Abstract
A new myosin motor-like chitin synthase gene, chsVb, has been identified in the vascular wilt fungus Fusarium oxysporum f. sp. lycopersici. Phylogenetic analysis of the deduced amino acid sequence of the chsVb chitin synthase 2 domain (CS2) revealed that ChsVb belongs to class VII chitin synthases. The ChsVb myosin motor-like domain (MMD) is shorter than the MMD of class V chitin synthases and does not contain typical ATP-binding motifs. Targeted disrupted single (ΔchsVb) and double (ΔchsV ΔchsVb) mutants were unable to infect and colonize tomato plants or grow invasively on tomato fruit tissue. These strains were hypersensitive to compounds that interfere with fungal cell wall assembly, produced lemon-like shaped conidia, and showed swollen balloon-like structures in hyphal subapical regions, thickened walls, aberrant septa, and intrahyphal hyphae. Our results suggest that the chsVb gene is likely to function in polarized growth and confirm the critical importance of cell wall integrity in the complex infection process of this fungus.
In order to establish successful infection, fungal pathogens must overcome highly effective, constitutive physical and chemical barriers, employing a range of different infection strategies. These strategies may be specific to a particular fungal species according to the nature of the host surface, and within a single species, they may depend on the type of spores initiating the infection process. A number of important steps in the infection process are common to all strategies, including adhesion to the surface of the plant, penetration of the plant surface, and acquisition of nutrients from the plant cells (18). Hyphae of plant and animal pathogenic filamentous fungi navigate on the underlying surface topography by thigmotropism in order to locate points of weakened surface integrity to gain vulnerable sites for invasion (18, 15). These sites are penetrated mechanically by expansion of the growing hyphal tips. Furthermore, fungal hyphae have been predicted to resist, at their tips, an opposing resistant force exerted by 8% (wt/vol) agar (28). These forces are related to the cell turgor pressure acting against the surface of a substrate at cell expansion places. Thus, fungal morphogenesis is an essential component for host invasion (16) and, at the same time, for establishing the correct fungal cell wall biogenesis.
Chitin, a microfibrillar β-1,4-linked homopolymer of N-acetylglucosamine (GlcNAc) (5), is considered to be a relatively minor but structurally important component of fungal cell walls. While in yeast (Saccharomyces cerevisiae), chitin constitutes 1 to 2% of the total dry weight, for filamentous fungi, the chitin content has been reported to reach up to 10 to 20% (5). Chitin seems to be present in all eukaryote kingdoms except Plantae. Thus, chitin and its biosynthesis are potentially interesting targets for the discovery of novel fungicides against phytopathogenic fungi. Chitin synthesis on the plasma membrane occurs by the extrusion of nascent chains into the cell wall space to form the skeleton layer of this extraordinary rigid structure (6). When synthesis is disrupted, the fungal wall shows characteristic alterations in composition and architecture, becoming disorganized and therefore unable to resist internal turgor pressure (20, 24, 38, 45). Chitin synthases (CHS) are the enzymes implicated in chitin synthesis, and in fungi, they constitute a great family of isozymes grouped into two divisions that branch into seven classes based on amino acid sequence similarities (29, 31, 34, 35). Furthermore, each fungal species contains a number of CHS belonging to different divisions and classes (35). In the tomato pathogen Fusarium oxysporum f. sp. lycopersici, five genes implicated in chitin synthesis, chs1, chs2, chs3, chs7, and chsV, have been isolated and characterized (24, 26). Chs1, Chs2, Chs3, and ChsV belong to classes I, II, III, and V, respectively. Meanwhile, Chs7 is orthologous to a class IV CHS-related activity from S. cerevisiae (43). In F. oxysporum, an intact cell wall structure has been implicated in the plant-host interaction (37). In fact, chitin has been shown to play an important role in the pathotypic behavior toward tomato plants (Lycopersicon esculentum). Moreover, in F. oxysporum, shown to be an emerging opportunistic human pathogen that causes mortality in immunocompromised mammalian model mice, the disruption of the chsV gene provoked a very fast mortality or “fast killing” in immunocompetent and immunosuppressed mice (32). Thus, the absence of class V CHS demonstrates that this enzyme plays different roles in the pathogenesis of plant and mammalian systems.
