Abstract
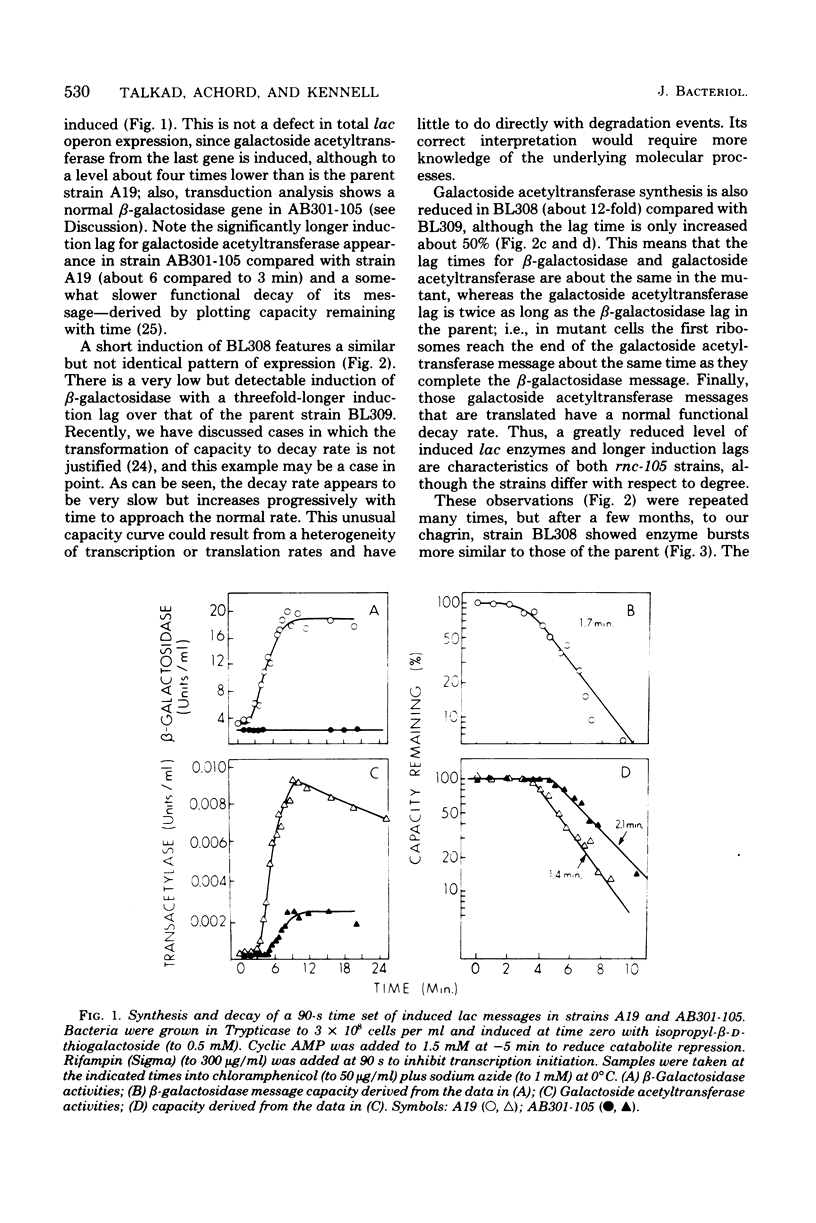
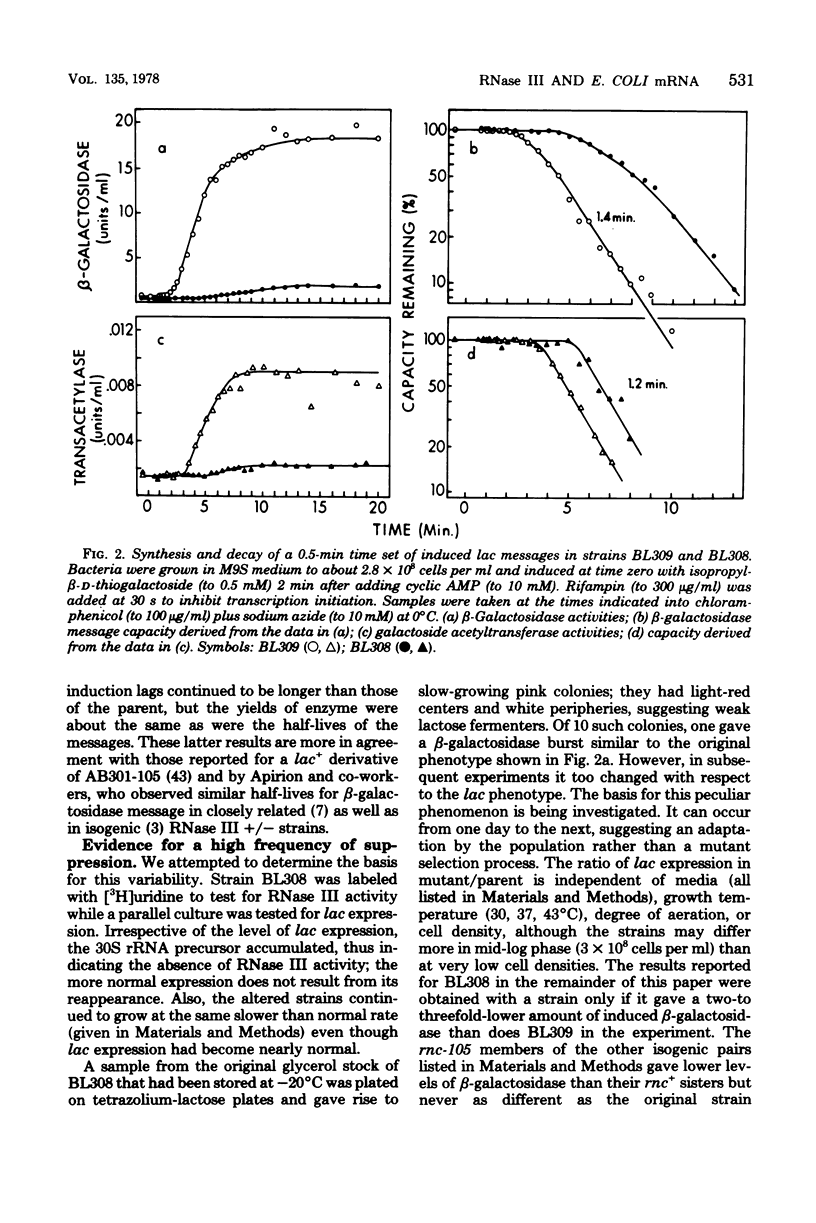
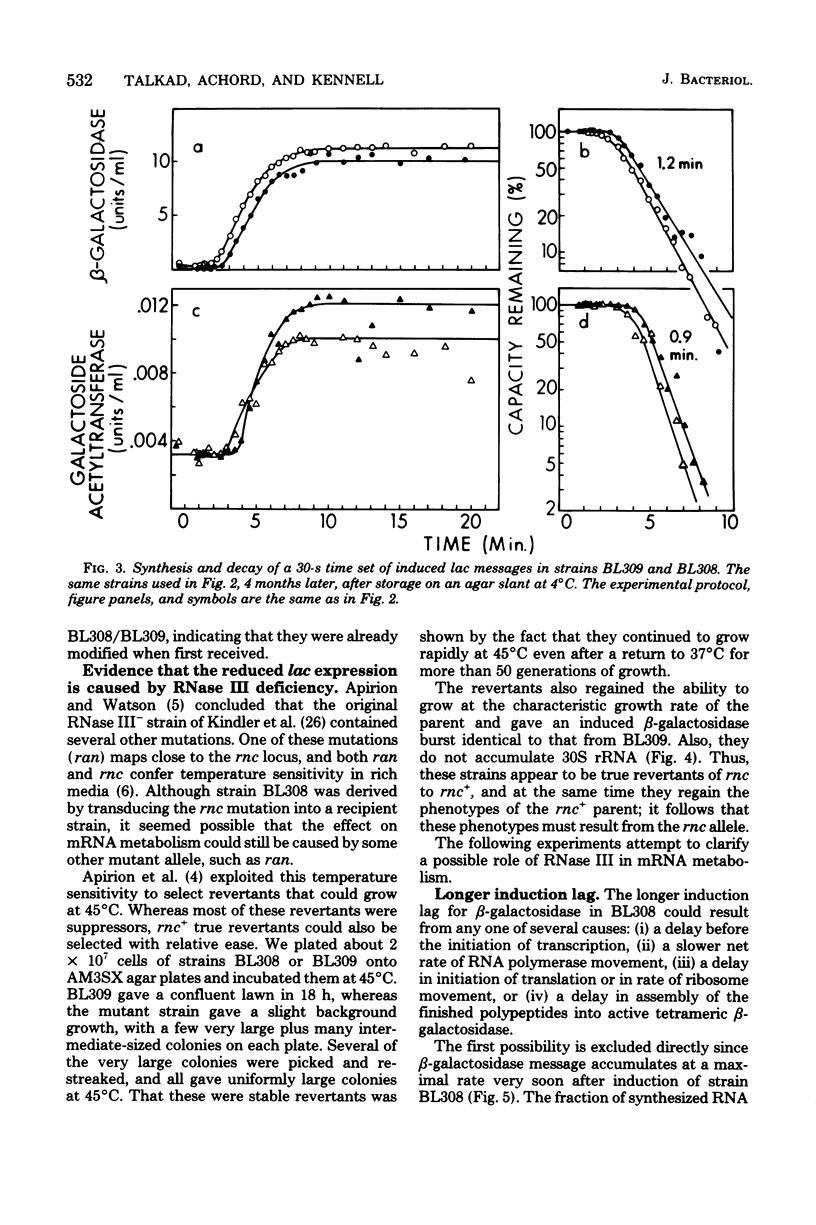
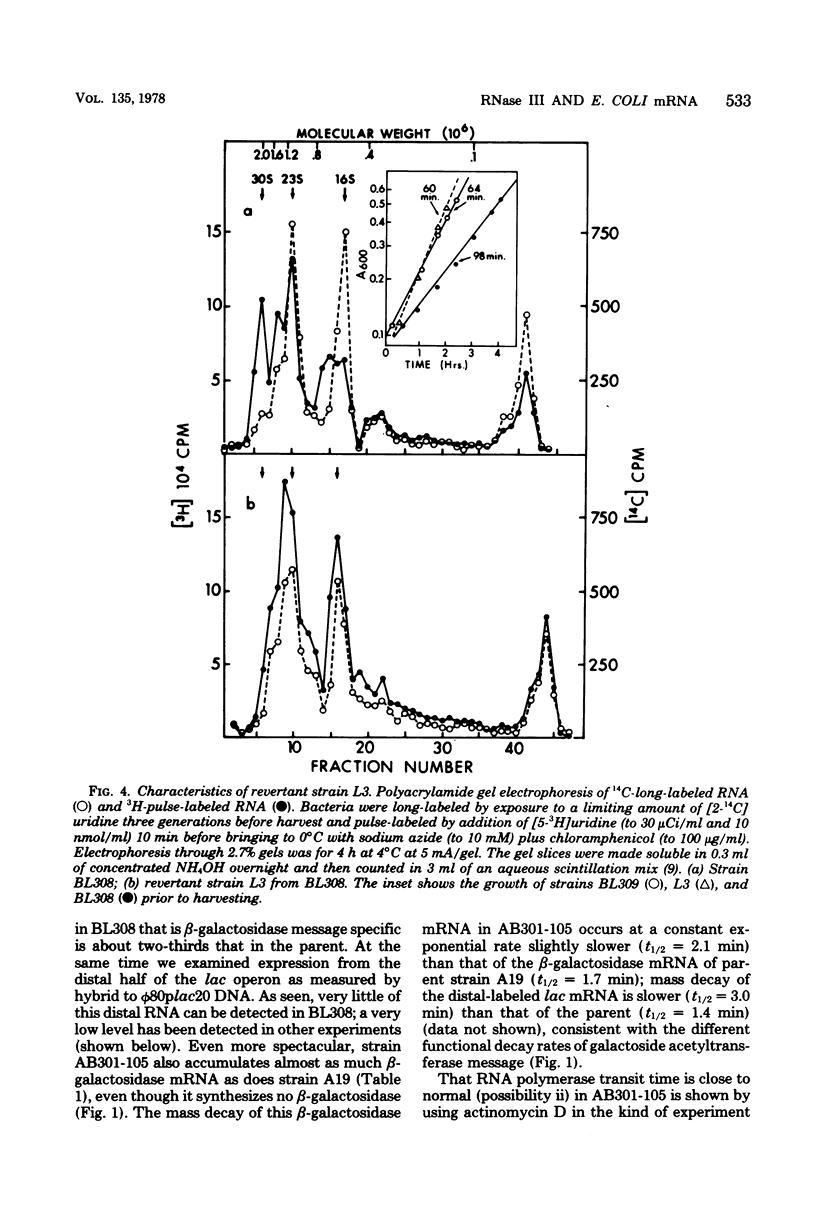
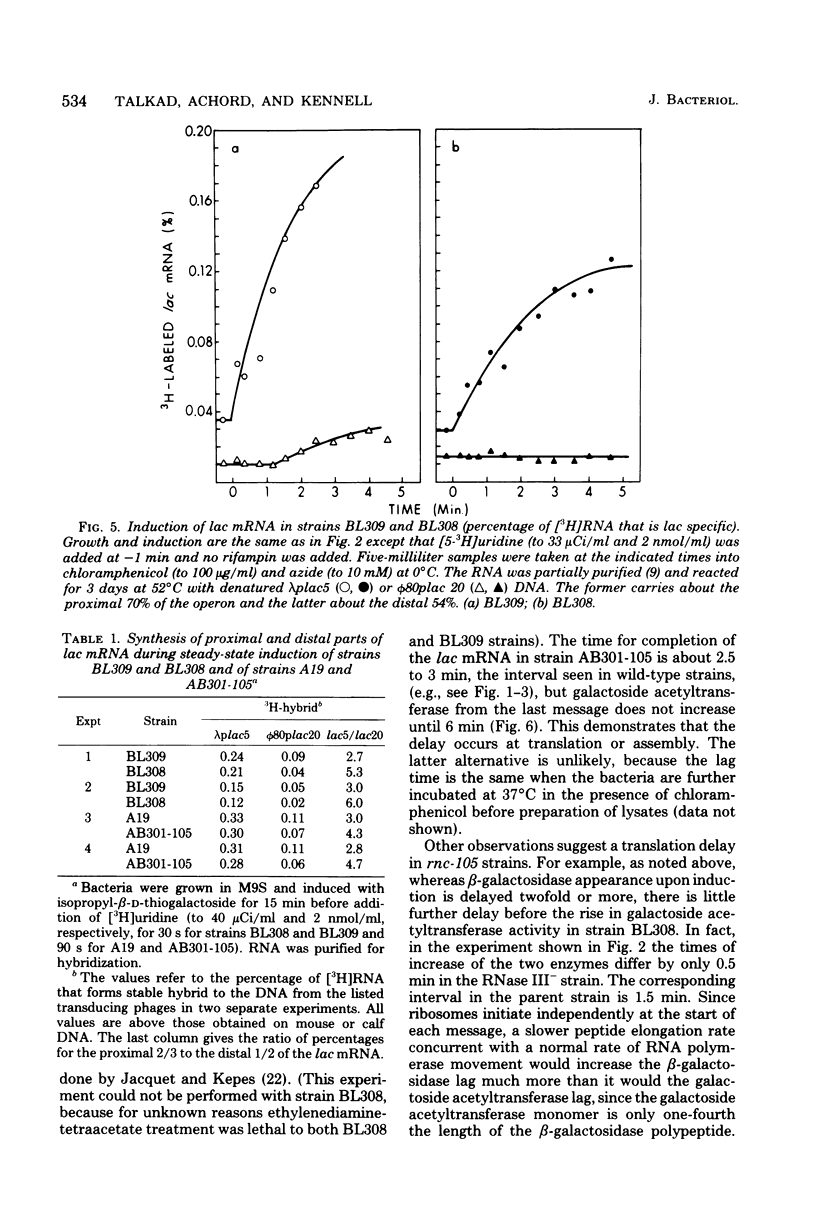
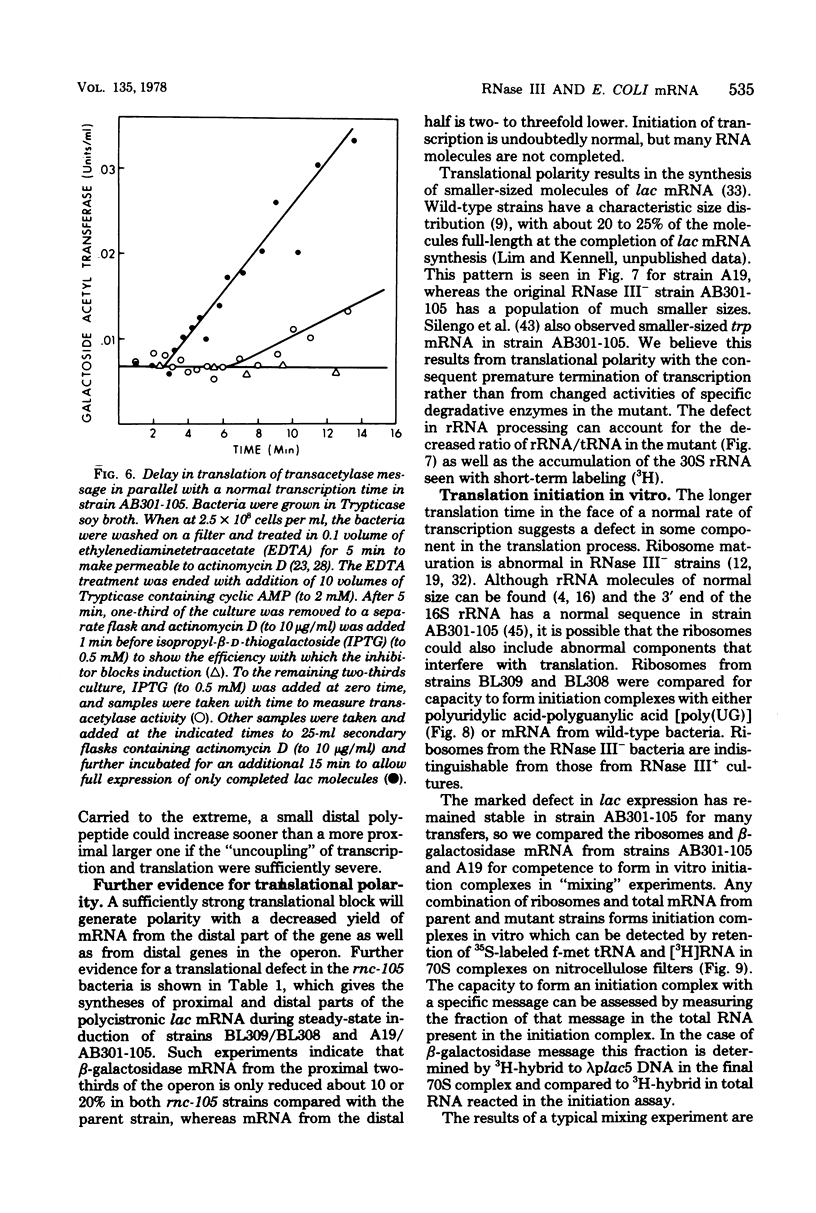
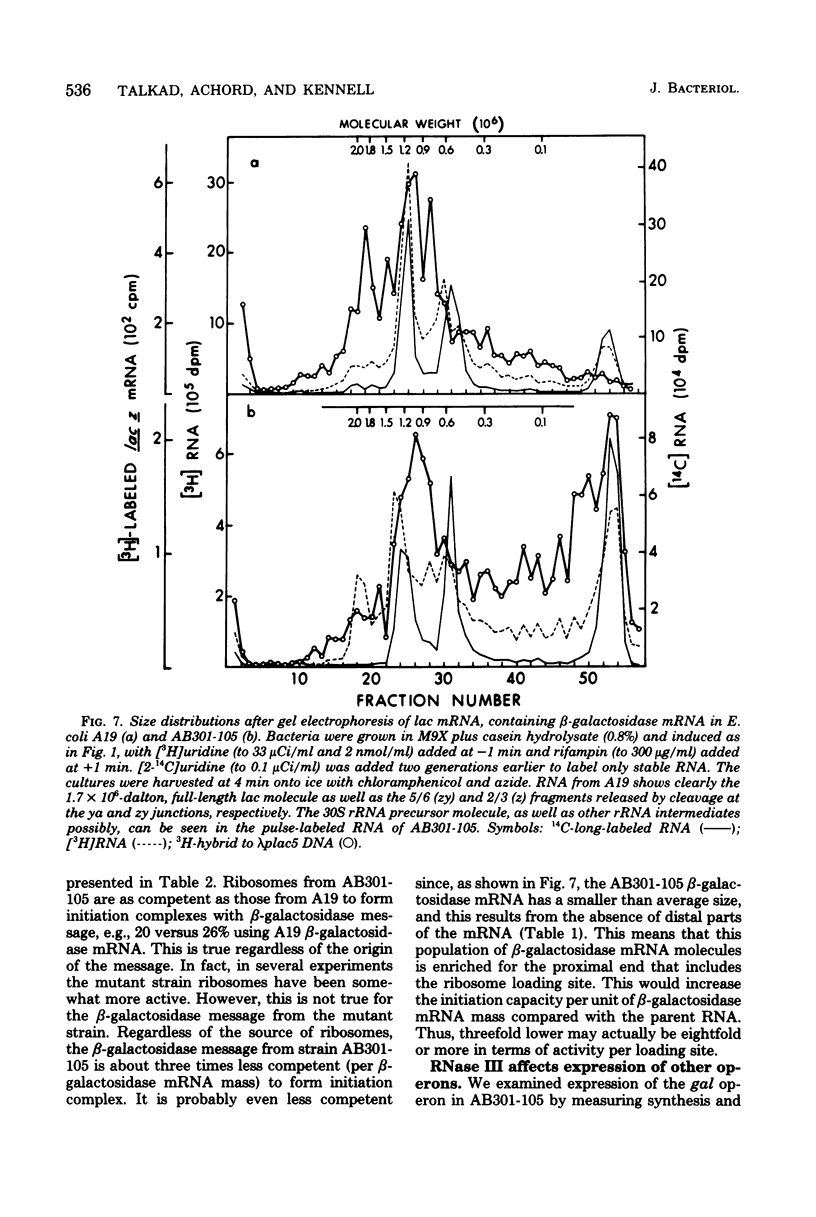
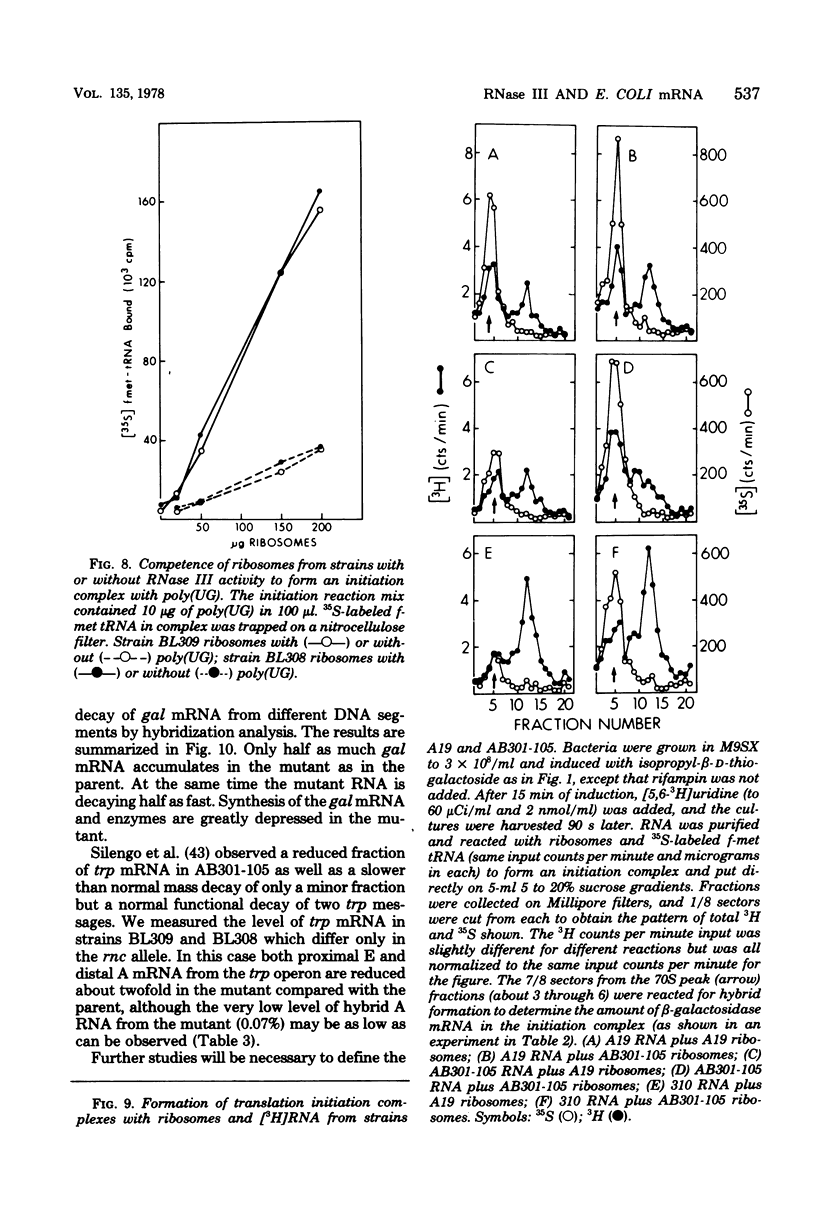
The metabolism of mRNA from the lactose (lac) operon of Escherichia coli has been studied in ribonuclease (RNase) III-deficient strains (rnc-105). The induction lag for beta-galactosidase from the first gene was twice as long, and enzyme synthesis was reduced 10-fold in one such mutant compared with its isogenic rnc+ sister; in the original mutant strain AB301-105, synthesis of beta-galactosidase was not even detectable, although transduction analysis revealed the presence of a normal lac operon. This defect does not reflect a loss of all lac operon activity galactoside acetyltransferase from the last gene was synthesized even in strain AB301-105 but at a rate several times lower than normal. Hybridization analyses suggested that both the frequency of transcription initiation and the time to transcribe the entire operon are normal in rnc-105 strains. The long induction lag was caused by a longer translation time. This defect led to translational polarity with reduced amounts of distal mRNA to give a population of smaller-sized lac mRNA molecules. All these pleiotropic effects seem to result from RNase III deficiency, since it was possible to select revertants to rnc+ that grew and expressed the lac operon at normal