suppurative cholangitis, pancreatic abscess, or infected necrotizing pancreatitis were excluded from enrollment. Patients who received intraoperative antibacterial irrigants or peritoneal antibacterial agents, such as antibiotic impregnated sponges, were also excluded. In addition, patients with scores for acute physiology and chronic health evaluation (APACHE) (10) of >30, neutropenia (absolute neutrophil count of <500/mm3), hepatic disease, or calculated creatinine clearances of <41 ml/min/1.73 m2 and those on immunosuppressive therapy were not enrolled. In the phase 3 trials, patients with known Proteus and Pseudomonas infections were excluded. Patients in whom Pseudomonas was identified from baseline culture could be continued on tigecycline at the discretion of the investigator. For the phase 2 trial, parenteral nonstudy antibiotic therapy was not allowed after the baseline culture was obtained. For the phase 3 trials, only one dose of antibiotic (or an antibiotic combination) was allowed after the baseline culture was obtained. Wounds could be irrigated with sterile water, normal saline, or a topical antiseptic. Antifungal and antiviral agents, ophthalmic aminoglycosides, and oral vancomycin were permitted in the study population.
Patients in the phase 2 trial received open-label i.v. tigecycline (100-mg loading dose followed by 50 mg every 12 h [100/50 mg]) for up to 14 days. The dosing regimens in both phase 3 trials were tigecycline 100/50 mg every 12 h (with placebo doses given on an alternate every-12-h schedule) versus imipenem-cilastatin (500 mg) i.v. every 6 hours administered for up to 14 days. Patients in the phase 3 program were stratified at the time of randomization for APACHE II scores of ≤15 or >15 and then randomly assigned (at a 1:1 ratio) to receive either i.v. tigecycline or a comparator. For all patients, PK samples were collected prior to the first dose on day 1 and then on either the day of or the day before discharge from the hospital, at time zero (predose), at the end of the infusion, and 3 h and 6 h after the start of the infusion. Home delivery of antibiotics was permitted to allow early discharge from the hospital.
Patient- and disease-related descriptors were recorded during the screening visit. It was assumed that the values of demographic characteristics recorded at baseline remained constant for the duration of the trial. Baseline microorganisms were collected and sent to a central laboratory (Covance Central Laboratories, Indianapolis, IN) for identification and susceptibility testing, following approved CLSI guidelines.
Efficacy was assessed using both clinical and microbiological criteria at the test-of-cure (TOC) visit, which was at least 2 weeks after the last dose of study medication. Clinically evaluable patients received at least 5 days of tigecycline treatment, unless the patient was declared a failure after at least eight doses of study medication. Clinical responses were categorized as cure (resolution of intra-abdominal infection without additional surgical or radiological intervention or additional antibiotic therapy used to cure the infection), failure (additional surgical or radiological intervention and/or additional antimicrobial therapy required or death), or indeterminate (extenuating circumstances that precluded classification as either a cure or failure). Indeterminate clinical responses were not considered in this analysis.
Microbiological efficacy was evaluated at the pathogen level since, as anticipated, the majority of intra-abdominal infections were polymicrobial. A response of eradication, persistence, or indeterminate outcome was assigned for each pathogen isolated at baseline. A successful microbiological response was defined as documented or presumed to indicate eradication of the pathogen(s) present at baseline from culture specimens obtained at the TOC visit or the last available culture during therapy. An unsuccessful microbiological response was defined as documented or presumed to indicate persistence of the baseline pathogen(s). Those patients who were clinically evaluable and had a baseline culture from the infected site with at least one identified causative pathogen susceptible to the study drug were classified as microbiologically evaluable. As with the clinical outcome, patients with indeterminate responses were excluded from the microbiologically evaluable population.
Population PK model.
Serum samples for PK analysis were collected, and the tigecycline dose, date, and time of each infusion were recorded. Samples were frozen at −70°C until analyzed for tigecycline concentrations by using a validated liquid chromatography-tandem mass spectrometry assay with a lower limit of quantitation of 10 ng/ml. The overall precision and accuracy for the quality control samples (1,500 ng/ml, 200 ng/ml, and 25 ng/ml) were in the range of 0.9 to 12% and 93 to 110% (16).
