Abstract
Cryptococcus neoformans produces vesicles containing its major virulence factor, the capsular polysaccharide glucuronoxylomannan (GXM). These vesicles cross the cell wall to reach the extracellular space, where the polysaccharide is supposedly used for capsule growth or delivered into host tissues. In the present study, we characterized vesicle morphology and protein composition by a combination of techniques including electron microscopy, proteomics, enzymatic activity, and serological reactivity. Secretory vesicles in C. neoformans appear to be correlated with exosome-like compartments derived from multivesicular bodies. Extracellular vesicles manifested various sizes and morphologies, including electron-lucid membrane bodies and electron-dense vesicles. Seventy-six proteins were identified by proteomic analysis, including several related to virulence and protection against oxidative stress. Biochemical tests indicated laccase and urease activities in vesicles. In addition, different vesicle proteins were recognized by sera from patients with cryptococcosis. These results reveal an efficient and general mechanism of secretion of pathogenesis-related molecules in C. neoformans, suggesting that extracellular vesicles function as “virulence bags” that deliver a concentrated payload of fungal products to host effector cells and tissues.
Cryptococcus neoformans is the causative agent of cryptococcosis and a major pathogen for immunosuppressed individuals (39). Cryptococcosis is rampant in Africa and Asia in association with human immunodeficiency virus infection, where it is associated with high morbidity and mortality (5). C. neoformans provides a unique model in cell biology studies because it is the only eukaryotic pathogen with a polysaccharide capsule, a structure that is essential for virulence (26, 32, 39). The major capsular polysaccharide, glucuronoxylomannan (GXM), is released extracellularly during infection and induces a number of deleterious effects to the host, including interference with phagocytosis, inhibition of leukocyte migration to infected tissues, and modulation of cytokine production (reviewed in reference 32). It also represents a potential component for vaccines (13) and is the target of potentially therapeutic antibodies (7).
The mechanisms of secretion of macromolecules by fungal cells have not been well elucidated. Fungal cells are encased in a rigid, pore-containing cell wall consisting of polysaccharides, proteins, and pigments (14, 35). Different studies demonstrate that structures with molecular masses higher than 1,000 kDa can cross the cell wall and reach the extracellular milieu (26, 27, 47). GXM, for instance, has an average molecular mass ranging from 1.7 × 106 to 7 × 106 Da (27). Several studies by our group and others indicate that C. neoformans synthesizes GXM intracellularly and then transports the polysaccharide to the extracellular space for assembly into a capsule (17, 20, 45, 54, 55). The mechanism by which capsular polysaccharide is synthesized inside the cell and exported to the extracellular environment for capsule assembly and release is a central question in cryptococcal cell biology.
We have recently described how GXM-containing vesicles accumulate in supernatants of C. neoformans cultures (45). These extracellular vesicles contain bilayered membranes enriched with key fungal lipids, such as glucosylceramide and sterols. This observation led to the proposal that extracellular export of GXM is accomplished by vesicular transport (45). The existence of a vesicular transport mechanism raises the possibility that other fungal molecules could be released using the same cellular apparatus. Indeed, a secretion mutant of C. neoformans that accumulates secretory vesicles in the cytoplasm has severe defects in protein secretion (54). We therefore hypothesized that vesicular extracellular secretion in C. neoformans is a general mechanism used for the trans-cell-wall transport of protein, lipid, and carbohydrate components to the extracellular environment.
In the present work, we used microscopic, serological, biochemical, and proteomic approaches to analyze the extracellular vesicles of C. neoformans. Electron microscopy suggests that vesicle secretion derives from the traffic of multivesicle-like compartments to the cell surface. The results indicate that C. neoformans extracellular vesicles represent a heterogeneous population of “virulence bags” containing numerous molecules associated with fungal survival and host pathogenicity.
MATERIALS AND METHODS
Strains.
The C. neoformans isolates used in this study included strains H99 (serotype A, wild type), Cap 67 (derived from serotype D and lacking a GXM capsule), and 2E-TU and 2E-TUC (serotype D, LAC1 gene deletion and reconstituted laccase mutants) (46). Yeast cells were cultivated in a minimal medium composed of dextrose (15 mM), MgSO4 (10 mM), KH2PO4 (29.4 mM), glycine (13 mM), and thiamine-HCl (3 μM). Fungal cells were cultivated for 48 h at 30°C, with continuous shaking. Strain Cap 67 was used for electron microscopy. Strains 2E-TU and 2E-TUC were used for laccase assays. Strain H99 was used for all other experiments. Proteomic analyses were performed using strains Cap 67, H99, and 2E-TUC.
Vesicle purification.
Isolation of extracellular vesicles was done using the protocol described by Rodrigues et al. (45). Briefly, cell-free culture supernatants were obtained by sequential centrifugation at 4,000 and 15,000 × g (15 min, 4°C). These supernatants contained vesicles and were concentrated by approximately 20-fold using an Amicon ultrafiltration system (cutoff of 100 kDa). The concentrate was again centrifuged at 4,000 and 15,000 × g (15 min, 4°C) and then at 100,000 × g for 1 h at 4°C. The supernatants were discarded, and pellets were washed by five sequential suspension and centrifugation steps, each consisting of 100,000 × g for 1 h at 4°C with 0.1 M Tris-buffered saline. To remove extravesicular GXM contamination, vesicles were subjected to passage through a column packed with cyanogen bromide-activated Sepharose coupled to a monoclonal antibody to GXM, as described previously (45). Fractions that were not bound to the monclonal antibody-containing column were again centrifuged at 100,000 × g. The resulting pellets were then suspended in fixative solution for electron microscopy analysis or prepared for biochemical, proteomic, and Western blotting analyses, as described below.
TEM.
For transmission electron microscopy (TEM), C. neoformans cells were grown in minimal medium, serially washed in phosphate-buffered saline (PBS), and fixed for 1 h in 0.1 M sodium cacodylate buffer (pH 7.2) containing 4% paraformaldehyde and 2% glutaraldehyde. Cells were then infiltrated for 2 h in a solution containing 25% polyvinylpyrrolidone and 2.1 M sucrose and then rapidly frozen by immersion in liquid nitrogen. They were transferred to a cryo-ultramicrotome (Ultracut; Reichert), and cryosections were obtained in a temperature range of −70 to −90°C. The material was collected with a sucrose loop and transferred to Formvar-carbon-coated grids. Specimens were observed in a Zeiss 900 transmission electron microscope operating at 80 kV.
Pellets obtained after centrifugation of cell-free supernatants at 100,000 × g were fixed with 2% glutaraldehyde in 0.1 M cacodylate at room temperature for 2 h and then incubated overnight in 4% formaldehyde, 1% glutaraldehyde, and 0.1% PBS. The samples were incubated for 90 min in 2% osmium, serially dehydrated in ethanol, and embedded in Spurrs epoxy resin. Thin sections were obtained on a Reichart Ultracut UCT and stained with 0.5% uranyl acetate and 0.5% lead citrate. Samples were observed in a JEOL 1200EX transmission electron microscope operating at 80 kV.
Biochemical detection of enzymatic activities.
