Abstract
A structure-based approach was used to design RNA-binding zinc fingers that recognize the HIV-1 Rev response element (RRE). An arginine-rich α-helix from HIV-1 Rev was engineered into the zinc finger framework, and the designed fingers were shown to bind specifically to the RRE with high affinity and in a zinc-dependent manner, and display cobalt absorption and CD spectra characteristic of properly folded fingers. The results indicate that a monomeric zinc finger can recognize a specific nucleic acid site and that the α-helix of a finger can be used to recognize the major groove of RNA as well as DNA. The RRE-binding zinc fingers demonstrate how structure-based approaches may be used in the design of potential RNA-binding therapeutics and provide a framework for selecting RNA-binding fingers with desired specifications.
Zinc fingers are compact protein domains composed of an α-helix and β-sheet held together by a single zinc ion (1–4). Tandem arrays of zinc fingers are commonly used to recognize specific DNA sequences through insertion of α-helices into the DNA major groove (3, 5–9), and novel DNA-binding fingers have been designed by using the structures of DNA-protein complexes as guides (7, 10–13). Some zinc finger proteins also bind specific RNA sites (14–18) but relatively little is known about their modes of recognition. TFIIIA is a nine-finger protein that binds specifically to both DNA and RNA but uses different sets of fingers and different features of the major groove in each case (16, 19). Other zinc fingers have been identified that bind to DNA-RNA hybrids (20).
To learn more about zinc finger-RNA interactions and to test our ability to use structural information to design RNA-binding proteins, we sought to create a zinc finger that specifically recognizes the Rev-binding site (Rev response element, RRE) of HIV-1. The Rev protein contains an arginine-rich RNA-binding domain that, as an isolated peptide, forms a marginally stable α-helix that binds the RRE with an affinity proportional to its helical content (21). We reasoned that placing the helix within a zinc finger scaffold would substantially enhance its stability and thereby its RRE-binding affinity, because isolated zinc fingers can fold into stable metal-dependent structures (2, 22, 23). Here we demonstrate that hybrid zinc finger-Rev (ZF-Rev) peptides fold in a zinc-dependent manner and bind specifically to the RRE. The results provide evidence that monomeric zinc fingers can recognize specific nucleic acid sites and that, as for DNA, zinc finger α-helices can bind in the major groove of RNA provided that the groove is sufficiently wide to accommodate an α-helix.
MATERIALS AND METHODS
Metal Binding, Folding, and RNA Binding in Vitro.
Peptides were synthesized on an Applied Biosystems model 432A peptide synthesizer, capped with an acetyl or succinyl group at the N terminus and an amide group at the C terminus, and purified by reverse-phase C4 HPLC. Molecular weights were confirmed by laser desorption MS, and concentrations were determined from amino acid analysis (University of Michigan Protein and Carbohydrate Structure Facility, Ann Arbor). Peptides were: ZF1-Rev, ac-RPYACPVESCDRRFSTRQARRNHRRRHRRG-am; ZF2-Rev, ac-KPFQCRICMRNFSTRQARRNHRRRHRRG-am; ZF2-Rev (C5,8S), ac-KPFQSRISMRNFSTRQARRNHRRRHRRG-am; Rev14 (helical), suc-AAAATRQARRNRRRRRRRAAAAR-am; and Rev14 (H21,H25), suc-AAAATRQARRNHRRRHRRAAAAR-am.
Cobalt absorption spectra were measured on an Aviv model 14DS spectrophotometer at 25°C in 20 mM Tris⋅HCl buffer (pH 7.0) degassed with argon. For each peptide, three sets of spectra were recorded from 800 to 300 nm at 5-nm intervals with a recording time of 2 sec at each interval, and data from the three scans were averaged. Baseline spectra of the reduced peptide and CoCl2 were subtracted. CD spectra were measured on an Aviv model 62DS spectropolarimeter at 4°C in degassed 10 mM Tris⋅HCl buffer (pH 7.5) in the absence or presence of ZnCl2. Five sets of spectra were recorded from 300 to 200 nm at 1-nm intervals with a recording time of 1 sec at each interval, and data from the five scans were averaged. Mean molar ellipticity (θ) was calculated per amino acid residue, and helical content was estimated from the value at 222 nm after baseline subtraction (21).