Tigecycline exposure measures for each patient in the cIAI clinical trials were generated from a previously developed two-compartment population PK model with zero-order input and first-order elimination, where tigecycline clearance was positively related to increasing body weight, creatinine clearance, and male gender (21). This model was developed using data from 174 subjects and 195 patients (a total of 3,056 serum concentrations) in all three phases of development. The 24-hour steady-state AUC (AUCss0-24), as the primary drug exposure measure, was predicted for all patients by using Bayesian estimation.
Exposure-response analysis of efficacy.
Data from patients with tigecycline exposure measurements and those classified as both clinically and microbiologically evaluable at the TOC visit were analyzed. Bayesian parameter estimates from the final tigecycline PK model were used to generate individual AUCss0-24/MIC ratios.
Prior to the statistical analyses, patients were clinically reviewed and classified into one of five predefined cohorts based on identification of a causative organism(s) isolated at baseline (Table 1). All other organisms isolated at baseline were considered nonpathogenic and were therefore excluded from this analysis. Patient cohorts were prospectively established to create more-homogenous pathogen populations for analysis. Consideration was given to the predominance of Enterobacteriaceae and B. fragilis as key pathogens in these infections. The classification system for cohorts, along with a summary of microbiological and clinical outcomes for patients included in the analysis, is shown in Table 2.
TABLE 1.
Pathogens considered in the cIAI analysis
| Pathogen |
|---|
| Gram positive |
| S. agalactiae |
| S. anginosus |
| S. constellatus |
| S. intermedius |
| S. aureus |
| Gram negative |
| Escherichia coli |
| Citrobacter spp. |
| Enterobacter spp. |
| Klebsiella spp. |
| Anaerobic |
| B. fragilis |
| C. perfringens |
| Peptostreptococcus anaerobius |
| Peptostreptococcus magnus |
| Peptostreptococcus micros |
| Prevotella melaninogenica |
TABLE 2.
Summary of cohort classification system and microbiological and clinical outcomes
| Cohort(s) | Pathogen criterion | MIC range | No. of patients (no. of pathogens) | No. (%) of microbiological curesc | No. (%) of clinical curesd |
|---|---|---|---|---|---|
| 1 | E. coli only | 0.12-1 | 34 (35) | 35 (100) | 32 (94) |
| 2 | ≥1 GNa | 0.12-1 | 16 (24) | 22 (92) | 14 (88) |
| 3 | ≥1 GNa + ≥1 anaerobe | 0.06-4 | 21 (47) | 39 (83) | 16 (76) |
| 4 | ≥1 GNa + ≥1 GPb | 0.06-1 | 21 (50) | 44 (88) | 18 (86) |
| 5 | Other | 0.004-4 | 31 (60) | 54 (90) | 27 (87) |
| 1+2+3 | ≥1 GNa ± ≥1 anaerobe | 0.06-4 | 71 (106) | 96 (91) | 62 (87) |
Gram-negative Enterobacteriaceae organism.
Gram-positive organism.
Pathogen-level microbiological response.
Patient-level clinical response.
Cohort 1 included patients with monomicrobial infections due to E. coli, a significant pathogen in cIAI. Cohort 2 included patients with other monomicrobial infections due to Enterobacteriaceae or polymicrobial gram-negative infections, including Klebsiella spp., Enterobacter spp., and Citrobacter spp. with or without E. coli. Patients with at least one gram-negative pathogen (Enterobacteriaceae) plus at least one anaerobic pathogen at baseline were grouped into cohort 3. Finally, cohort 4 consisted of both gram-positive and gram-negative mixed infections and cohort 5 contained those patients with other monomicrobial or polymicrobial infections. Sample size and distribution of outcomes within each cohort were prospectively evaluated to determine whether exploratory or statistical analyses could be performed or if cohorts needed to be combined to create a more robust patient population. The presence of baseline Pseudomonas aeruginosa was analyzed as a covariate.
Patient- and disease-related descriptors collected at baseline were also evaluated as potential predictors of microbiological and clinical efficacy. The demographics included age and country or region of treatment. The disease-related descriptors included clinical diagnosis of infection and presence (none, single, or multiple) and size (<10 ml, 10 to 100 ml, or >100 ml) of abscess. Occurrences of fecal contamination and peritonitis were also recorded. The probability of the primary surgical or radiological procedure being successful in controlling the source of infection was independently assessed by a surgical review board (<25%, 25 to 49%, 50 to 74%, 75 to 95%, or >95%). Calculated baseline APACHE II score was also evaluated.
Statistical analysis.