Laccase and urease activity in vesicle preparations was assayed spectrophotometrically. Acid phosphatase, a cryptococcal protein that mediates adhesion of cryptococci to epithelial cells and whose secretion has been associated with vesicle production (8, 54), was also assayed. Pellets obtained after centrifugation at 100,000 × g were suspended in PBS and serially diluted in media appropriate for the reactions catalyzed by laccase, urease, or acid phosphatase. The laccase reaction medium corresponded to 0.2% (10 mM) l-DOPA in PBS, while the medium for urease activity contained 4% urea, 0.02% yeast extract, 0.002% phenol red, 0.273% KH2PO4, and 0.285% Na2HPO-4. For phosphatase determination, the reaction medium consisted of acetate buffer (pH 5.0) supplemented with 5 mg/ml p-nitrophenyl phosphate. Vesicle suspensions were incubated overnight at room temperature and protected from the light. Reactions were quantified by reading at 450 (laccase), 405 (phosphatase), or 540 (urease) nm with a Multiscan mass spectrometer (MS) (Labsystem, Helsinki, Finland). The amount of vesicles in each system was assumed to be related to the protein concentration in each vesicular suspension. The protein concentration at the starting dilution in each system corresponded to 0.3 μg/ml. Enzymatic assays were repeated at least three times, producing similar results.
Western blot analysis.
Vesicles were suspended in loading buffer (1% sodium dodecyl sulfate [SDS], 10% glycerol, 10 mM Tris-Cl [pH 6.8], 1 mM 2-mercaptoethanol, and 0.05 mg/ml bromophenol blue) and a final amount of proteins corresponding to 8 μg was loaded onto 12% SDS-polyacrylamide gel electrophoresis (PAGE) gel. Separated proteins were transferred to nitrocellulose membranes, which were sequentially blocked in PBS containing 1% bovine serum albumin and incubated for 1 h at room temperature with pooled sera from 10 individuals diagnosed with cryptococcosis based on positive serum reactivity in latex agglutination tests for cryptococcal GXM. Alternatively, the membranes were incubated with pooled normal human sera from healthy volunteers with no previous diagnosis of any systemic mycosis. Pooled sera were used at a 1:100 dilution in PBS-bovine serum albumin. After extensive washing, the membranes were incubated in the presence of a peroxidase-labeled anti-human immunoglobulin antibody followed by immunodetection by chemiluminescence (Pierce). To exclude the possibility of nonspecific recognition of samples by secondary antibodies, samples were incubated directly with the peroxidase-labeled antihuman antibody, producing negative results (not shown). Serological analyses were repeated twice, with similar results.
Protein identification by liquid chromatography-tandem mass spectrometry.
Purified vesicles were suspended in 400 mM NH4HCO3 (40 μl) containing 8 M urea, and 50 mM dithiothreitol (10 μl) was then added for reduction of disulfide bonds. After incubation at 50°C for 15 min, the cysteine residues were alkylated through the addition of 100 mM iodoacetamide (10 μl), followed by incubation for 15 min at room temperature under protection from the light. The final concentration of urea was then adjusted to 1 M by the addition of high-performance liquid chromatography (HPLC)-grade water (Sigma). The mixture was supplemented with 4 μg sequencing-grade trypsin (Promega) and digested overnight at 37°C. The resulting sample was purified in reverse-phase ZipTip columns (POROS R2 50; Applied Biosystems) as described by Jurado et al. (24). Released peptides were then fractionated on a strong cation-exchange ZipTip column (POROS HS 50 resin; Applied Biosystems), preequilibrated with 25% acetonitrile (ACN)-0.5% formic acid (FA). After loading the peptide mixture, the column was washed with 25% ACN-0.5% FA and the peptides were eluted with the same solution supplemented with NaCl concentrations ranging from 0 to 500 mM. Each fraction was dried in a vacuum centrifuge (Eppendorf) and again purified by reverse-phase chromatography in POROS R2 50 ZipTip columns. Fractions were finally suspended in 0.05% trifluoracetic acid (30 μl). An 8-μl aliquot of each fraction was loaded into a C18 trap column (1 μl C18; OPTI-PAK). The separation was performed on a capillary reverse-phase column (Acclaim; LC Packings [3 μm C18, 75 μm by 25 cm]) connected to a nanoHPLC system (nanoLC 1D Plus; Eksigent). Peptides were eluted with increasing concentrations of ACN (0 to 40%) in 0.1% FA during 100 min and directly analyzed in an electrospray-linear ion trap MS equipped with a nanospray source (LTQ XL; Thermo Fisher). MS spectra were collected in centroid mode at the 400 to 1,700 m/z range, and the five most abundant ions were subjected twice to collision-induced dissociation with 35% normalized collision energy, before being dynamically excluded for 120 s.
All tandem MS spectra from peptides with 600 to 4,000 Da, more than 100 counts, and at least 15 fragments were converted into DTA files using Bioworks v.3.3.1 (Thermo). The DTA files were submitted to a database search using TurboSequest (15), available in Bioworks, and the C. neoformans protein database, available at www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans. Common contaminant sequences (retrieved from GenBank at http://www.ncbi.nlm.nih.gov/ and the International Protein Index at http://www.ebi.ac.uk/IPI) and 100,000 randomly generated sequences were used to supplement the C. neoformans database. The database search parameters included (i) trypsin cleavage in both peptide termini with one missed cleavage site allowed; (ii) carbamidomethylation of cysteine residues as a fixed modification; (iii) oxidation of methionine residues as a variable modification; and (iv) 2.0 and 1.0 Da for peptide and fragment mass tolerance, respectively. To ensure the quality of protein identification, the false-positive rate (FPR) was estimated using the TurboSequest output and the following formula: FPR = number of proteins matching random sequences/total number of proteins.
The FPR was calculated after applying the following filters in Bioworks: distinct peptides (for exclusion of redundant hits); DCn, ≥0.1; protein probability, ≤1 × 10−3; and Xcorr, ≥1.5, 2.2, and 2.7 for singly, doubly, and triply charged peptides, respectively. When necessary, protein consensus scores were also applied to limit the number of false-positive hits. All data sets showed an FPR lower than 3.2%.
RESULTS
The extracellular vesicles produced by C. neoformans consist of a complex population.
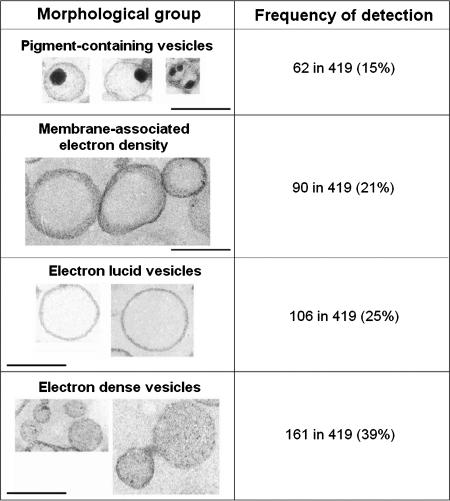
The individual analysis of micrographs of 419 secreted vesicles produced by C. neoformans revealed considerable morphological variation. Using morphological criteria, we identified four main vesicle groups (Fig. 1). The groups included electron-dense and electron-lucid vesicles, vesicular structures with membrane-associated electron-dense regions, and vesicles containing hyper-dense structures resembling a dark pigment.
FIG. 1.
Electron microscopic appearance (left panels) and prevalence (right panels) of the four major vesicle morphological groups observed in preparations of extracellular vesicles from C. neoformans. The total population analyzed consisted of 419 different vesicles. Scale bars, 200 nm.
Pathogenesis-related molecules are present in the extracellular vesicles.