For RNA-binding assays, internally labeled RNAs were prepared by using T7 RNA polymerase, and gel shift assays were performed by incubating peptide and RNA at 4°C in 10 μl of binding mixtures containing 10 mM Hepes-KOH (pH 7.5), 100 mM KCl, 1 mM MgCl2, 25 μM ZnCl2, 1 mM DTT, 50 μg/ml competitor Escherichia coli tRNA (Sigma), and 10% glycerol. To determine relative binding affinities, 1- to 5-nM radio-labeled RNAs were titrated with peptide, peptide-RNA complexes were resolved on polyacrylamide gels, and free RNA and RNA-peptide complexes were quantitated by using a Molecular Dynamics PhosphorImager.
RNA-Binding Assays in Vivo.
ZF-Rev hybrids were fused to HIV-1 Tat by cloning synthetic oligonucleotides into the EagI and XhoI sites of pSV2tat72, creating fusions after amino acid 49 of Tat (24). Peptide sequences were: Rev14 (nonhelical), TRQARRNRRRRRRR; Rev14 (helical), AAAATRQARRNRRRRRRRAAAAR; Rev14 (H21,H25), AAAATRQARRNHRRRHRRAAAAR; ZF1-Rev, RPYACPVESCDRRFSTRQARRNHRRRHRRG; ZF2-Rev, KPFQCRICMRNFSTRQARRNHRRRHRRG; and alanine-based ZF-Rev, APFACAACAAAFSTRQARRNHRRRHRRG. Activation was measured by cotransfecting 50 ng of an HIV-1 long terminal repeat (LTR)-RRE IIB-chloramphenicol acetyltransferase (CAT) reporter plasmid (21) with 1–50 ng Tat expression plasmids into HeLa cells by using Lipofectin. Total plasmid DNA was adjusted to 1 μg with pUC19. CAT activities were assayed after 48 hr by using an appropriate amount of cell extract (21), and activities were quantitated by using a Molecular Dynamics PhosphorImager. ZF-Rev fusions to λ N were constructed by cloning synthetic oligonucleotides into the BsmI and NcoI sites of pBRptacN* (25), creating fusions before amino acid 20 of N and replacing the RNA-binding domain. Peptide sequences were: Rev14 (helical), MATRQARRNRRRRRRRAAAA; Rev14 (H21,H25), MATRQARRNHRRRHRRAAAA; ZF1-Rev, MARPYACPVESCDRRFSTRQARRNHRRRHRRAAAA; ZF2-Rev, MAKPFQCRICMRNFSTRQARRNHRRRHRRAAAA; and alanine-based ZF-Rev, MAAPFACAACAAAFSTRQARRNHRRRHRRAAAA. Transcriptional antitermination was measured by transforming plasmids into E. coli strain N567 containing a pACYC-derived reporter plasmid in which the λ nut site was replaced by RRE IIB (25). Bacteria were grown at 34°C for 24–48 hr on tryptone agar plates containing 0.05 mg/ml ampicillin, 0.02 mg/ml chloramphenicol, 0.08 mg/ml 5-bromo-4-chloro-3-indolyl β-d-galactoside, and 50 mM isopropyl β-d-thiogalactoside, and blue color colony was estimated visually by using several N-fusion proteins and corresponding reporters as controls (25).
RESULTS
Design of ZF-Rev Peptides.