rates. However, the rnc-105 isogenic strains (but not AB301-105) also changed very easily to give a more normal rate of beta-galactosidase synthesis without regaining RNase III activity or a faster growth rate. The basis for this reversion is not known; it may represent a "phenotypic suppression" rather than result from a stable genetic change. Such suppressor effects could account for earlier reports of a noninvolvement of RNase III in mRNA metabolism in deliberately selected lac+ rnc-105 strains. The ribosomes from rnc-105 strains were as competent as ribosomes from rnc+ strains to form translation initiation complexes in vitro. However, per mass, beta-galactosidase mRNA from AB301-105 was at least three times less competent to form initiation complexes than was A19 beta-galactosidase mRNA. RNase III may be important in the normal cell to prepare lac mRNA for translation initiation. A defect at this step could account for all the observed changes in lac expression. A potential target within a secondary structure at the start of the lac mRNA is considered. Expression of many operons may be affected by RNase III activity; gal and trp operon expressions were also abnormal in RNase III- strains.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achord D., Kennell D. Metabolism of messenger RNA from the gal operon of Escherichia coli. J Mol Biol. 1974 Dec 15;90(3):581–599. doi: 10.1016/0022-2836(74)90236-8. [DOI] [PubMed] [Google Scholar]
- Anderson E. H. Growth Requirements of Virus-Resistant Mutants of Escherichia Coli Strain "B". Proc Natl Acad Sci U S A. 1946 May;32(5):120–128. doi: 10.1073/pnas.32.5.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apirion D., Neil J., Watson N. Consequences of losing ribonuclease III on the Escherichia coli cell. Mol Gen Genet. 1976 Mar 22;144(2):185–190. doi: 10.1007/BF02428107. [DOI] [PubMed] [Google Scholar]
- Apirion D., Neil J., Watson N. Revertants from RNase III negative strains of Escherichia coli. Mol Gen Genet. 1976 Dec 8;149(2):201–210. doi: 10.1007/BF00332890. [DOI] [PubMed] [Google Scholar]
- Apirion D., Watson N. Analysis of an Escherichia coli strain carrying physiologically compensating mutations one of which causes an altered ribonuclease 3. Mol Gen Genet. 1974;132(2):89–104. doi: 10.1007/BF00272175. [DOI] [PubMed] [Google Scholar]
- Apirion D., Watson N. Mapping and characterization of a mutation in Escherichia coli that reduces the level of ribonuclease III specific for double-stranded ribonucleic acid. J Bacteriol. 1975 Oct;124(1):317–324. doi: 10.1128/jb.124.1.317-324.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apirion D., Watson N. Ribonuclease III is involved in motility of Escherichia coli. J Bacteriol. 1978 Mar;133(3):1543–1545. doi: 10.1128/jb.133.3.1543-1545.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apirion D., Watson N. Unaltered stability of newly synthesized RNA in strains of Escherichia coli missing a ribonuclease specific for double-stranded RNA. Mol Gen Genet. 1975;136(4):317–326. doi: 10.1007/BF00341716. [DOI] [PubMed] [Google Scholar]