The microscopic analysis illustrated in Fig. 1 suggested that pigment synthesis could occur in vesicular structures. We therefore evaluated whether laccase, a cryptococcal enzyme characterized as a virulence factor responsible for melanin synthesis (46), was present in vesicular preparations. Laccase activity was detected in vesicle preparations from wild-type cells (Fig. 2A). Enzyme activity was absent in vesicles from a C. neoformans mutant lacking the ability to produce LAC1, while vesicular structures from a laccase-reconstituted strain had enzyme activity similar to wild-type cells. These results, combined with a recent report by our group (45), indicated that extracellular vesicles in C. neoformans contain at least three virulence determinants: GXM (reviewed in reference 32), glucosylceramides (44), and laccase (46). We therefore evaluated whether other molecules related to pathogenesis were present in the C. neoformans vesicular structures.
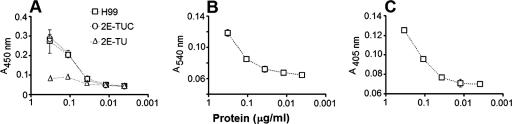
FIG. 2.
Laccase (A), urease (B), and phosphatase (C) activities are associated with extracellular vesicles in C. neoformans. (A). Vesicles purified from culture supernatants of strains H99 and the serotype D laccase mutants 2E-TUC (complemented strain) and 2E-TU (LAC1 deletion strain) were incubated in the presence of l-DOPA and analyzed spectrophotometrically. Vesicles purified from the supernatants of strain H99 were also incubated in urease (B) and phosphatase (C) reaction media, followed by spectrophotometric determination of enzyme activity.
We examined the activities of urease, a well-described extracellular virulence factor of C. neoformans (11), and acid phosphatase, an adhesion-related molecule (8) suggested to be released to the extracellular space in secretory vesicles (54). Dose-dependent activities of both enzymes were observed when the extracellular vesicles were incubated with urease and phosphatase substrates (Fig. 2B and C).
Vesicle proteins are recognized by sera from cryptococcosis patients.
Vesicle proteins were separated by SDS-PAGE and analyzed by reactivity with human sera by immunoblotting (Fig. 3). No significant reactivity was observed using sera from normal individuals. However, when the vesicle sample was probed with a pool of sera from cryptococcosis patients, seven major bands were observed, with relative molecular masses corresponding to 131, 101, 67, 48, 38, 27, and 19 kDa. Diffuse areas of serological reaction were also observed in the molecular mass ranges of 50 to 65 and 70 to 100 kDa. These results suggest that vesicle-related cryptococcal proteins are produced during human infection and also that potentially effective protein immunogens are present in extracellular vesicles. Unfortunately, the identification of the proteins recognized by the sera of cryptococcosis patients has been hampered by the lack of sufficient amount of vesicular material needed for liquid chromatography-tandem MS sequencing from SDS-PAGE gel bands.
FIG. 3.
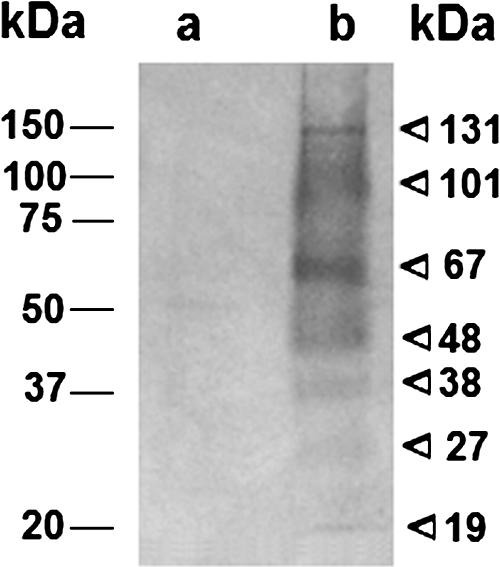
Vesicle-associated proteins are recognized by sera from cryptococcosis patients. Vesicle-associated proteins (a and b) were separated by SDS-PAGE and incubated with pooled sera from healthy individuals (a) or cryptococcosis patients (b). Molecular masses for standard (left values) or vesicle (right values) proteins are indicated.
Proteomic analysis of the C. neoformans extracellular vesicles.
To further characterize the protein components of the secreted vesicles, the material purified by centrifugation was enzymatically digested, fractionated by chromatographic methods, and analyzed by liquid chromatography-tandem MS. Overall, 76 different proteins were identified (Table 1). The yield of protein detection varied considerably, depending on the presence of capsular structures, suggesting that the polysaccharides in extracellular fractions might interfere with protein digestion, as previously suggested for Aspergillus oryzae (38). Accordingly, no proteins were detected by proteomic approaches in preliminary analysis of vesicles from the highly encapsulated strain 24067 (data not shown). Twenty different proteins were identified using strain H99, which presents smaller capsules than isolate 24067. A higher number of proteins (n = 26) was identified using the encapsulated strain 2E-TUC, but protein characterization was maximum using the acapsular mutant Cap 67 (70 proteins). The results obtained with each strain were combined and are summarized in Table 1. A complete, detailed list of proteins analyzed by mass spectrometry is available (see Table S1 in the supplemental material). Chaperones, including heat shock proteins (Hsp70 and Hsp90) and superoxide dismutase, signal transduction regulators, antioxidant and cytosolic proteins, and enzymes were identified. Of note, nuclear proteins such as histones and ribosomal proteins were also identified, as previously described in vesicles secreted by mammalian cells (1, 42). Overall, 27 of the 76 proteins identified in the C. neoformans vesicles were already reported as vesicular proteins in mammalian exosomes (1, 21, 28, 42, 48, 49), including a ubiquitin-related protein. Figure 4 shows identified C. neoformans vesicle proteins distributed according to their functions in fungal cells.
TABLE 1.