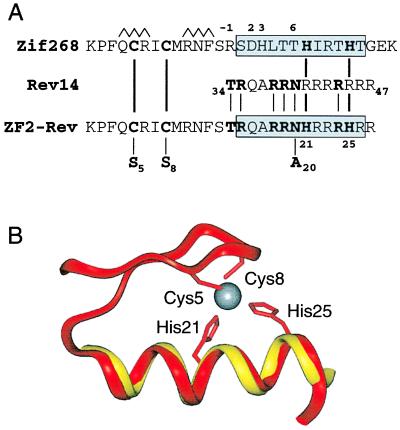
As an isolated peptide, the arginine-rich RNA-binding domain of Rev forms a relatively unstable α-helix that can be partially stabilized by adding chemical blocking groups or alanine residues to the N and C termini (21). Because specific RRE-binding affinity is proportional to α-helix content, we wanted to lock the Rev peptide into a fully folded state that would bind RNA with high affinity and could be expressed in vivo. Because zinc fingers fold as independent structural modules (2, 22, 23) and contain DNA-binding α-helices, they appeared to provide an ideal scaffold in which to engineer a stable RNA-binding helix. By using the crystal structure of the Zif268 protein-DNA complex (3), which contains three zinc fingers, and the NMR structure of a Rev peptide complexed to the RRE IIB hairpin (a high-affinity Rev binding site) (26), we aligned the sequences of the two helices, initially by using finger 2 from Zif (Fig. 1A), to determine whether the two histidines required for zinc binding could be located within the Rev helix without changing the six amino acids required for RRE recognition (21, 26, 27). An alignment was found in which the N-terminal threonine of Rev was placed at position −1 of the zinc finger helix, allowing replacement of two nonessential arginines in Rev with histidines. The backbone atoms of the two helices superimposed with a rms deviation of 0.84 Å (Fig. 1B). When positioned in the RRE site, the β-sheet portion of the finger was located outside the RNA major groove, and no steric clashes were observed (Fig. 2A). A similar arrangement was possible with Zif finger 1 (not shown). The hybrid zinc fingers (ZF2-Rev is shown in Fig. 2B) were designed with shorter helices than the Rev peptide because the C-terminal alanines needed to stabilize the isolated Rev helix were presumed to be unnecessary in the zinc finger context.
Figure 1.
Design of ZF-Rev peptides. (A) The sequence of Rev14 (24) was aligned with the α-helix of Zif268 zinc finger 2 (boxed) such that the histidines required for metal binding and the amino acids required for RRE recognition (shown in bold and indicated by lines) did not overlap. The numbering above the Zif268 α-helix indicates positions (−1, +2, +3, and +6, relative to the start of the helix) typically used for DNA binding, and the β-strands of the finger are indicated by open triangles. The putative α-helix in the hybrid ZF2-Rev is boxed. Positions of cysteine-to-serine mutations and an asparagine-to-alanine mutation are shown. A similar alignment was used to construct a fusion to Zif zinc finger 1 (ZF1-Rev; not shown). (B) Overlap between the α-helical segment of Zif finger 2 (magenta) (3) and residues 34–47 of the Rev peptide (yellow) (26). Backbone heavy atoms were superimposed from Ser-47 to Thr-58 of the finger and from Arg-35 to Arg-46 of the Rev peptide by using insight ii software (Biosym Technologies, San Diego) on a Silicon Graphics workstation. The rms deviation between all 36 superimposed atoms was 0.84 Å, and a similar rms deviation (0.93 Å) was obtained by using Zif finger 1 (not shown). The metal-coordinating cysteine and histidine residues and zinc ion in the finger are shown.
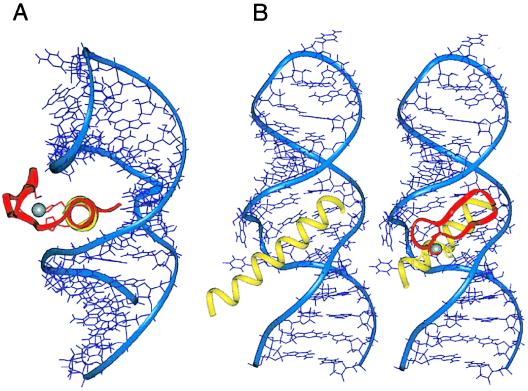
Figure 2.
(A) Proposed orientation of a zinc finger bound to RRE IIB. The α-helices of Rev (yellow) and Zif finger 2 (magenta) were superimposed and the aligned zinc finger was docked against the RNA. The view down the α-helical axes shows that the β-sheet portion of the finger emerges from the major groove and can accommodate the peptide backbone and side chains without steric clashes to the RNA. (B) Comparison of the Rev peptide- and ZF2-Rev-RRE complexes. The α-helix of Zif finger 2 was replaced by residues 34–47 of Rev, with two histidine substitutions (yellow). The β-sheet portion of the finger and the two cysteine ligands are shown in magenta and the zinc ion in white.
Metal Binding and Folding of ZF-Rev Peptides.