- Blundell M., Kennell D. Evidence for endonucleolytic attack in decay of lac messenger RNA in Escherichia coli. J Mol Biol. 1974 Feb 25;83(2):143–161. doi: 10.1016/0022-2836(74)90385-4. [DOI] [PubMed] [Google Scholar]
- Crouch R. J. Ribonuclease 3 does not degrade deoxyribonucleic acid-ribonucleic acid hybrids. J Biol Chem. 1974 Feb 25;249(4):1314–1316. [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Barnes W. M., Reznikoff W. S. Genetic regulation: the Lac control region. Science. 1975 Jan 10;187(4171):27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Effect of RNAase III, cleavage on translation of bacteriophage T7 messenger RNAs. J Mol Biol. 1975 Dec 15;99(3):487–499. doi: 10.1016/s0022-2836(75)80140-9. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Processing transcription, and translation of bacteriophage T7 messenger RNAs. Brookhaven Symp Biol. 1975 Jul;(26):267–276. [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs are generated by site-specific cleavages. Proc Natl Acad Sci U S A. 1973 May;70(5):1559–1563. doi: 10.1073/pnas.70.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenheimer P., Watson N., Apirion D. Multiple pathways for primary processing of ribosomal RNA in Escherichia coli. J Biol Chem. 1977 May 10;252(9):3064–3073. [PubMed] [Google Scholar]
- Gilbert W., Maxam A. The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg D., Steitz J. A. The 30 S ribosomal precursor RNA from Escherichia coli. A primary transcript containing 23 S, 16 S, and 5 S sequences. J Biol Chem. 1975 Jul 25;250(14):5647–5654. [PubMed] [Google Scholar]