Protein components of C. neoformans extracellular vesiclesa
| Hit no. | Accession no./identification | Function (reference) | Strain(s) |
|---|---|---|---|
| Pathogenesis/immune response | |||
| 1 | CNAG_01727.1/heat shock 70-kDa protein 2* (644 aa, 69.53 kDa) | Chaperone | Cap67, 2E-TUC, H99 |
| 2 | CNAG_03891.1/60-kDa chaperonin* (582 aa, 61.36 kDa) | Chaperone | Cap67 |
| 3 | CNAG_06150.1/heat shock protein 90* (700 aa, 79.15 kDa) | Chaperone | Cap67 |
| 4 | CNAG_00334.1/heat shock protein 70* (615 aa, 67.09 kDa) | Chaperone, C. neoformans immunogen (25) | Cap67, 2E-TUC, H99 |
| 5 | CNAG_02292.1/superoxide dismutase copper chaperone* (288 aa, 30.32 kDa) | Antioxidant defense, C. neoformans virulence (9, 34) | Cap67 |
| 6 | CNAG_02944.1/acid phosphatase (552 aa, 59.32 kDa) | Adhesion of C. neoformans to epithelial cells (8) | Cap67, 2E-TUC |
| 7 | CNAG_02801.1/thioredoxin (105 aa, 11.44 kDa)* | Protection against oxidative stress, survival in the oxidative environment of macrophages, C. neoformans virulence (29) | Cap67, H99 |
| 8 | CNAG_05847.1/thioredoxin reductase (372 aa, 39.84 kDa) | C. neoformans viability (30) | Cap67 |
| 9 | CNAG_06917.1/thiol-specific antioxidant protein (234 aa, 25.90 kDa) | C. neoformans virulence, resistance to nitric oxide and peroxide (31) | H99, 2E-TUC, Cap67 |
| 10 | CNAG_00575.1/catalase A (683 aa, 76.42 kDa) | C. neoformans antioxidant defense (22) | 2E-TUC |
| 11 | CNAG_03322.1/UDP-glucuronic acid decarboxylase Uxs1p (411 aa, 46.47 kDa) | Converts UDP-glucuronic acid to UDP-xylose, capsule synthesis in C. neoformans (3) | Cap67 |
| 12 | CNAG_04969.1/UDP-glucose dehydrogenase (469 aa, 51.36 kDa) | C. neoformans growth at 37°C and capsule biosynthesis (33) | Cap67, H99 |
| Signal transduction | |||
| 13 | CNAG_04762.1/Ras2 (364 aa, 39.47 kDa) | Mating and high-temp growth in C. neoformans (52) | Cap67 |
| 14 | CNAG_03315.1/Rho1* (198 aa, 21.68 kDa) | GTP-binding protein, cell wall integrity in C. neoformans (19) | Cap67 |
| 15 | CNAG_05235.1/14-3-3 protein* (257 aa, 28.98 kDa) | Cell cycle regulation in C. neoformans (53) | Cap67, H99, 2E-TUC |
| 16 | CNAG_04577.1/nucleoside-diphosphate kinase (216 aa, 23.36 kDa) | Exchange of phosphate groups between different nucleoside diphosphates | Cap67 |
| 17 | CNAG_03052.1/protein phosphatase type 2C (560 aa, 59.40 kDa) | Stress response and cell cycle regulation | Cap67 |
| Ribosomal proteins* | |||
| 18 | CNAG_06447.1/60S ribosomal protein l17 (183 aa, 20.49 kDa) | Ribosomal protein | Cap67, H99 |
| 19 | CNAG_03283.1/60S ribosomal protein L24 (L30) (151 aa, 16.89 kDa) | Ribosomal protein | Cap67 |
| 20 | CNAG_00232.1/60S ribosomal protein l30-1 (l32) (114 aa, 12.33 kDa) | Ribosomal protein | 2E-TUC |
| 21 | CNAG_05762.1/ribosomal protein P2 (112 aa, 11.07 kDa) | Ribosomal protein | Cap67 |
| 22 | CNAG_00771.1/ribosomal protein L35 (128 aa, 14.67 kDa) | Ribosomal protein | Cap67 |
| 23 | CNAG_00779.1/ribosomal protein L27 (137 aa, 15.76 kDa) | Ribosomal protein | Cap67 |
| 24 | CNAG_06605.1/ribosomal protein S2 (257 aa, 27.62 kDa) | Ribosomal protein | Cap67 |
| 25 | CNAG_02144.1/60S ribosomal protein l1-a (l10a) (225 aa, 25.50 kDa) | Ribosomal protein | Cap67, 2E-TUC |
| 26 | CNAG_04114.1/40S ribosomal protein S0 (293 aa, 31.46 kDa) | Ribosomal protein | Cap67 |
| 27 | CNAG_01480.1/ribosomal protein L12 (166 aa, 17.55 kDa) | Ribosomal protein | Cap67 |
| 28 | CNAG_06633.1/40S ribosomal protein S15 (151 aa, 17.12 kDa) | Ribosomal protein | Cap67 |
| 29 | CNAG_00656.1/60S ribosomal protein l7 (251 aa, 28.25 kDa) | Ribosomal protein | Cap67 |
| 30 | CNAG_00034.1/60S ribosomal protein l9 (192 aa, 21.17 kDa) | Ribosomal protein | Cap67 |
| 31 | CNAG_06095.1/ribosomal protein L13 (206 aa, 23.31 kDa) | Ribosomal protein | Cap67 |
| 32 | CNAG_04011.1/60S ribosomal protein l37a (93 aa, 10.07 kDa) | Ribosomal protein | Cap67 |
| 33 | CNAG_02818.1/ribosomal protein S11 (410 aa, 44.31 kDa) | Ribosomal protein | Cap67 |
| Sugar and lipid metabolism | |||
| 34 | CNAG_06699.1/glyceraldehyde-3-phosphate dehydrogenase* (340 aa, 36.28 kDa) | Glucose metabolism, secreted enzyme | Cap67, 2E-TUC |
| 35 | CNAG_00061.1/citrate synthase* (465 aa, 51.02 kDa) | Energy metabolism | Cap67, 2E-TUC |
| 36 | CNAG_05653.1/malate synthase* (539 aa, 59.96 kDa) | Glucose metabolism | 2E-TUC |
| 37 | CNAG_03072.1/phosphopyruvate hydratase (enolase)* (434 aa, 46.44 kDa) | Cell wall constituent in Candida (40) | Cap67, H99, 2E-TUC |
| 38 | CNAG_04659.1/pyruvate decarboxylase* (624 aa,) | Decarboxylation of pyruvic acid to acetaldehyde and carbon dioxide | Cap67, H99 |
| 39 | CNAG_03225.1/malate dehydrogenase* (339 aa, 35.58 kDa) | Conversion of malate into oxaloacetate, major antigen in P. brasiliensis (12) | Cap67 |
| 40 | CNAG_02620.1/phosphogluconate dehydrogenase* (decarboxylating) (492 aa, 53.85 kDa) | Pentose phosphate pathway | Cap67, 2E-TUC |
| 41 | CNAG_03920.1/isocitrate dehydrogenase* (453 aa, 50.60 kDa) | Citric acid cycle | Cap67 |
| 42 | CNAG_02100.1/fatty-acid synthase complex protein* (1,439 aa, 157.47 kDa) | Fatty acid biosynthetic process | Cap67 |
| 43 | CNAG_06140.1/long-chain fatty acid transporter (111 aa, 11.91 kDa) | Fatty acid biosynthetic process | Cap67 |
| 44 | CNAG_06572.1/pyruvate dehydrogenase e1 component alpha subunit, mitochondrial precursor (414 aa, 45.66 kDa) | Conversion of pyruvate to acetyl coenzyme A and CO2 | Cap67 |
| Nuclear proteins | |||
| 45 | CNAG_01648.1/histone H4* (104 aa, 11.40 kDa) | DNA assembly | H99, Cap67, 2E-TUC |
| 46 | CNAG_05221.1/histone H2A* variant (139 aa, 14.70 kDa) | DNA assembly | Cap67, 2E-TUC |
| Protein/amino acid metabolism | |||
| 47 | CNAG_00370.1/ubiquitin-carboxy extension protein fusion* (130 aa, 14.64 kDa) | Posttranslational protein modification | H99, 2E-TUC |