All data processing, data cleanup, database creation, and statistical analyses were performed using SAS software, version 8.2 (19). Classification-and-regression-tree (CART) analysis was performed using S-Plus, version 6.2 (12). Exploratory analyses of microbiological and clinical response were conducted to identify relationships between outcome, exposure measurements, patient demographic characteristics, and comorbidities. CART analyses were performed to determine breakpoints in exposure measures stratified by response for each cohort. The numbers of cures and failures within each CART-identified category were computed. Categories with fewer than five cures or failures were combined with adjacent categories to allow the groups to be tested as a predictor of probability of response by using logistic regression.
Logistic regression analyses were used to determine whether exposure measures and patient covariates were statistically significant predictors of clinical and microbiological response. In the case of multiple observations per patient, generalized estimating equations were employed (8). Univariate analyses were followed by multivariable modeling, utilizing a backward elimination procedure with a level of significance of 0.05 to identify predictor variables with statistically significant influences on outcomes. Goodness of fit of the logistic regression model was assessed using the Hosmer-Lemeshow test, and the predictive ability of the model was assessed using the area under the receiver operating characteristic (ROC) curve (9).
RESULTS
Population PK parameters.
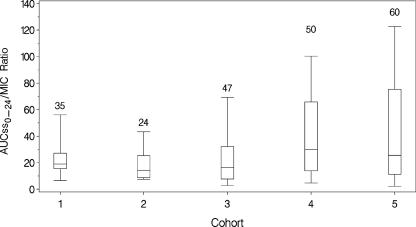
Exposure estimates were generated using the previously noted population PK model for tigecycline (21). Significant differences between the PK parameters for patients enrolled in the phase 2 and phase 3 trials were not observed. The ranges of age, weight, and creatinine clearance for the phase 3 patients were within the ranges studied in phase 2. Individual predicted AUCss0-24 values were calculated for each evaluable cIAI patient. For all patients included in the efficacy analyses (n = 123; 40 received 50 mg per day, and all others received 100 mg per day), the mean AUCss0-24 was 6.08 (standard deviation [SD], 2.48) μg·h/ml, with a range of 2.88 to 22.6 μg·h/ml. Similar ranges of AUCss0-24 values were observed across each cohort. Across all cohorts, the mean AUCss0-24/MIC ratio was 29.8 (SD, 75.1), with a range of 0.97 to 802. In Fig. 1, the AUCss0-24/MIC ratios are shown for the five patient cohorts. Greater variability in this measure was observed in cohorts 4 and 5.
FIG. 1.
AUCss0-24/MIC ratios for the five patient cohorts. The boxes represent the 25th to 75th percentiles of AUC/MIC ratio. The whiskers extend from the 5th to 95th percentiles of AUC/MIC ratio. The number above each box represents the number of observations in each box.
Exposure-response analysis of efficacy. (i) Data.
The data set for the exposure-response analyses included 123 evaluable patients, with 216 pathogens (Table 2). Forty (33%) patients were from the phase 2 trial, and 83 (67%) patients were from the phase 3 trials. Patient demographic characteristics are presented in Table 3. The mean age was 45 (SD, 18) years, with a range of 18 to 85 years, and the mean (SD) weight was 77 (SD, 17) kg, with a range of 45 to 138 kg. Sixty-three percent of patients were male, 74% were Caucasian, and 90% were treated in either North America or Europe. The mean baseline APACHE II score was 6. A total of 10 (8%) patients had P. aeruginosa pathogens isolated at baseline, of which 80% were microbiologically eradicated. In addition, the majority (58%) of patients evaluated were diagnosed with complicated appendicitis, followed by 12% with complicated cholecystitis, 11% with intra-abdominal, hepatic, or splenic abscesses, and 9% with peritonitis due to perforation of the small or large intestines. Supplementary data regarding disease severity were collected for the 83 patients in the phase 3 trials. Of these patients, 65 (78%) had a single abscess. Totals of 38 (46%) and 29 (35%) patients had abscesses ranging from 10 to 100 ml and greater than 100 ml, respectively. Only 25 (24%) patients were found to have fecal contamination, whereas 48 (58%) were diagnosed with peritonitis. The probability of primary surgical success was assessed as greater than 95% in 54 (65%) patients, between 75 and 95% in 19 (23%) patients, and less than 75% in the remaining 10 (12%) patients.
TABLE 3.