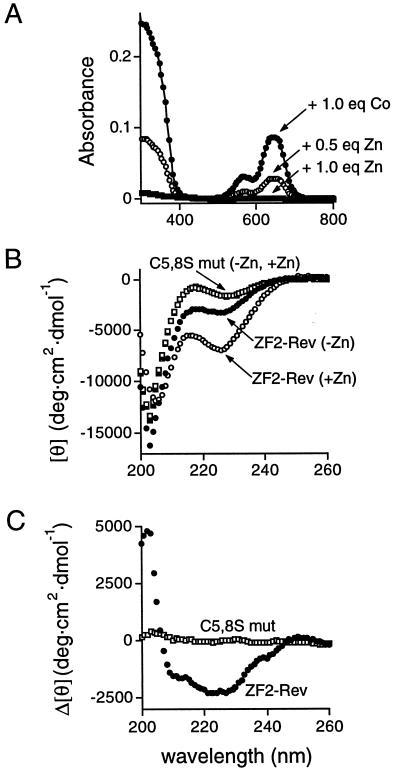
To test whether the modeled ZF-Rev peptides fold into zinc fingers, we measured absorption and CD spectra of synthetic peptides in the absence and presence of metal ions. Upon addition of stoichiometric cobalt, ZF2-Rev displayed a visible absorption spectrum (Fig. 3A) characteristic of a tetrahedrally bound cobalt complex coordinated by two cysteine and two histidine ligands (2). Adding stoichiometric zinc, which produces no absorbance in the visible region when complexed to a zinc finger, eliminated the absorbance of the cobalt complex (Fig. 3A), as expected for competitive, high-affinity zinc binding. Also as expected, a mutant peptide in which the two metal-coordinating cysteines were replaced by serines (C5,8S) showed no cobalt absorbance (data not shown). CD spectra (Fig. 3B) indicated that ZF2-Rev has some α-helical content in the absence of zinc (≈9%) and increases helicity upon zinc addition (to ≈19%), producing a CD spectrum similar to that observed with other DNA-binding fingers (2, 22, 28). In contrast, the C5,8S mutant displayed little helical content in the absence of zinc (≈3%) and no change upon zinc addition (Fig. 3B). The presence of some helix in the absence of zinc is somewhat unusual for zinc finger peptides and may reflect an unusually high helical propensity of the Rev sequence (21). Nonetheless, CD difference spectra showed clear zinc-dependent helix formation for ZF2-Rev but not for the double-cysteine mutant (Fig. 3C). Cobalt absorption and CD experiments with ZF1-Rev produced similar results (data not shown), further indicating that the ZF-Rev peptides have metal-binding and structural characteristics of correctly folded zinc fingers.
Figure 3.
Cobalt absorption spectroscopy and metal-dependent peptide folding. (A) Absorption spectra of ZF2-Rev (140 μM) in the presence of stoichiometric CoCl2 (●), or in the presence of 210 μM CoCl2 plus 70 μM (○) or 140 μM (■) ZnCl2 competitor. The spectra shown are characteristic of tetrahedral metal coordination in a monomeric zinc finger; red-shifted spectra have been observed with misfolded zinc finger dimers (29) and we observed similar shifts at low metal stoichiometries (data not shown), suggesting that incorrect complexes may be populated initially. (B) CD spectra of ZF2-Rev and a double-cysteine mutant (C5,8S): reduced ZF2-Rev in the absence of metal (●), ZF2-Rev in the presence of stoichiometric ZnCl2 (○), ZF2-Rev (C5,8S) in the absence of metal (■), and ZF2-Rev (C5,8S) in the presence of stoichiometric ZnCl2 (□). Spectra were recorded at 4°C with peptide concentrations of 30–60 μM. (C) Difference CD spectra of ZF2-Rev (●) and ZF2-Rev (C5,8S) (□), calculated by subtracting spectra in the absence of zinc from spectra in the presence of zinc (from B). The difference spectrum of ZF2-Rev shows two minima (at 208 and 222 nm) characteristic of α-helix formation whereas the spectrum of the cysteine mutant (see B) indicates little structure and no metal-dependent folding. Because ZF2-Rev showed some helix formation in the absence of zinc and had a higher helical content than ZF2-Rev (C5,8S), we performed several experiments to rule out the presence of adventitious zinc ions. We observed little change in the CD spectra by using buffers extensively treated with Chelex 100 chelating resin or in the presence of excess chelators (EDTA or 1,10-phenanthroline). Furthermore, careful titrations with cobalt and zinc showed no change in the visible absorption spectra after addition of stoichiometric CoCl2 and competition with stoichiometric amounts of ZnCl2, suggesting that little or no zinc was prebound to the peptide. In addition, 1D and 2D proton NMR spectra indicate that the peptide is unstructured in the absence of ZnCl2 and becomes ordered upon titration of stoichiometric ZnCl2 (W. Gmeiner, D.J.M., and A.D.F., unpublished data).