- Hercules K., Schweiger M., Sauerbier W. Cleavage by RNase 3 converts T3 and T7 early precursor RNA into translatable message. Proc Natl Acad Sci U S A. 1974 Mar;71(3):840–844. doi: 10.1073/pnas.71.3.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto F. Evidence for premature termination of transcription of the tryptophan operon in polarity mutants of Escherichia coli. Nature. 1970 Oct 17;228(5268):232–235. doi: 10.1038/228232a0. [DOI] [PubMed] [Google Scholar]
- Jacquet M., Kepes A. Initiation, elongation and inactivation of lac messenger RNA in Escherichia coli studied studied by measurement of its beta-galactosidase synthesizing capacity in vivo. J Mol Biol. 1971 Sep 28;60(3):453–472. doi: 10.1016/0022-2836(71)90181-1. [DOI] [PubMed] [Google Scholar]
- KEPES A. KINETICS OF INDUCED ENZYME SYNTHESIS. DETERMINATION OF THE MEAN LIFE OF GALACTOSIDASE-SPECIFIC MESSENGER RNA. Biochim Biophys Acta. 1963 Oct 15;76:293–309. [PubMed] [Google Scholar]
- Kennell D., Simmons C. Synthesis and decay of messenger ribonucleic acid from the lactose operon of Escherichia coli during amino-acid starvation. J Mol Biol. 1972 Oct 14;70(3):451–464. doi: 10.1016/0022-2836(72)90552-9. [DOI] [PubMed] [Google Scholar]
- Kennell D., Talkad V. Messenger RNA potential and the delay before exponential decay of messages. J Mol Biol. 1976 Jun 14;104(1):285–298. doi: 10.1016/0022-2836(76)90014-0. [DOI] [PubMed] [Google Scholar]
- Kindler P., Keil T. U., Hofschneider P. H. Isolation and characterization of a ribonuclease 3 deficient mutant of Escherichia coli. Mol Gen Genet. 1973 Oct 16;126(1):53–59. doi: 10.1007/BF00333481. [DOI] [PubMed] [Google Scholar]