Summary statistics of patient demographic characteristics
| Demographic characteristic | Summary statistics (n = 123) |
|---|---|
| Age (yrs) | |
| Mean (SD) | 45.0 (17.7) |
| Median | 42.6 |
| Range | 18-85 |
| Weight (kg) | |
| Mean (SD) | 77.1 (17.0) |
| Median | 75 |
| Range | 45-138 |
| Baseline APACHE II score | |
| Mean (SD) | 6.07 (3.87) |
| Median | 6 |
| Range | 0-25 |
| No. (%) of patients by gender | |
| Male | 78 (63) |
| Female | 45 (37) |
| No. (%) of patients by ethnicity | |
| Caucasian | 91 (74) |
| Black | 5 (4) |
| Hispanic | 25 (20) |
| Other | 2 (2) |
| No. (%) of patients by region of treatment | |
| Europe | 61 (50) |
| North America | 49 (40) |
| Latin America | 10 (8) |
| Other | 3 (2) |
(ii) Microbiological response.
A summary of microbiological outcomes is presented in Table 2. Cohort 1 included a total of 34 patients, with 35 pathogens, classified as having monomicrobial E. coli infections at baseline (1 patient had 2 different strains of E. coli). All pathogens in this cohort were eradicated. Approximately 14%, 60%, 23%, and 3% of E. coli organisms had MICs of 0.12, 0.25, 0.5, and 1 μg/ml, respectively. Cohort 2 included a total of 16 patients (24 pathogens), classified as having monomicrobial or polymicrobial gram-negative infections at baseline. The range of baseline MICs for patients in cohort 2 was identical to that for cohort 1, with approximately 4%, 29%, 42%, and 25% of baseline pathogens having MICs of 0.12, 0.25, 0.5, and 1 μg/ml, respectively. Two pathogens within this cohort were not eradicated at the TOC visit: the Citrobacter freundii complex, with an MIC of 0.5 μg/ml, and K. pneumoniae, with an MIC of 1 μg/ml. Twenty-one patients, with at least 1 gram-negative pathogen plus at least 1 anaerobic pathogen at baseline (a total of 47 pathogens), were classified into cohort 3. The inclusion of anaerobic organisms increased the range of MICs from 0.06 to 4 μg/ml. From three separate patients, eight baseline pathogens failed to be eradicated in this cohort: four of these were B. fragilis organisms, with MICs ranging from 2 to 4 μg/ml, and the remaining gram-negative pathogens (three E. coli and one K. oxytoca pathogen) had MICs between 0.12 and 0.5 μg/ml. Cohort 4 included a total of 21 patients (50 pathogens), classified as having at least one gram-negative pathogen plus at least one gram-positive pathogen at baseline. The majority (74%) of pathogens represented in this cohort had MICs of ≤0.25 μg/ml. Finally, cohort 5 included a total of 31 patients (60 pathogens), classified as having all other monomicrobial or polymicrobial pathogens at baseline.
After the classification of cohorts was completed, each category was examined for sample size and distribution of responses to determine if exploratory or statistical analyses could be performed. Although cohort 1 was of primary interest and had 34 patients with monomicrobial E. coli infections, no meaningful statistical analyses could be performed on this group alone, since all organisms were eradicated. The next step was to determine if adding the other monomicrobial or polymicrobial gram-negative pathogens in cohort 2 would provide sufficient sample size and outcome to warrant a valid analysis. Although there were 16 patients in cohort 2, only 2 patients had microbiological failures. Combining cohorts 1, 2, and 3, however, resulted in a total of 71 patients, with 106 pathogens and 10 microbiological failures. This was the group where statistical analyses began since there was an adequate distribution of cures and failures as well as sufficient sample size. Since the majority of patients in this combined group had polymicrobial infections, longitudinal logistic regression analyses defined by multiple pathogens, using generalized estimating equations, were required. An additional analysis, using similar analytical methods, was performed to evaluate the impact of sequentially adding mixed gram-positive and gram-negative infections in cohort 4 and the other infections in cohort 5.
Several variables could not be evaluated in the logistic regression analysis. The indicator variable for Pseudomonas aeruginosa at baseline could not be formally evaluated, due to the small number of failures. In the phase 3 studies, presence and size of abscess(es), presence of fecal contamination, presence of peritonitis, and predicted success of primary surgical procedure were also not evaluated as categorical covariates, due to the small sample size and distribution of outcomes. Additionally, investigation using multivariable logistic regression models was not evaluated, due to the limited sample size and, thus, the inability to obtain precise and accurate parameter estimates.