Interestingly, recent studies indicate the importance of the myosin motor-like CHS, exclusive to filamentous fungi species, not only for the maintenance of cell wall integrity but also for direct or indirect implications in pathotype (14, 23, 24, 44). The isolation and characterization of new myosin CHS in fungi have shown the existence of two different versions of the myosin motor-like domain (MMD). Therefore, some authors propose the classification of class V CHS in two groups. One group, named class V or class V, subgroup A, includes CHS with a larger version of the MMD containing a characteristic ATP-binding site motif (3). The second group, named class V, subgroup B, or class VI or class VII, depending on the authors, comprises CHS with a shorter MMD lacking ATP-binding site motifs (3, 7, 9, 31).
In the present study, we describe the characterization and targeted inactivation of a new F. oxysporum CHS gene, named chsVb, classified as class VII according to Niño-Vega et al. (31). The construction of single (ΔchsVb) or double (ΔchsV ΔchsVb) knockout mutants showed that ChsVb is required for full virulence of this pathogenic fungus.
MATERIALS AND METHODS
Fungal isolates and culture conditions.
For designations and genotypes of the different strains used in this study, see Table S1 in the supplemental material. F. oxysporum f. sp. lycopersici strain 4287 (race 2) was obtained from J. Tello, Universidad de Almería, Spain, and stored at −80°C, with 30% glycerol as a microconidial suspension (12). The chsV mutant strain has been isolated and described previously (24). For microconidium production, cultures were grown in potato dextrose broth (PDB) (Difco, Detroit-MI) at 28°C, with shaking at 170 rpm. The pathotypes of the isolates were periodically confirmed by plant infection assays.
For phenotypic analysis of colony growth, 5 μl water containing 2.5 × 105 freshly obtained microconidia were transferred to 1.5% (wt/vol) agar plates of synthetic medium (SM) containing 1% (wt/vol) glucose as the carbon source and 0.1% (wt/vol) NaNO3 as the nitrogen source (12). When needed, SM was supplemented with 5-bromo-4-chloro-3-indolylphosphate (BCIP), Congo red, calcofluor white (CFW), sodium dodecyl sulfate (SDS), and menadione at the indicated concentrations (all from Sigma-Aldrich Química, Spain). After inoculation, plates were maintained in the dark at 28°C.
Nucleic acid manipulations, cloning, and analysis of the chsVb gene.
Total RNA and genomic DNA were extracted from F. oxysporum mycelium as described previously (1, 8). Southern and northern blot analyses as well as probe labeling were carried out as previously reported (12) by using the nonisotopic digoxigenin labeling kit (Roche Diagnostics S.L., Spain). For reverse transcription-PCR (RT-PCR) analysis, multiple-time-sampling total RNA was isolated from mycelia grown on PDB from 0 to 24 h or from mycelia grown on SM or SM supplemented with 1.2 M sorbitol for 12 h and then reverse transcribed into first-strand cDNA with Moloney murine leukemia virus reverse transcriptase (Invitrogen S.A., Spain) by using a poly(dT) antisense primer. For normalization, initial RNA templates and the subsequent synthesized first-strand cDNAs were quantified to use equal amounts of both RNA and cDNA from each sample. For PCR amplification, primers chs1-8 and chs1-22 for chs1, chs2-12 and chs2-23 for chs2, chs3-12 and chs3-18 for chs3, chsV-8 and chsV-35X for chsV, and chsVb-18 and chsVb-19 for chsVb were used (see Table S2 in the supplemental material). All the samples were amplified during identical numbers of cycles. Controls included amplification of the actin gene by using primers act-1 and act-2 (see Table S2 in the supplemental material) for normalization of the PCR conditions and F. oxysporum genomic DNA for comparison with the intron-containing amplified bands.
Cloning of the chsVb gene (EF673037) was performed by PCR amplification of F. oxysporum 4287 genomic DNA with the primer pair chsVb-6 and chsVb-7 (see Table S2 in the supplemental material), derived from Fusarium graminearum genome sequence (www.broad.mit.edu/annotation/genome/fusarium_group) with homology to the chsV gene of F. oxysporum (AF484941). The amplified DNA fragment was used to screen a λ-EMBL3 genomic library of F. oxysporum f. sp. lycopersici strain 4287 as described in standard protocols (36). Sequencing of both DNA strands was performed at the SIPI-Universidad de Córdoba by using the Dyedeoxy Terminator cycle sequencing kit (PE Biosystems) on an ABI Prism 377 genetic analyzer apparatus (Applied Biosystems). DNA and protein sequence databases were searched using the BLAST algorithm (2) at the National Center for Biotechnology Information (NCBI, Bethesda, MD).