- Kramer R. A., Rosenberg M., Steitz J. A. Nucleotide sequences of the 5' and 3' termini of bacteriophage T7 early messenger RNAs synthesized in vivo: evidence for sequence specificity in RNA processing. J Mol Biol. 1974 Nov 15;89(4):767–776. doi: 10.1016/0022-2836(74)90051-5. [DOI] [PubMed] [Google Scholar]
- Leive L., Kollin V. Synthesis, utilization and degradation of lactose operon mRNA in Escherichia coli. J Mol Biol. 1967 Mar 14;24(2):247–259. doi: 10.1016/0022-2836(67)90330-0. [DOI] [PubMed] [Google Scholar]
- Lozeron H. A., Anevski P. J., Apirion D. Antitermination and absence of processing of the leftward transcript of coliphage lambda in the RNAase III-deficient host. J Mol Biol. 1977 Jan 15;109(2):359–365. doi: 10.1016/s0022-2836(77)80039-9. [DOI] [PubMed] [Google Scholar]
- Maizels N. M. The nucleotide sequence of the lactose messenger ribonucleic acid transcribed from the UV5 promoter mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3585–3589. doi: 10.1073/pnas.70.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D. E., Mosteller R. D., Yanofsky C. Dynamics of synthesis, translation, and degradation of trp operon messenger RNA in E. coli. Cold Spring Harb Symp Quant Biol. 1969;34:725–740. doi: 10.1101/sqb.1969.034.01.082. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Silengo L., Schlessinger D. Synthesis of a large precursor to ribosomal RNA in a mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3361–3365. doi: 10.1073/pnas.70.12.