The CART technique identified several breakpoints in the distribution of AUCss0-24/MIC ratios for each cohort. Due to the sensitivity of the procedure, the tree model was trimmed to four terminal nodes. Terminal breakpoints of 8.49 (cohort 2 alone), 6.96, and 11.07 (cohorts 1, 2, and 3 combined) were identified. The breakpoints were then evaluated for the numbers of microbiological outcomes above and below each point. The numbers of microbiological cures and failures within each category of AUCss0-24/MIC ratio, as defined by the breakpoints, were computed. Ten failures were observed in the combined cohort 1, 2, and 3 data set. The breakpoint at 6.96 had a better distribution of cures and failures around the breakpoint and was of primary interest. In the 71 patients (106 pathogens) from cohorts 1, 2, and 3 combined, 10 pathogens fell below the CART-identified breakpoint of 6.96, of which 4 (40%) were classified as microbiological failures. Ninety-six pathogens fell at or above the CART-identified breakpoint of 6.96, of which 6 (6.25%) were microbiological failures. Evaluation of the CART-identified breakpoint of 11.07 resulted in 36 pathogens below and 75 pathogens at or above this breakpoint. Of the 10 microbiological failures, 7 fell below and 7 fell at or above the breakpoint 11.07, yielding a greater proportion of failures below the breakpoint. Therefore, only the terminal breakpoint of 6.96 was utilized in the subsequent logistic regression analyses.
Univariate logistic regression models assessing the impacts of AUCss0-24/MIC ratio and patient covariates on the probability of microbiological response were evaluated. For the combined cohort 1, 2, and 3 data set, AUCss0-24/MIC ratio above the CART-identified breakpoint of 6.96 and race (Caucasian versus all others) were found to be statistically significant predictors of microbiological response (P values of 0.0004 and 0.0290, respectively). The odds ratio was 26.4 (95% confidence interval, 3.9 to 177.5) for AUCss0-24/MIC ratios above the breakpoint of 6.96. However, caution must be used when interpreting the model results evaluating the AUCss0-24/MIC breakpoint due to the small number of failures observed above this point. The AUCss0-24/MIC ratio as a continuous covariate was of borderline statistical significance (P = 0.0568) but provided the most informative model for these data; as the AUCss0-24/MIC ratio increased, the model-predicted probability of microbiological success increased. The odds ratio of 1.055 for the AUCss0-24/MIC ratio indicated that for a 1-unit increase in the ratio, a patient was 5.5% more likely to have a successful microbiological response.
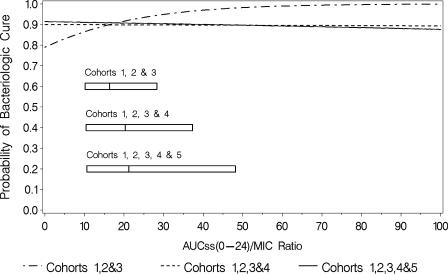
For the final logistic regression model for microbiological response (Fig. 2) in cohorts 1, 2, and 3, the Hosmer-Lemeshow goodness-of-fit statistic was 9.78 with 7 degrees of freedom (P = 0.2016), and the area under the ROC curve was 0.70, indicating an adequate and predictive model. At the median AUCss0-24/MIC ratio of 15.6 for cohorts 1, 2, and 3 combined, the model-predicted probability of microbiological success was 0.896.
FIG. 2.
Final logistic regression models for microbiological response for each cohort. The boxes represent the 25th to 75th percentiles of AUC/MIC ratio for each group of cohorts.
Additional analyses were done to include cohorts 4 and 5 and justify the use of the cohort classification methodology. Sequential addition of cohorts 4 and 5 to the combined cohort 1, 2, and 3 data set, however, occluded the ability to detect any exposure-response relationship (Fig. 2). The only discernible relationship was detected when the more homogenous combined cohort 1, 2, and 3 data set was evaluated.
Clinical response.
Table 2 provides a summary of clinical response for each cohort. Due to the limited sample size and distribution of clinical cures and failures, formal statistical analysis could be performed only on the combined cohort 1, 2, and 3 data set. As in the microbiological response analysis, the indicator variable for Pseudomonas aeruginosa at baseline could not be evaluated in the logistic regression analysis, due to the small number of failures. Likewise, the disease-related descriptors (abscess size, fecal contamination, peritonitis, and probability of surgical success) could not be evaluated as categorical covariates, due to the small sample size as well as the distribution of outcomes.