Metal-Dependent RNA Binding in Vitro.
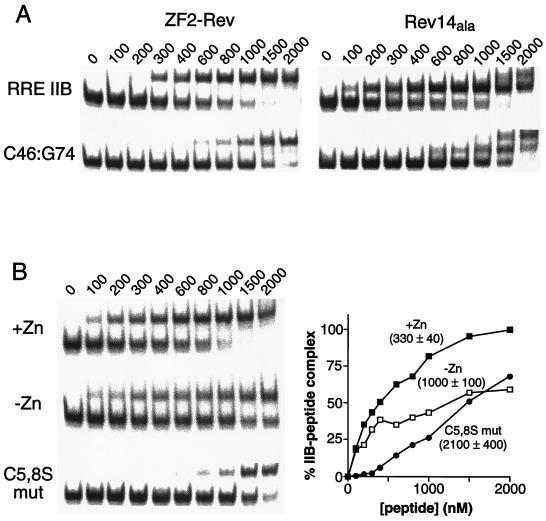
We next measured the RNA-binding affinities of synthetic peptides for the RRE IIB site by using gel shift assays. In the presence of zinc, ZF2-Rev bound with an affinity similar to that of an α-helical Rev peptide containing flanking alanines that stabilize the Rev helix (apparent Kds were 330 ± 40 versus 330 ± 20 nM; Fig. 4A). Previous experiments have shown that adding flanking alanines to the Rev sequence increases helical content to ≈50% (21). Both peptides bound with ≈5-fold lower affinity to a C46-G74 mutant (which reverses a base pair critical for recognition; refs. 21, 26, and 27), confirming that the designed finger binds RRE IIB with an affinity and specificity similar to a helical Rev peptide. Two experiments indicate that RNA binding is zinc dependent. First, the affinity of ZF2-Rev was ≈3-fold higher in the presence of zinc than in the absence of zinc (Fig. 4B), correlating with the 2- to 3-fold increase in helical content determined by CD. Second, the double-cysteine mutant, which contains even less helix and shows no zinc-dependent helix formation, bound RRE IIB ≈7-fold less tightly than ZF2-Rev (Fig. 4B), further indicating that specific RNA binding is coupled to zinc-dependent folding of the finger and stabilization of the α-helix.
Figure 4.
RRE IIB binding by ZF2-Rev in vitro. (A) Gel shift assays of ZF2-Rev and Rev14ala peptides with wild-type RRE IIB RNA and a C46-G74 mutant (that reverses a base pair critical for recognition; refs. 21, 26, and 27) at the peptide concentrations (nM) indicated. ZF2-Rev binds with a similar affinity and specificity as the helical Rev peptide; apparent binding constants are substantially tighter in the absence of competitor tRNA (21, 33). (B, Left) Zinc-dependent RRE IIB RNA binding: ZF2-Rev in the presence of 25 μM ZnCl2 (■), in the absence of zinc and with addition of 0.5 mM EDTA (□), and ZF2-Rev (C5,8S) in the presence of 25 μM ZnCl2 (●). (Right) Binding curves quantitated by PhosphorImager analyses and apparent dissociation constants (nM) estimated by fitting to standard binding equations.
RNA-Binding Activities of ZF-Rev Peptides in Vivo.