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastushok C., Kennell D. Residual polarity and transcription-translation coupling during recovery from chloramphenicol or fusidic acid. J Bacteriol. 1974 Feb;117(2):631–640. doi: 10.1128/jb.117.2.631-640.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel M., Herzberg M., Greenshpan H. Initiator protein dependent binding of messenger RNA to the ribosome. Cold Spring Harb Symp Quant Biol. 1969;34:261–275. doi: 10.1101/sqb.1969.034.01.032. [DOI] [PubMed] [Google Scholar]
- Richardson J. P., Grimley C., Lowery C. Transcription termination factor rho activity is altered in Escherichia coli with suA gene mutations. Proc Natl Acad Sci U S A. 1975 May;72(5):1725–1728. doi: 10.1073/pnas.72.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D., Dickson E., Dunn J. J. A nucleotide sequence from a ribonuclease III processing site in bacteriophage T7 RNA. Proc Natl Acad Sci U S A. 1977 Mar;74(3):822–826. doi: 10.1073/pnas.74.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D., Dunn J. J. Ribonucleic acid processing activity of Escherichia coli ribonuclease III. J Biol Chem. 1975 Apr 25;250(8):3050–3056. [PubMed] [Google Scholar]
- Robertson H. D., Webster R. E., Zinder N. D. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968 Jan 10;243(1):82–91. [PubMed] [Google Scholar]
- Rosenberg M., Kramer R. A. Nucleotide sequence surrounding a ribonuclease III processing site in bacteriophage T7 RNA. Proc Natl Acad Sci U S A. 1977 Mar;74(3):984–988. doi: 10.1073/pnas.74.3.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Kramer R. A., Steitz J. A. T7 early messenger RNAs are the direct products of ribonuclease III cleavage. J Mol Biol. 1974 Nov 15;89(4):777–782. doi: 10.1016/0022-2836(74)90052-7. [DOI] [PubMed] [Google Scholar]