Of the 123 patients eligible for the exposure-response analysis for clinical efficacy, a total of 71 patients were included in cohorts 1, 2, and 3 combined, and 62 (87%) patients had successful clinical responses. Univariate logistic regression models assessing the impacts of AUCss0-24/MIC ratio and patient covariates on the probability of clinical response were evaluated (data not shown). AUCss0-24/MIC ratio above the CART-identified breakpoint of 6.96 and baseline APACHE II score were both statistically significant predictors of clinical response (P values of 0.0399 and 0.0279, respectively). The AUCss0-24/MIC ratio as a continuous covariate was not statistically significant (P = 0.0740) but was still considered to be the most informative model for these data. An increase in AUCss0-24/MIC ratio was associated with an increased model-predicted probability of clinical success. A 1-unit increase in the AUCss0-24/MIC ratio resulted in a patient being 9.8% more likely to have a successful clinical response. The Hosmer-Lemeshow goodness-of-fit statistic for the final logistic regression model for cohorts 1, 2, and 3 was 10.66 with 8 degrees of freedom (P = 0.2214), and the area under the ROC curve was 0.71, indicating an adequate and predictive model (data not shown). At the median AUCss0-24/MIC ratio of 15.6 for cohorts 1, 2, and 3, the model-predicted probability of clinical success was 0.888.
DISCUSSION
Quantifying the impact of antibiotic therapy for cIAI can be challenging, as these infections are frequently polymicrobial. These are the first analyses to demonstrate exposure-response relationships for antibiotic efficacy in cIAI. In an effort to create a more homogenous patient population for the microbiological exposure-response analyses, each individual patient eligible for analysis was evaluated and grouped into cohorts based upon their baseline pathogen(s), as previously described. Patients grouped into cohorts 1, 2, and 3 were considered to be the most homogenous population, with sufficient numbers of cures and failures for statistical analysis. Patients in these cohorts had baseline pathogens of primary interest: monomicrobial infections with E. coli, other monomicrobial or polymicrobial infections with a gram-negative pathogen(s) (Klebsiella spp., Enterobacter spp., and/or Citrobacter spp. with or without E. coli), or polymicrobial infections with a gram-negative pathogen(s) plus one or more key anaerobes (B. fragilis, Prevotella melaninogenica, Clostridium perfringens, or Peptostreptococcus spp.), respectively. Since there were no microbiological failures in cohort 1 and only two failures in cohort 2, analyses were performed with cohorts 1, 2, and 3 combined in order to achieve sufficient sample size and distribution of cures and failures, despite the fact that from a microbiological perspective, these microorganisms are not alike and have different methods of causing disease in patients.
It must be remembered that the numbers of clinical and microbiological failures were very small and that the exposure/response analyses are post hoc. Additional studies may be necessary to confirm the findings of this study. However, it must be understood that the number of patients required for a more robust analysis is very large. Over 800 patients were enrolled in the clinical studies that supported this exposure/response analysis.
The AUCss0-24/MIC ratio was analyzed as both a continuous variable and a categorical variable, with a CART-identified breakpoint of 6.96. When monomicrobial E. coli, the target gram-negative pathogen, and mixed gram-negative and anaerobic infections (cohorts 1, 2, and 3) were considered simultaneously, these analyses detected a marginal relationship between exposure and microbiological response for the continuous AUCss0-24/MIC ratio and a significant relationship for the categorical (breakpoint at 6.96) AUCss0-24/MIC ratio. One point to consider in this analysis is the impact of B. fragilis on microbiological outcome and the subsequent affect on breakpoint determination: four of the eight microbiological failures in cohort 3 were observed in patients with B. fragilis isolates, with MICs ranging from 2 to 4 μg/ml. Race (Caucasian versus all others) was also found to be a significant predictor of microbiological response in univariate analyses. The importance of this finding is unknown and may require further investigation. Race, however, was not found to be important for clinical success. Finally, the model-predicted probability of microbiological success was 0.896 for cohorts 1, 2, and 3, with a median AUCss0-24/MIC ratio of 15.6. This is concordant with the observed 91% microbiological success rate for cohorts 1, 2, and 3, lending credence to the predictive ability of the final logistic regression model. When all cohorts were analyzed together, however, the ability to detect any semblance of an exposure-response relationship was eliminated. This can be explained by the small numbers of failures and by the heterogeneity in the total patient and pathogen populations.