| 48 | CNAG_02500.1/endoplasmic reticulum-associated protein, catabolism-related protein (552 aa, 60.97 kDa), similar to fungal calnexin | Retention of unfolded or unassembled N-linked glycoproteins in the endoplasmic reticulum | Cap67, H99 |
| 49 | CNAG_00441.1/inosine 5-monophosphate dehydrogenase (545 aa, 57.85 kDa) | Oxidation of inosine 5′-monophosphate to xanthosine 5′-monophosphate | Cap67 |
| 50 | CNAG_01890.1/5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase (764 aa, 85.32 kDa) | Amino acid metabolism, cobalamin-independent methionine synthase | Cap67, H99 |
| 51 | CNAG_05725.1/acetohydroxy acid reductoisomerase (402 aa, 44.31 kDa) | Amino acid metabolism | Cap67 |
| 52 | CNAG_00457.1/glutamine synthetase (359 aa, 39.49 kDa) | Amino acid metabolism | Cap67 |
| 53 | CNAG_00785.1/translation initiation factor* (402 aa, 45.13 kDa) | Protein synthesis | Cap67 |
| 54 | CNAG_00930.1/argininosuccinate synthase (430 aa, 47.34 kDa) | Amino acid metabolism | Cap67 |
| 55 | CNAG_04601.1/glycine hydroxymethyltransferase (500 aa, 54.39 kDa) | Amino acid metabolism | Cap67, H99 |
| 56 | CNAG_00450.1/3-isopropylmalate dehydrogenase (374 aa, 39.85 kDa) | Amino acid metabolism | Cap67 |
| 57 | CNAG_00992.1/homocitrate synthase (490 aa, 53.51 kDa) | Amino acid metabolism | Cap67 |
| 58 | CNAG_06125.1/translation elongation factor 1α* (461 aa, 50.32 kDa) | Protein synthesis | Cap67, H99, 2E-TUC |
| 59 | CNAG_06908.1/aminomethyltransferase, mitochondrial precursor (338 aa, 34.92 kDa) | Amino acid metabolism | Cap67 |
| Plasma membrane proteins | |||
| 60 | CNAG_06101.1/ADP/ATP carrier (314 aa, 33.77 kDa) | ATP/ADP translocation, multipass membrane protein | Cap67, 2E-TUC |
| 61 | CNAG_06400.1/plasma membrane H(+)-ATPase (999 aa, 108.47 kDa) | Proton pump | Cap67 |
| 62 | CNAG_03058.1/Hmp1 protein (115 aa, 11.74 kDa) | Similar to α-catenins, proteins found in complexes with cadherin cell adhesion | 2E-TUC |
| 63 | CNAG_04758.1/ammonium transporter (497 aa, 53.04 kDa) | Multipass membrane protein | Cap67 |
| Cytoskeleton proteins | |||
| 64 | CNAG_00483.1/actin* (378 aa, 42.02 kDa) | Cytoskeleton protein | Cap67, 2E-TUC, H99 |
| 65 | CNAG_03787.1/α-tubulin* (449 aa, 49.67 kDa) | Cytoskeleton protein | Cap67 |
| 66 | CNAG_01840.1/tubulin β chain* (452 aa, 49.99 kDa) | Cytoskeleton protein | Cap67 |
| Miscellaneous | |||
| 67 | CNAG_01577.1/glutamate dehydrogenase (452 aa, 49.16 kDa) | Urea synthesis | Cap67, 2E-TUC |
| 68 | CNAG_05750.1/ATP synthase α chain, mitochondrial precursor (541 aa, 58.02 kDa) | ATP synthesis | Cap67, H99, 2E-TUC |
| 69 | CNAG_05918.1/ATP synthase β chain* (548 aa, 58.63 kDa) | ATP synthesis | Cap67, 2E-TUC |
| 70 | CNAG_02974.1/voltage-dependent ion-selective channel (293 aa, 30.61 kDa) | Membrane transport | Cap67, 2E-TUC, H99 |
| 71 | CNAG_01539.1/inositol-3-phosphate synthase (559 aa, 61.34 kDa) | Metabolism of inositol-containing molecules | Cap67 |
| 72 | CNAG_05909.1/electron transporter, transferring electrons within CoQH2-cytochrome c reductase complex (320 aa, 34.97 kDa) | Mitochondrial protein, electron transport | 2E-TUC |
| 73 | CNAG_00799.1/hypothetical protein similar to glycosyl hydrolase family 5 protein from Stigmatella aurantiaca (790 aa, 85.25 kDa) | Cell wall assembly (putative) | 2E-TUC |
| 74 | CNAG_03537.1/FA and derivative metabolism-related protein (1022 aa, 108.90 kDa) | FA metabolism | Cap67, H99 |
| 75 | CNAG_00965.1/probable transketolase (688 aa, 74.28 kDa) | NADPH metabolism | Cap67, H99 |
| 76 | CNAG_05759.1/acetyl coenzyme A carboxylase (2238 aa, 247.95 kDa) | Synthesis of malonyl coenzyme A | Cap67 |
Biological functions of the proteins detected by proteomics are referenced when an association of the molecule with the biology or pathogenesis of C. neoformans or other fungal pathogens was previously described. General functions in other models were obtained from the ExPASy Proteomics Server (http://ca.expasy.org/). The number of amino acids (aa) and molecular mass values are presented for each protein. Twenty-five proteins with unknown functions were characterized but not presented in this table. For detailed information, see the supplemental material. Proteins that were already characterized in extracellular vesicles produced by mammalian cells are marked with an asterisk. For details, see references 1, 21, 28, 42, 48, and 49.
FIG. 4.
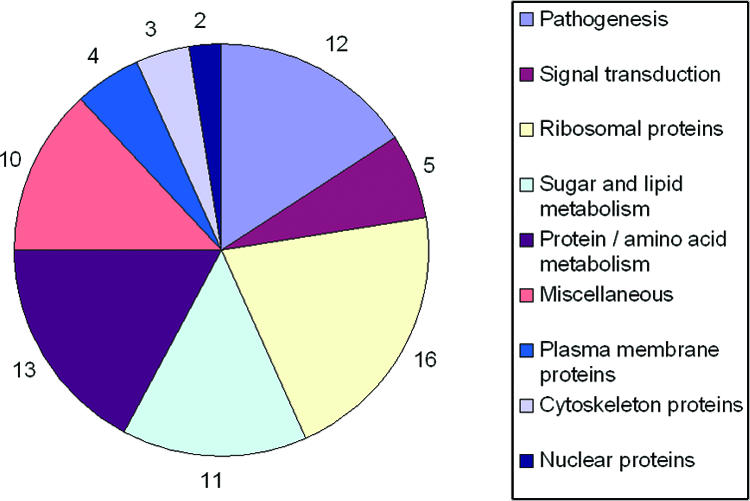
Functional classification of the C. neoformans vesicle proteins. The number of proteins found for each class is shown. Unidentified proteins are not shown. For details, see Table S1 in the supplemental material.
Intracellular distribution of vesicles in C. neoformans.
Mammalian exosomes consist of secreted vesicles that originate from key compartments of the endocytic pathway, namely multivesicular bodies (MVBs) (51). Since the profile of protein detection in the C. neoformans vesicles resembled those found in mammalian exosomes, ultrathin sections of yeast cells were searched for the presence of MVB-like structures. Vesicular structures with dimensions similar to those found at the extracellular fractions were observed inside vacuole-like compartments (Fig. 5), resembling endosome-derived MVBs (51). These vesicle-containing vacuoles were detected in all the sections analyzed, independently of the technique used for TEM (data not shown). Bilayered membranes were present at the vacuole-like structures, which were sometimes found in fusion with the plasma membrane. These results suggest that vesicle secretion in C. neoformans may derive from MVBs or vesicle-containing vacuoles.
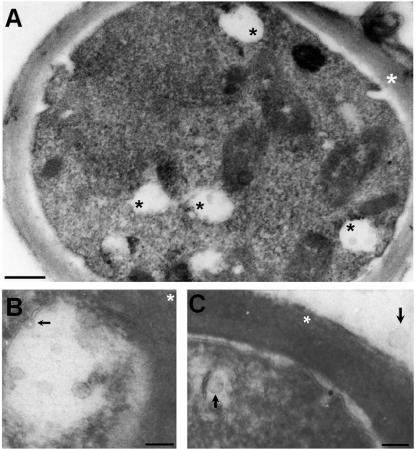
FIG. 5.