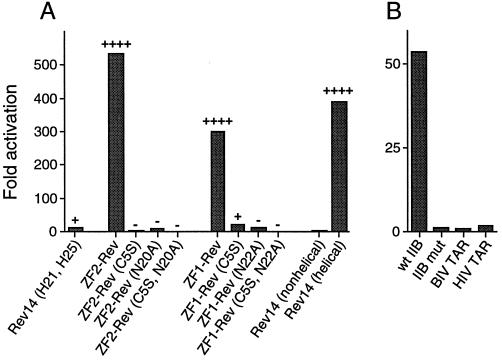
To test whether the ZF-Rev hybrids are able to bind the RRE in the environment of a mammalian cell, we fused ZF1-Rev or ZF2-Rev fingers to the activation domain of HIV-1 Tat and measured the ability of the Tat fusions to activate transcription of an HIV-1 LTR-CAT reporter containing the RRE IIB site in HeLa cells (21). The designed ZF1-Rev and ZF2-Rev peptides displayed strong activities on the RRE reporter whereas a Rev peptide containing the two histidines but without the rest of the zinc finger framework [Rev14 (H21,H25)] showed little activity (Fig. 5A). For comparison, the extent of activation by the hybrid fingers was similar to that observed when the wild-type Rev helix was stabilized by the addition of flanking alanine residues [compare Rev14 (nonhelical) and Rev14 (helical); Fig. 5A]. It is interesting that the Rev14 (H21,H25) peptide is inactive when not in a finger context even though it contains flanking alanines and likely has some helical content. Indeed, CD experiments indicate that the histidine-containing and wild-type peptides have similar α-helical content when placed in the alanine context (≈18%; data not shown), yet the histidine peptide requires the zinc finger context for RRE binding in vivo.
Figure 5.
ZF-Rev binding to RRE IIB in vivo. (A) ZF-Rev peptides were fused to the activation domain of HIV-1 Tat, and activation of an HIV-1 LTR-CAT reporter containing RRE IIB (21) was measured in HeLa cells (bars). ZF-Rev peptides also were fused to the bacteriophage λ N protein and transcriptional antitermination of an RRE IIB-lacZ reporter (25) was scored in E. coli (+ or − above bars). For CAT assays, 10 ng of each Tat-fusion plasmid was cotransfected with 50 ng of IIB reporter plasmid, and fold activation refers to the ratio of activities generated by the Tat fusion proteins to the activity in the absence of Tat. For β-galactosidase scoring, ++++ represents the darkest blue colonies and − represents white colonies. (B) Activity of ZF2-Rev was determined by CAT assays as in A by using reporter plasmids containing the wild-type IIB RRE site, a C46-G74 mutant that reverses a base pair critical for recognition (21, 26, 27), or the bovine immunodeficiency virus (BIV) or HIV-1 TAR hairpins.
The activity of the ZF-Rev peptides in vivo requires a specific RNA-binding site, an intact zinc finger structure, and specific RRE-binding residues. ZF-Rev peptides were inactive on reporters containing a mutant RRE IIB site (the C46-G74 mutant that reverses a critical base pair) or heterologous RNA-binding sites (Fig. 5B), indicating that the interaction is highly specific. Substitution of a single cysteine residue required for zinc binding, or an asparagine that makes an essential contact to a G:A base pair, eliminated activity (Fig. 5A), indicating that both the finger structure and RRE-binding residues are required. The Rev helix also was placed in the context of a simplified zinc finger containing alanines at all positions not essential for finger folding (29) and weak but measurable RRE-binding activity was observed (data not shown), further indicating that the Rev helix can be accommodated by different finger frameworks.
We constructed an additional series of fusion proteins in which ZF-Rev peptides were fused to the N terminus of the bacteriophage λ N protein and measured the ability of the N fusions to antiterminate transcription of an RRE IIB-lacZ reporter in bacteria (25). Results virtually identical to those obtained in the mammalian system were seen with ZF1-Rev, ZF2-Rev, and the mutant peptides in bacteria (Fig. 5A), indicating that the designed ZF-Rev peptides fold correctly and specifically recognize the RRE IIB site in vitro, in eukaryotic cells, and in prokaryotic cells.
DISCUSSION
We have demonstrated that the zinc finger motif can provide an excellent scaffold for stabilizing a monomeric α-helix and presenting the helix to an RNA major groove. This work also demonstrates the feasibility of using structure-based approaches to design a sequence-specific RNA-binding protein. It remains to be determined whether similar strategies can be used to design zinc fingers that recognize RNA sites other than the RRE, which has evolved to bind a monomeric helix. To date, only two other examples have been reported in which monomeric helices are used to recognize RNA. The bacteriophage λ and P22 N proteins each contain an arginine-rich helix that binds to the terminal loop of an RNA hairpin (30, 31), but in both cases the helices are bent and are unlikely to be accommodated within a zinc finger framework. Although it may not be possible at this time to directly design other RNA-binding zinc fingers, our results suggest that monomeric fingers are likely to be good starting points for selection experiments by using the RRE or other RNA sites. Recent phage display experiments have identified two-finger sequences that recognize 5S rRNA and RRE IIB sites (17, 18), highlighting the versatility of the zinc finger motif in RNA recognition.