- Schwartz T., Craig E., Kennell D. Inactivation and degradation of messenger ribnucleic acid from the lactose operon of Escherichia coli. J Mol Biol. 1970 Dec 14;54(2):299–311. doi: 10.1016/0022-2836(70)90431-6. [DOI] [PubMed] [Google Scholar]
- Schweitz H., Ebel J. P. A study of the mechanism of action of E. coli ribonuclease 3. Biochimie. 1971;53(5):585–593. doi: 10.1016/s0300-9084(71)80014-7. [DOI] [PubMed] [Google Scholar]
- Silengo L., Nikolaev N., Schlessinger D., Imamoto F. Stabilization of mRNA with polar effects in an Escherichia coli mutant. Mol Gen Genet. 1974;134(1):7–19. doi: 10.1007/BF00332808. [DOI] [PubMed] [Google Scholar]
- Smith T. F., Sadler J. R. The nature of lactose operator constitive mutations. J Mol Biol. 1971 Jul 28;59(2):273–305. doi: 10.1016/0022-2836(71)90051-9. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Steitz J. A. The 3' terminal oligonucleotide of E. coli 16S ribosomal RNA: the sequence in both wild-type and RNase iii- cells is complementary to the polypurine tracts common to mRNA initiator regions. Nucleic Acids Res. 1975 Jun;2(6):787–798. doi: 10.1093/nar/2.6.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Genetic mapping of a mutation that causes ribonucleases III deficiency in Escherichia coli. J Bacteriol. 1975 Oct;124(1):307–316. doi: 10.1128/jb.124.1.307-316.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]