TEM of C. neoformans suggesting the presence of cytoplasmic vacuole-containing vesicles reminiscent of exosome-like structures. (A) Overview of a C. neoformans cell with different cytoplasmic vacuoles containing vesicles (black asterisks). The white asterisk indicates the cell wall. Scale bar, 500 nm. A magnified view of the vesicle-containing vacuoles is shown. Panel B demonstrates that these structures are surrounded by a bilayered membrane, which sometimes invaginates (arrow). A close association with the cell wall (white asterisk) was observed, suggesting fusion with the plasma membrane. Scale bar, 200 nm. (C) Intracellular and extracellular vesicles (black arrows) have similar dimensions. Scale bar, 200 nm.
DISCUSSION
The secretion of virulence factors is a mechanism used by different pathogens to cause damage to host cells (6). In C. neoformans, secreted virulence factors include superoxide dismutase (9), phospholipase B (10), urease (11), and GXM, a high-molecular-weight polysaccharide (reviewed in references 26 and 32). GXM is synthesized in intracellular compartments (17, 20, 54) and transported through the cell wall in secretory vesicles (45). The virulence regulator glucosylceramide (44) is also present in the cryptococcal extracellular vesicles (45), but the presence of other molecules in these membrane compartments is unknown. In this context, we hypothesized that vesicular secretion in C. neoformans could be a general secretory mechanism, possibly used for the delivery of different molecules related to pathogenic mechanisms to the extracellular space.
Polysaccharide-containing vesicles in C. neoformans appeared to be associated with the Golgi apparatus-derived secretory pathway (54). Golgi apparatus-derived secretory vesicles, however, are expected to fuse with the plasma membrane and release their internal content to the extracellular space (41). In C. neoformans, this model would satisfactorily explain how GXM reaches the periplasmic space, although it does not explain how the polysaccharide would reach the outer layer of the cell wall to be incorporated into the growing capsule. Furthermore, such a mechanism could not account for the origin of the GXM-containing extracellular vesicles that we recently described. In fact, the existence of GXM-containing extracellular vesicles implies the existence of a vesicle secretion mechanism whereby there is no fusion of secretory vesicles and the plasma membrane. In this context, we examined whether the secretory vesicles of C. neoformans could have a relationship with exosomes, which are defined as non-plasma-membrane-derived vesicles (18).
Exosomes are the only type of bioactive vesicles originating from an intracellular compartment, named MVBs on the basis of their morphology (18, 51). These intracellular compartments are derived from endosomes and have well-known functions as intermediates in the degradation of proteins internalized from the cell surface or sorted from the trans-Golgi network (18, 51). Internal vesicles of MVBs are generated by budding from the limiting membrane into the lumen of endosomes (51). In the degradation pathway, MVBs fuse with lysosomes. However, in several hematopoietic and nonhematopoietic cells, MVBs fuse with the plasma membrane, resulting in the release of internal vesicles to the extracellular milieu as exosomes (18, 51). In this context, we evaluated whether MVB-like compartments were present in C. neoformans cells. In fact, several vacuole-like compartments containing vesicles with dimensions similar to those of exosomes and the C. neoformans extracellular vesicles were observed. Some of these vesicle-containing vacuoles were in close association with the plasma membrane and the cell wall, suggesting that the release of extracellular vesicles to the extracellular space in C. neoformans involves MVB-like compartments. Since endosomes and MVBs can be connected to the trans-Golgi secretory pathway (51), we speculate that the previously described Golgi apparatus-derived vesicles containing GXM (54) are linked to MVBs in C. neoformans, which would result in the release of polysaccharide-containing vesicles into the periplasmic space. In accordance with this supposition, treatment of C. neoformans with brefeldin A, an inhibitor of the Golgi apparatus-derived transport of molecules, results in a significant inhibition of capsule expression (23).
Fungal proteins frequently have more than a single function and are found in different cellular locations (2, 4, 16, 37). For example, histones have been described as being present at the cell wall of Histoplasma capsulatum, where they are targeted by antifungal antibodies (37). Glyceraldehyde-3-phosphate dehydrogenase, a major protein component of the glycolytic pathway, is present in the cell wall of Paracoccidioides brasiliensis, where it participates in the pathogenic processes mediating the adhesion of yeast cells to host cells and the extracellular matrix (2). In the same model, the mitochondrial protein Mdj1 was detected not only in the mitochondria, where it is apparently sorted, but also in the cell wall (4). Proteomic analysis of the C. neoformans vesicles revealed a complex protein composition that included chaperone and membrane, cytoplasmic, and even nuclear and mitochondrial proteins. Several of these proteins were similar to those described in mammalian exosomes, which usually contain cytoplasmic proteins such as elongation factors, tubulin, actin, actin-binding proteins, annexins, and Rab protein, molecules responsible for signal transduction, and heat-shock proteins such as Hsp70 and Hsp90 (1, 21, 28, 42, 48, 49). Sorting of cytosolic proteins into exosomes is normally explained by a random engulfment of small portions of cytosol during the inward budding process of MVBs (51). These observations, together with morphological data, could support the supposition that the extracellular vesicles produced by C. neoformans are exosome-like structures.
Protein composition and morphological analyses indicate that the C. neoformans extracellular vesicles are not a uniform population. Vesicles with clearly different electron densities were observed, and some of them were observed to carry pigment-like structures. The observation of electron-dense spots in the inner vesicle compartments suggests the presence of the molecular machinery necessary for the synthesis of melanin, a pigment that has been concretely associated with the virulence of C. neoformans (46). Melanin is autopolymerized from the oxidation of diphenolic compounds by the enzyme laccase (36). In this context, we incubated the vesicular suspension with the laccase substrate l-DOPA, which demonstrated laccase activity in C. neoformans vesicles. The finding of laccase in vesicles that are possibly derived from MVB has an intriguing parallel in mammalian systems where tyrosinase-containing melanosomes are shed from melanocytes after synthesis from early endosomal vesicles (43). However, laccase was not detected by the proteomic approach. This observation is probably a false-negative result related to a low protein concentration, since pigmented vesicles are the less-abundant fraction in the vesicle population (15%). A relatively low protein concentration could also explain why urease is also detectable by a sensitive enzymatic colorimetric assays but not by proteomic approaches. Hence, we postulate that the proteins identified here may be only a subset of the total proteins found in vesicles.
We identified several virulence-related molecules in C. neoformans vesicles. This group of molecules includes well-known virulence factors such as GXM and glucosylceramide, which were characterized as vesicle components in a previous study (45). In the present study, we demonstrate the presence of several other components associated with virulence in vesicular fractions, such as enzymes related to capsule synthesis (3), urease (11), laccase (46), acid phosphatase (8), heat shock proteins (25), and several antioxidant proteins such as superoxide dismutase (9, 34), thioredoxin (29), thioredoxin reductase (30), thiol-specific antioxidant protein (31), and catalase A (22). Some of the vesicle proteins were recognized by sera from cryptococcosis patients, suggesting that these proteins are produced during human infection. The combined presence of lipids, pigments, polysaccharides, and virulence-related and immunogenic proteins suggests that C. neoformans uses vesicular secretion as a single mechanism to deliver virulence factors into the fungal extracellular space. Since vesicle production has been observed in vivo and during macrophage infection (45), we suggest that the C. neoformans extracellular vesicles function as “virulence factor delivery bags” that could expressively influence the interaction of fungal cells with the host. Different types of vesicles may carry different types of toxic payloads. Clearly, the presence of numerous virulence-associated components in vesicular preparations would allow C. neoformans to deliver a toxic concentrated payload to target cells such as predatory amoebae and macrophages. Vesicle secretion could also presumably occur in phagosomal spaces and allow delivery of toxic payloads to cells that have ingested cryptococcal cells. Vesicular delivery of concentrated virulence-associated components could be significantly more effective in damaging toxic cells than if such components were secreted separately and had to reach target cells through diffusion. In this context, it has been recently suggested by our group that vesicle secretion occurs during infection of host macrophages (45). This observation could be related to the known ability of C. neoformans to secrete GXM during intracellular infection of phagocytes (50), which results in host cell toxicity and release of fungal cells to extracellular host sites.