Phylogenetic analyses.
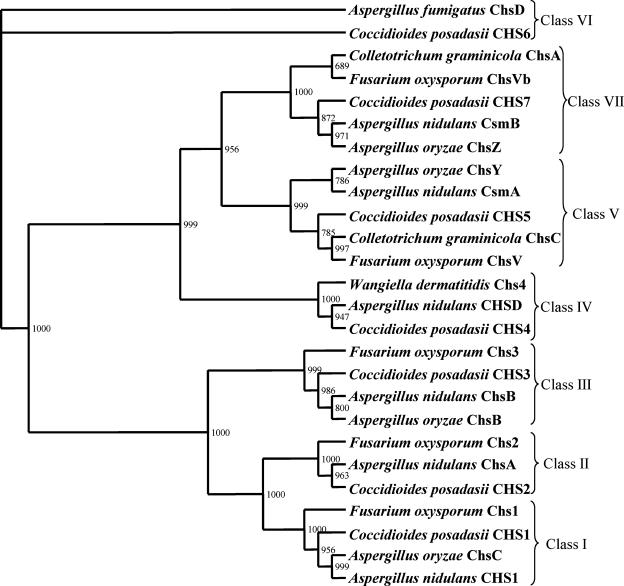
The predicted amino acid sequence of F. oxysporum ChsVb was submitted to the Conserved Domain Database (see the NCBI Web page at www.ncbi.nlm.nih.gov/structure/cdd/wrpsb.cgi) to localize its conserved CS2 amino acid domain. This search identified a CS2 domain represented by amino acids 1055 to 1590. Thus, the conserved CS2 amino acid domains from this protein as well as the previously described synthases Chs1, Chs2, Chs3, and ChsV from F. oxysporum (24, 26) and the fungal CHS related to classes I to VII, reported by Mandel et al. (25), were aligned with ClustalW algorithm (42). The phyml 2.4.4 program (17) was used to perform a 1,000 nonparametric bootstrap phylogenetic analysis of the resulting alignment of 455 amino acid characters with the maximum likelihood method after optimization of the settings by the ModelGenerator program, version 0.84 (22). The analysis was performed using the rtREV substitution model (10), with a gamma distribution parameter alpha of 1.89 and a proportion of invariable sites of 0.04. The phylogenetic relationship between CHS sequences was depicted in a phylogenetic tree constructed using the TreeView 1.6.6 program (33) (Fig. 1).
FIG. 1.
Phylogenetic tree of 26 fungal CHS, representing seven classes of these enzymes. Sequences from the conserved CS2 amino acid domain found in all CHS were aligned using ClustalW. Numbers at the branch points represent bootstrap values based on 1,000 repeated samplings.
Construction of gene disruption vectors and fungal transformation.
A vector for simultaneous replacement of both chsV and chsVb was constructed with an adapted, cloning-free, PCR-based allele replacement and bipartite gene-targeting substrate protocols adapted from those of Erdeniz et al. (13) and Nielsen et al. (30). This method employed two vectors, pDchsV (24) and pDchsVb (developed during this work), each one harboring the hygromycin B resistance (Hygr) cassette interrupting the open reading frame (ORF) of the corresponding gene. Two independent fragments were amplified using the primer pairs chsV-25 and hph-6 for pDchsV and chsVb-27 and hph-12 for pDchsVb (see Table S1 in the supplemental material). Two segments were generated, one containing a 1,640-bp fragment corresponding to the 3′ end of the Hygr cassette adjacent to 710 bp of the chsV ORF, and the other one containing a 1,394-bp fragment corresponding to the 5′ end of the Hygr cassette adjacent to 1,102 bp of the chsVb ORF. Thus, both amplified segments were overlapping in 390 bp of the Hygr cassette and neither contained the complete cassette, involving the requirement of a homologous recombination event in order to generate the complete and functional Hygr.
This bipartite gene-targeting substrate was used for cotransformation of F. oxysporum wild-type protoplasts, according to a protocol described previously (12). Hygr transformants were routinely subjected to two consecutive rounds of single sporing and stored as microconidia at −80°C.
Optical, fluorescence, and transmission electron microscopy (TEM).