Patient demographic factors, severity of illness, and surgical data were also examined in the microbiological efficacy analysis for cohorts 1, 2, and 3. APACHE II is one method of evaluating severity of illness and outcome (10) and has been validated in surgical patients with intra-abdominal infections (4, 17). Age, gender, and baseline APACHE II scores, however, were not statistically significant predictors of microbiological response. An inability to obtain adequate surgical control of the source of infection has been associated with an increased risk of adverse outcomes (3, 22), but this and other surgical covariates examined could not be evaluated categorically, due to the small sample size and distribution of outcomes. Finally, since P. aeruginosa is intrinsically resistant to tigecycline, the presence of this organism at baseline was considered to determine if there was a direct effect on outcome. The small number of failures associated with baseline P. aeruginosa, however, precluded evaluation of this covariate.
An exposure-response analysis for clinical outcome was also performed. For cohorts 1, 2, and 3 combined, the CART-identified AUCss0-24/MIC breakpoint of 6.96 was a statistically significant predictor of clinical outcome, but the AUCss0-24/MIC ratio as a continuous covariate was not found to be predictive. It is important to note that the CART-identified AUCss0-24/MIC breakpoint of 6.96 was consistent across both clinical and microbiological outcomes. As anticipated, one difference between the clinical and microbiological analyses was the ability of baseline APACHE II scores to effectively predict clinical response.
In conclusion, heterogeneous patient and/or pathogen populations can hinder the detection of exposure-response analyses, especially when small datasets are analyzed. It was hypothesized that the ability to detect these relationships would increase with patient and infecting-pathogen-population homogeneity, and pathogen cohorts were prospectively created to test this hypothesis. This hypothesis was previously tested in an analysis of patients treated with tigecycline for complicated skin and skin structure infections, with noteworthy results (21). Using a similar methodology adapted for patients with cIAI, a prospective procedure for pathogen classification was followed, and PK-PD relationships for microbiological and clinical outcome were detected only when cohorts were evaluated on the basis of key cIAI pathogens, namely, Enterobacteriaceae and B. fragilis. Creating homogenous populations based on target organisms may be essential for identifying exposure-response relationships.
Acknowledgments
This analysis was supported, in part, by a grant from Wyeth Research.
Footnotes
Published ahead of print on 22 October 2007.
REFERENCES
- 1.Babinchak, T., E. Ellis-Grosse, N. Dartois, G. M. Rose, and E. Loh. 2005. The efficacy and safety of tigecycline for the treatment of complicated intra-abdominal infections: analysis of pooled clinical trial data. Clin. Infect. Dis. 41(Suppl. 5):S354-S367. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett, J. G., A. B. Onderdonk, T. Louie, D. L. Kasper, and S. L. Gorbach. 1978. A review. Lessons from an animal model of intra-abdominal sepsis. Arch. Surg. 113:853-857. [DOI] [PubMed] [Google Scholar]
- 3.Christou, N. V., P. S. Barie, E. P. Dellinger, J. P. Waymack, and H. H. Stone. 1993. Surgical Infection Society intra-abdominal infection study. Prospective evaluation of management techniques and outcome. Arch. Surg. 128:193-198. [DOI] [PubMed] [Google Scholar]
- 4.Dellinger, E. P., M. J. Wertz, J. L. Meakins, J. S. Solomkin, M. D. Allo, R. J. Howard, and R. L. Simmon. 1985. Surgical infection stratification system for intra-abdominal infection. Multicenter trial. Arch. Surg. 120:21-29. [DOI] [PubMed] [Google Scholar]
- 5.Exposure-Response Working Group. April 2003, posting date. Guidance for industry: exposure-response relationships—study design, data analysis, and regulatory applications. U.S. Department of Health and Human Services, Rockville, MD. http://www.fda.gov/cber/gdlns/exposure.htm.
- 6.Gorbach, S. L. 1991. Antimicrobial prophylaxis for appendectomy and colorectal surgery. Rev. Infect. Dis. 13(Suppl. 10):S815-S820. [DOI] [PubMed] [Google Scholar]
- 7.Gorbach, S. L. 1993. Intraabdominal infections. Clin. Infect. Dis. 17:961-965. [DOI] [PubMed] [Google Scholar]
- 8.Hardin, J. W., and J. M. Hilbe. 2003. Generalized estimating equations. Chapman & Hall/CRC, Boca Raton, FL.