In summary, we report the identification of numerous virulence-associated components in C. neoformans vesicle preparations. The similarities in the protein content of C. neoformans vesicles and mammalian exosome-like structures combined with electron microscopic morphological evidence of exosome-like structures in cryptococcal cells led us to propose that the extracellular vesicles originate from fungal exosomes. The complexity of vesicle populations with respect to their morphology and cargo suggest numerous new avenues for the investigation of their role in virulence and cryptococcal cell biology.
Supplementary Material
Acknowledgments
M.L.R. and L.N. are supported by grants from the Brazilian agencies CNPq, CAPES, FAPESP, and FAPERJ. A.C. is supported by NIH grants AI033142, AI033774, AI052733, and HL059842. I.C.A. is supported by NIH grant 5G12RR008124 (to the Border Biomedical Research Center [BBRC]/University of Texas at El Paso [UTEP]). J.D.N. is supported by NIH AI056070-01A2 and AI52733. We are thankful to the Biomolecule Analysis Core Facility/BBRC/UTEP, supported by NIH/NCRR grant 5G12RR008124. D.L.O. is a Ph.D. student at the Instituto de Bioquimica Medica, UFRJ.
We thank Kildare Miranda and the Albert Einstein College of Medicine Analytical Imaging Facility staff for help with the electron microscopy. We are also indebted to Fabio Gozzo (Laboratorio Nacional de Luz Sincrontron, Campinas, Brazil) for the 100,000 random sequences used for statistics in the proteomic analysis.
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aoki, N., S. Jin-no, Y. Nakagawa, N. Asai, E. Arakawa, N. Tamura, T. Tamura, and T. Matsuda. 2007. Identification and characterization of microvesicles secreted by 3T3-L1 adipocytes: redox- and hormone-dependent induction of milk fat globule-epidermal growth factor 8-associated microvesicles. Endocrinology 1483850-3862. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa, M. S., S. N. Baó, P. F. Andreotti, F. P. de Faria, M. S. S. Felipe, L. dos Santos Feitosa, M. J. S. Mendes-Giannini, and C. M. d. A. Soares. 2006. Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect. Immun. 74382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bar-Peled, M., C. L. Griffith, and T. L. Doering. 2001. Functional cloning and characterization of a UDP-glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc. Natl. Acad. Sci. USA 9812003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batista, W. L., A. L. Matsuo, L. Ganiko, T. F. Barros, T. R. Veiga, E. Freymüller, and R. Puccia. 2006. The PbMDJ1 gene belongs to a conserved MDJ1/LON locus in thermodimorphic pathogenic fungi and encodes a heat shock protein that localizes to both the mitochondria and cell wall of Paracoccidioides brasiliensis. Eukaryot. Cell 5379-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bicanic, T., and T. S. Harrison. 2004. Cryptococcal meningitis. Br. Med. Bull. 7299-118. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall, A., and L. A. Pirofski. 2003. The damage-response framework of microbial pathogenesis. Nat. Rev. Microbiol. 117-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadevall, A., W. Cleare, M. Feldmesser, A. Glatman-Freedman, D. L. Goldman, T. R. Kozel, N. Lendvai, J. Mukherjee, L.-A. Pirofski, J. Rivera, A. L. Rosas, M. D. Scharff, P. Valadon, K. Westin, and Z. Zhong. 1998. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 421437-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collopy-Junior, I., F. F. Esteves, L. Nimrichter, M. L. Rodrigues, C. S. Alviano, and J. R. Meyer-Fernandes. 2006. An ectophosphatase activity in Cryptococcus neoformans. FEMS Yeast Res. 61010-1017. [DOI] [PubMed] [Google Scholar]
- 9.Cox, G. M., T. S. Harrison, H. C. McDade, C. P. Taborda, G. Heinrich, A. Casadevall, and J. R. Perfect. 2003. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 71173-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, G. M., H. C. McDade, S. C. Chen, S. C. Tucker, M. Gottfredsson, L. C. Wright, T. C. Sorrell, S. D. Leidich, A. Casadevall, M. A. Ghannoum, and J. R. Perfect. 2001. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol. Microbiol. 39166-175. [DOI] [PubMed] [Google Scholar]
- 11.Cox, G. M., J. Mukherjee, G. T. Cole, A. Casadevall, and J. R. Perfect. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect. Immun. 68443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Da Fonseca, C. A., R. S. Jesuino, M. S. Felipe, D. A. Cunha, W. A. Brito, and C. M. Soares. 2001. Two-dimensional electrophoresis and characterization of antigens from Paracoccidioides brasiliensis. Microbes Infect. 3535-542. [DOI] [PubMed] [Google Scholar]
- 13.Datta, K., and L. A. Pirofski. 2006. Towards a vaccine for Cryptococcus neoformans: principles and caveats. FEMS Yeast Res. 6525-536. [DOI] [PubMed] [Google Scholar]
- 14.De Souza Pereira, R., and J. Geibel. 1999. Direct observation of oxidative stress on the cell wall of Saccharomyces cerevisiae strains with atomic force microscopy. Mol. Cell. Biochem. 20117-24. [DOI] [PubMed] [Google Scholar]
- 15.Eng, J. K., A. L. McCormack, and J. R. Yates III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5976-989. [DOI] [PubMed] [Google Scholar]
- 16.Esnault, K., B. el Moudni, J.-P. Bouchara, D. Chabasse, and G. Tronchin. 1999. Association of a myosin immunoanalogue with cell envelopes of Aspergillus fumigatus conidia and its participation in swelling and germination. Infect. Immun. 671238-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldmesser, M., Y. Kress, and A. Casadevall. 2001. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology 1472355-2365. [DOI] [PubMed] [Google Scholar]
- 18.Fevrier, B., and G. Raposo. 2004. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 16415-421. [DOI] [PubMed] [Google Scholar]
- 19.Fuchs, B. B., G. P. Tegos, M. R. Hamblin, and E. Mylonakis. 2007. Susceptibility of Cryptococcus neoformans to photodynamic inactivation is associated with cell wall integrity. Antimicrob. Agents Chemother. 512929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Rivera, J., Y. C. Chang, K. J. Kwon-Chung, and A. Casadevall. 2004. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatti, J. L., S. Metayer, M. Belghazi, F. Dacheux, and J. L. Dacheux. 2005. Identification, proteomic profiling, and origin of ram epididymal fluid exosome-like vesicles. Biol. Reprod. 721452-1465. [DOI] [PubMed] [Google Scholar]
- 22.Giles, S. S., J. E. Stajich, C. Nichols, Q. D. Gerrald, J. A. Alspaugh, F. Dietrich, and J. R. Perfect. 2006. The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryot. Cell 51447-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, G., B. R. Steen, T. Lian, A. P. Sham, N. Tam, K. L. Tangen, and J. W. Kronstad. 2007. Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathog. 3e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurado, J. D., E. D. Rael, C. S. Lieb, E. Nakayasu, W. K. Hayes, S. P. Bush, and J. A. Ross. 2007. Complement inactivating proteins and intraspecies venom variation in Crotalus oreganus helleri. Toxicon 49339-350. [DOI] [PubMed] [Google Scholar]