Recognition of the RRE by Rev requires two purine-purine base pairs to help widen the RNA major groove, allowing deep penetration of the arginine-rich α-helix (26, 27). Our results are consistent with major groove binding of the zinc finger helix, perhaps reflecting a general mode of zinc finger docking to nucleic acids as previously suggested based on mutagenesis of α-helical residues in the RNA-binding fingers of TFIIIA (17). The structures of zinc finger-DNA complexes (3, 5–9) show that the α-helices usually make base-specific contacts in the major groove, but substantial differences are observed in the orientation of the helices and some fingers bind outside the groove through nonspecific interactions (5, 9). Most of the canonical DNA-binding fingers use side chains located at positions −1, +2, +3, and +6 of the helix to make base-specific contacts (Fig. 1A) whereas we presume that the residues known to be important for Rev binding, located at positions −1, +1, +4, +5, +6, and +10, are used by the ZF-Rev peptides. The orientation of the Rev helix in the major groove is quite different from that observed in canonical zinc finger-DNA complexes, with the helix buried deeply in the major groove and forming an extensive RNA-protein interface (26, 27). This arrangement is likely to be preserved in the ZF-Rev complexes, which show the same requirement for asparagine at position +6 (Fig. 5A) and for the G46:C74 base pair (Figs. 4A and 5B) as the Rev peptide-RRE IIB complex (21). NMR structures of the Rev peptide complex (26, 27) show a clear interaction between Asn-40 and the G:A base pair in the major groove and a possible interaction between Arg-44 and G46.
In addition to likely differences in positioning of the helix in the major groove, the results highlight other features that may differ between zinc finger-DNA and zinc finger-RNA recognition. To date it has been observed that multiple fingers are required for sequence-specific DNA recognition, in part because side chains from adjacent fingers are used to contact a single base pair and perhaps also because multiple fingers, as well as the intervening linker, help establish the orientation of helix binding (8). In contrast, the tertiary architecture of an RNA can be sufficient to provide a defined framework for orienting a monomeric helix within the groove, as in the case of the RRE-helix interaction (21). Furthermore, given the wide diversity of RNA tertiary structure, it seems plausible that others parts of a zinc finger not typically used in DNA binding can be used to contact RNA, either in conjunction with the helix or perhaps even by using the β-sheet as the primary binding surface. It will be instructive to engineer additional contacts into our ZF-Rev-RRE complexes to further probe which parts of a finger can contact RNA, and it will be particularly interesting to test whether interactions can be introduced from the β-turn, which is modeled to be close to the RRE (Fig. 2 A and B).
Further structure-based design may lead to the discovery of ZF-Rev variants with RRE-binding affinities and specificities higher than Rev. As observed for other types of RRE-binding peptides, the evolution of tighter binders may enhance their effectiveness as competitors of Rev binding and function in vivo (32). Given the apparent facility of zinc fingers to fold in different cellular environments, the zinc finger framework may provide an excellent means to express peptide-based inhibitors. It seems likely that future structure-based and combinatorial experiments with monomeric and multimeric zinc fingers will identify novel and interesting modes of RNA recognition and perhaps provide new ways to target specific RNA sites for therapeutic intervention.
Acknowledgments
We thank Bernhard Walberer for help with computer modeling, Donna Campisi and Colin Smith for helpful suggestions, and Raul Andino, Judith Frydman, Bill Gmeiner, Peter Walter, and members of the Frankel laboratory for comments on the manuscript. This work was supported by grants from the National Institutes of Health and the University of California Universitywide AIDS Research Program.