- 9.Harrell, F. E., Jr., K. L. Lee, R. M. Califf, D. B. Pryor, and R. A. Rosati. 1984. Regression modelling strategies for improved prognostic prediction 566. Stat. Med. 3:143-152. [DOI] [PubMed] [Google Scholar]
- 10.Knaus, W. A., J. E. Zimmerman, D. P. Wagner, E. A. Draper, and D. E. Lawrence. 1981. APACHE—acute physiology and chronic health evaluation: a physiologically based classification system. Crit. Care Med. 9:591-597. [DOI] [PubMed] [Google Scholar]
- 11.Lorber, B., and R. M. Swenson. 1975. The bacteriology of intra-abdominal infections. Surg. Clin. North Am. 55:1349-1354. [DOI] [PubMed] [Google Scholar]
- 12.MathSoft, Inc., Data Analysis Division. 2000. S-Plus 6.0 for UNIX Guide to Statistics, vol. 1 and 2. MathSoft, Inc., Seattle, WA.
- 13.McClean, K. L., G. J. Sheehan, and G. K. Harding. 1994. Intraabdominal infection: a review. Clin. Infect. Dis. 19:100-116. [DOI] [PubMed] [Google Scholar]
- 14.Meagher, A. K., J. A. Passarell, B. B. Cirincione, S. A. Van Wart, K. Liolios, T. Babinchak, E. J. Ellis-Grosse, and P. G. Ambrose. 2007. Exposure-response analyses of tigecycline efficacy in patients with complicated skin and skin-structure infections. Antimicrob. Agents Chemother. 51:1939-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meagher, A. K., P. G. Ambrose, T. H. Grasela, and E. J. Ellis-Grosse. 2005. The pharmacokinetic and pharmacodynamic profile of tigecycline. Clin. Infect. Dis. 41(Suppl. 5):S333-S340. [DOI] [PubMed] [Google Scholar]
- 16.Muralidharan, G., M. Micalizzi, J. Speth, D. Raible, and S. Troy. 2005. Pharmacokinetics of tigecycline after single and multiple doses in healthy subjects. Antimicrob. Agents Chemother. 49:220-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohmann, C., D. H. Wittmann, H. Wacha, et al. 1993. Prospective evaluation of prognostic scoring systems in peritonitis. Eur. J. Surg. 159:267-274. [PubMed] [Google Scholar]
- 18.Onderdonk, A. B., R. B. Markham, D. F. Zaleznik, R. L. Cisneros, and D. L. Kasper. 1982. Evidence for T cell-dependent immunity to Bacteroides fragilis in an intra-abdominal abscess model. J. Clin. Investig. 69:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SAS Institute, Inc. 1999. SAS/STAT user's guide, version 8.2. SAS Institute, Inc., Cary, NC.
- 20.van Ogtrop, M. L., D. Andes, T. J. Stamstad, B. Conklin, W. J. Weiss, W. A. Craig, and O. Vesgo. 2000. In vivo pharmacodynamic activities of two glycylcyclines (GAR-936 and WAY 152,288) against various gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 44:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Wart, S. A., J. S. Owen, E. A. Ludwig, A. K. Meagher, J. M. Korth-Bradley, and B. B. Cirincione. 2006. Population pharmacokinetics of tigecycline in patients with complicated intra-abdominal or skin and skin structure infections. Antimicrob. Agents Chemother. 50:3701-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wacha, H., T. Hau, R. Dittmer, C. Ohmann, et al. 1999. Risk factors associated with intra-abdominal infections: a prospective multicenter study. Langenbecks Arch. Surg. 384:24-32. [DOI] [PubMed] [Google Scholar]
- 23.Weinstein, W. M., A. B. Onderdonk, J. G. Bartlett, T. J. Louie, and S. L. Gorbach. 1975. Antimicrobial therapy of experimental intra-abdominal sepsis. J. Infect. Dis. 132:282-286. [DOI] [PubMed] [Google Scholar]
- 24.Wilcox, M. H. 2005. Efficacy of tigecycline in complicated skin and skin structure infections and complicated intra-abdominal infections. J. Chemother. 17(Suppl. 1):23-29. [DOI] [PubMed] [Google Scholar]