- 25.Kakeya, H., H. Udono, S. Maesaki, E. Sasaki, S. Kawamura, M. A. Hossain, Y. Yamamoto, T. Sawai, M. Fukuda, K. Mitsutake, Y. Miyazaki, K. Tomono, T. Tashiro, E. Nakayama, and S. Kohno. 1999. Heat shock protein 70 (hsp70) as a major target of the antibody response in patients with pulmonary cryptococcosis. Clin. Exp. Immunol. 115485-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McFadden, D., O. Zaragoza, and A. Casadevall. 2006. The capsular dynamics of Cryptococcus neoformans. Trends Microbiol. 14497-505. [DOI] [PubMed] [Google Scholar]
- 27.McFadden, D. C., M. De Jesus, and A. Casadevall. 2006. The physical properties of the capsular polysaccharides from Cryptococcus neoformans suggest features for capsule construction. J. Biol. Chem. 2811868-1875. [DOI] [PubMed] [Google Scholar]
- 28.Mears, R., R. A. Craven, S. Hanrahan, N. Totty, C. Upton, S. L. Young, P. Patel, P. J. Selby, and R. E. Banks. 2004. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics 44019-4031. [DOI] [PubMed] [Google Scholar]
- 29.Missall, T. A., and J. K. Lodge. 2005. Function of the thioredoxin proteins in Cryptococcus neoformans during stress or virulence and regulation by putative transcriptional modulators. Mol. Microbiol. 57847-858. [DOI] [PubMed] [Google Scholar]
- 30.Missall, T. A., and J. K. Lodge. 2005. Thioredoxin reductase is essential for viability in the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4487-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Missall, T. A., J. M. Moran, J. A. Corbett, and J. K. Lodge. 2005. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsa1. Eukaryot. Cell 4202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monari, C., F. Bistoni, and A. Vecchiarelli. 2006. Glucuronoxylomannan exhibits potent immunosuppressive properties. FEMS Yeast Res. 6537-542. [DOI] [PubMed] [Google Scholar]
- 33.Moyrand, F., and G. Janbon. 2004. UGD1, encoding the Cryptococcus neoformans UDP-glucose dehydrogenase, is essential for growth at 37°C and for capsule biosynthesis. Eukaryot. Cell 31601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narasipura, S. D., J. G. Ault, M. J. Behr, V. Chaturvedi, and S. Chaturvedi. 2003. Characterization of Cu, Zn superoxide dismutase (SOD1) gene knock-out mutant of Cryptococcus neoformans var. gattii: role in biology and virulence. Mol. Microbiol. 471681-1694. [DOI] [PubMed] [Google Scholar]
- 35.Nimrichter, L., M. L. Rodrigues, E. G. Rodrigues, and L. R. Travassos. 2005. The multitude of targets for the immune system and drug therapy in the fungal cell wall. Microbes Infect. 7789-798. [DOI] [PubMed] [Google Scholar]
- 36.Nosanchuk, J. D., and A. Casadevall. 2006. Impact of melanin on microbial virulence and clinical resistance to antimicrobial compounds. Antimicrob. Agents Chemother. 503519-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nosanchuk, J. D., J. N. Steenbergen, L. Shi, G. S. Deepe, Jr., and A. Casadevall. 2003. Antibodies to a cell surface histone-like protein protect against Histoplasma capsulatum. J. Clin. Investig. 1121164-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oda, K., D. Kakizono, O. Yamada, H. Iefuji, O. Akita, and K. Iwashita. 2006. Proteomic analysis of extracellular proteins from Aspergillus oryzae grown under submerged and solid-state culture conditions. Appl. Environ. Microbiol. 723448-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perfect, J. R., and A. Casadevall. 2002. Cryptococcosis. Infect. Dis. Clin. N. Am. 16837-874, v-vi. [DOI] [PubMed] [Google Scholar]
- 40.Pitarch, A., A. Jimenez, C. Nombela, and C. Gil. 2006. Decoding serological response to Candida cell wall immunome into novel diagnostic, prognostic, and therapeutic candidates for systemic candidiasis by proteomic and bioinformatic analyses. Mol. Cell Proteomics 579-96. [DOI] [PubMed] [Google Scholar]
- 41.Ponnambalam, S., and S. A. Baldwin. 2003. Constitutive protein secretion from the trans-Golgi network to the plasma membrane. Mol. Membr. Biol. 20129-139. [DOI] [PubMed] [Google Scholar]
- 42.Potolicchio, I., G. J. Carven, X. Xu, C. Stipp, R. J. Riese, L. J. Stern, and L. Santambrogio. 2005. Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J. Immunol. 1752237-2243. [DOI] [PubMed] [Google Scholar]
- 43.Raposo, G., and M. S. Marks. 2007. Melanosomes—dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 8786-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rittershaus, P. C., T. B. Kechichian, J. C. Allegood, A. H. Merrill, Jr., M. Hennig, C. Luberto, and M. Del Poeta. 2006. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J. Clin. Investig. 1161651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodrigues, M. L., L. Nimrichter, D. O. Oliveira, S. Frases, K. Miranda, O. Zaragoza, M. Alvarez, A. Nakouzi, M. Feldmesser, and A. Casadevall. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 648-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salas, S. D., J. E. Bennett, K. J. Kwon-Chung, J. R. Perfect, and P. R. Williamson. 1996. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J. Exp. Med. 184377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmid, F., F. Separovic, B. M. McDougall, B. A. Stone, R. T. Brownlee, and R. J. Seviour. 2007. Characterisation of the extracellular polysaccharides produced by isolates of the fungus Acremonium. Carbohydr. Res. [Epub ahead of print.] [DOI] [PubMed]
- 48.Segura, E., S. Amigorena, and C. Thery. 2005. Mature dendritic cells secrete exosomes with strong ability to induce antigen-specific effector immune responses. Blood Cells Mol. Dis. 3589-93. [DOI] [PubMed] [Google Scholar]
- 49.Thery, C., M. Boussac, P. Veron, P. Ricciardi-Castagnoli, G. Raposo, J. Garin, and S. Amigorena. 2001. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 1667309-7318. [DOI] [PubMed] [Google Scholar]
- 50.Tucker, S. C., and A. Casadevall. 2002. Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proc. Natl. Acad. Sci. USA 993165-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Niel, G., I. Porto-Carreiro, S. Simoes, and G. Raposo. 2006. Exosomes: a common pathway for a specialized function. J. Biochem. (Tokyo) 14013-21. [DOI] [PubMed] [Google Scholar]
- 52.Waugh, M. S., C. B. Nichols, C. M. DeCesare, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148191-201. [DOI] [PubMed] [Google Scholar]
- 53.Woyke, T., M. E. Berens, D. B. Hoelzinger, G. R. Pettit, G. Winkelmann, and R. K. Pettit. 2004. Differential gene expression in auristatin PHE-treated Cryptococcus neoformans. Antimicrob. Agents Chemother. 48561-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoneda, A., and T. L. Doering. 2006. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol. Biol. Cell 175131-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaragoza, O., A. Telzak, R. A. Bryan, E. Dadachova, and A. Casadevall. 2006. The polysaccharide capsule of the pathogenic fungus Cryptococcus neoformans enlarges by distal growth and is rearranged during budding. Mol. Microbiol. 5967-83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.