ABBREVIATIONS
- RRE
Rev response element
- ZF-Rev
zinc finger-Rev
- CAT
chloramphenicol acetyltransferase
- LTR
long terminal repeat
References
- 1.Miller J, McLachlan A D, Klug A. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frankel A D, Berg J M, Pabo C O. Proc Natl Acad Sci USA. 1987;84:4841–4845. doi: 10.1073/pnas.84.14.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavletich N P, Pabo C O. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 4.Berg J M, Shi Y. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 5.Pavletich N P, Pabo C O. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 6.Fairall L, Schwabe J W, Chapman L, Finch J T, Rhodes D. Nature (London) 1993;366:483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 7.Kim C A, Berg J M. Nat Struct Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- 8.Wuttke D S, Foster M P, Case D A, Gottesfeld J M, Wright P E. J Mol Biol. 1997;273:183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- 9.Nolte R T, Conlin R M, Harrison S C, Brown R S. Proc Natl Acad Sci USA. 1998;95:2938–2943. doi: 10.1073/pnas.95.6.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rebar E J, Pabo C O. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 11.Choo Y, Sanchez G I, Klug A. Nature (London) 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 12.Jamieson A C, Kim S H, Wells J A. Biochemistry. 1994;33:5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- 13.Greisman H A, Pabo C O. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 14.Joho K E, Darby M K, Crawford E T, Brown D D. Cell. 1990;61:293–300. doi: 10.1016/0092-8674(90)90809-s. [DOI] [PubMed] [Google Scholar]
- 15.Theunissen O, Rudt F, Guddat U, Mentzel H, Pieler T. Cell. 1992;71:679–690. doi: 10.1016/0092-8674(92)90601-8. [DOI] [PubMed] [Google Scholar]
- 16.Clemens K R, Wolf V, McBryant S J, Zhang P, Liao X, Wright P E, Gottesfeld J M. Science. 1993;260:530–533. doi: 10.1126/science.8475383. [DOI] [PubMed] [Google Scholar]
- 17.Friesen W J, Darby M K. J Biol Chem. 1997;272:10994–10997. doi: 10.1074/jbc.272.17.10994. [DOI] [PubMed] [Google Scholar]
- 18.Friesen W J, Darby M K. Nat Struct Biol. 1998;5:543–546. doi: 10.1038/794. [DOI] [PubMed] [Google Scholar]
- 19.McBryant S J, Veldhoen N, Gedulin B, Leresche A, Foster M P, Wright P E, Romaniuk P J, Gottesfeld J M. J Mol Biol. 1995;248:44–57. doi: 10.1006/jmbi.1995.0201. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, Berg J M. Science. 1995;268:282–284. doi: 10.1126/science.7536342. [DOI] [PubMed] [Google Scholar]
- 21.Tan R, Chen L, Buettner J A, Hudson D, Frankel A D. Cell. 1993;73:1031–1040. doi: 10.1016/0092-8674(93)90280-4. [DOI] [PubMed] [Google Scholar]
- 22.Parraga G, Horvath S J, Eisen A, Taylor W E, Hood L, Young E T, Klevit R E. Science. 1988;241:1489–1492. doi: 10.1126/science.3047872. [DOI] [PubMed] [Google Scholar]
- 23.Lee M S, Gippert G P, Soman K V, Case D A, Wright P E. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- 24.Tan R, Frankel A D. Proc Natl Acad Sci USA. 1998;95:4247–4252. doi: 10.1073/pnas.95.8.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harada K, Martin S S, Frankel A D. Nature (London) 1996;380:175–179. doi: 10.1038/380175a0. [DOI] [PubMed] [Google Scholar]
- 26.Battiste J L, Mao H, Rao N S, Tan R, Muhandiram D R, Kay L E, Frankel A D, Williamson J R. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 27.Ye X, Gorin A, Ellington A D, Patel D J. Nat Struct Biol. 1996;3:1026–1033. doi: 10.1038/nsb1296-1026. [DOI] [PubMed] [Google Scholar]
- 28.Kim C A, Berg J M. Nature (London) 1993;362:267–270. doi: 10.1038/362267a0. [DOI] [PubMed] [Google Scholar]
- 29.Michael S F, Kilfoil V J, Schmidt M H, Amann B T, Berg J M. Proc Natl Acad Sci USA. 1992;89:4796–4800. doi: 10.1073/pnas.89.11.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Legault P, Li J, Mogridge J, Kay L E, Greenblatt J. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 31.Cai Z, Gorin A, Frederick R, Ye X, Hu W, Majumdar A, Kettani A, Patel D J. Nat Struct Biol. 1998;5:203–212. doi: 10.1038/nsb0398-203. [DOI] [PubMed] [Google Scholar]
- 32.Harada K, Martin S S, Tan R, Frankel A D. Proc Natl Acad Sci USA. 1997;94:11887–11892. doi: 10.1073/pnas.94.22.11887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan R, Frankel A D. Biochemistry. 1994;33:14579–14585. doi: 10.1021/bi00252a025. [DOI] [PubMed] [Google Scholar]