Optical microscopy analysis of ΔchsVb and ΔchsV ΔchsVb strains was performed with germlings grown on SM, with or without 1.2 M sorbitol, for 14 h by using the Nomarsky technique with a Leica DMR microscope (objective, 100×).
For fluorescence analysis, germlings grown on PDB for 14 h were transferred to a solution containing 3.7% formaldehyde, 50 mM phosphate buffer, pH 7.0, and 0.2% Triton X-100 for fixation and incubated at room temperature for 30 to 45 min (19). Staining was achieved by incubating 5 min at room temperature with 10 μg ml−1 CFW and 0.8 μg ml−1 4′,6-diamidino-2-phenylindole (DAPI) (both from Sigma). Germlings were then mounted in 10% phosphate buffer, pH 7.0, and 50% glycerol and screened by fluorescence microscopy using a 100× objective (Leica DMR).
For TEM, germlings were initially fixed overnight at 4°C in a mixture of 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.0, and then washed in buffer and postfixed in 1% osmium tetroxide at 4°C. After dehydration in an ethanol series, the samples were treated with propylene oxide and embedded in EMBed 812. After curing, the blocks were sectioned with a thickness of about 80 nm in an ultramicrotome and mounted on Cu grids. The samples were stained in 2% aqueous uranyl acetate for 2 min at 37°C and then transferred to Reynolds lead citrate for 3 min at room temperature. Micrographs were obtained using a Philips CM 10 electron microscope.
Virulence assays.
Tomato plant infection assays were performed as reported previously (12). Seeds from tomato cv. Monika were kindly provided by Novartis Seeds. Briefly, 2-week-old tomato seedlings were inoculated with F. oxysporum strains by immersing the roots in a suspension of 5 × 106 spores ml−1 planted in vermiculite and maintained in a growth chamber. At different times after inoculation, the severity of disease symptoms was recorded using an index ranging from 1 (healthy plant) to 5 (dead plant). Fifteen plants were used for each treatment. Assays for invasive growth on tomato fruits (cv. Daniela) were carried out as described previously (11).
RESULTS
Isolation, characterization, and phylogenetic relationship of F. oxysporum chsVb.
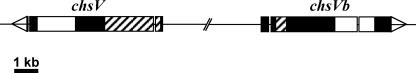
In order to identify class V CHS-related genes from F. oxysporum f. sp. lycopersici, the previously isolated gene chsV (24) was used to search for homologous sequences in the F. graminearum genomic database (www.broad.mit.edu/annotation/genome/fusarium_group) and, from these, the primers chsVb-6 and chsVb-7 were designed (see Table S1 in the supplemental material). PCR amplification of genomic DNA from F. oxysporum f. sp. lycopersici wild-type strain 4287 gave a fragment that was cloned and identified by DNA sequence analysis. A F. oxysporum genomic λ-EMBL3 library was probed with the PCR fragment to isolate the complete gene, allowing the identification of a novel CHS gene. The gene was designated chsVb, and the sequence data were submitted to the DDBJ/EMBL/GenBank databases (EF673037). The genes chsVb and chsV are arranged in a head-to-head configuration within the F. oxysporum genome at a distance of 4,219 bp between their translational start points (Fig. 2). This head-to-head configuration has previously been described for orthologue genes in other ascomycetes (25, 41).
FIG. 2.
Organization of genes chsV and chsVb in a head-to-head configuration within Fusarium oxysporum f. sp. lycopersici genome. The MMD (striped bars) and the typical CHS CS2 domain (white bars) are indicated. The orientation of transcription is shown by arrowheads. The distance between both translational start points is 4,219 bp.
The analysis of the complete DNA sequence revealed an ORF of 5,440 bp encoding a 1,780-amino-acid polypeptide organized in three exons interrupted by two introns. The first intron of 49 bp is 270 bp downstream of the putative start codon, and the second of 51 bp is 1,425 bp upstream of the stop codon. ChsVb has an N-terminal myosin-like domain between amino acids 180 to 331, in contrast to that in ChsV located between amino acids 19 to 779 (24). Furthermore, the full-length sequences of both proteins (ChsV and ChsVb) showed an overall identity of 33%, 62% identity along the catalytic domains, and only 28% identity along the MMDs. Low identity of MMDs has been previously described for these type of proteins. The absence of the ATP-binding motifs P-loop, switch I, and switch II is characteristic in class VII CHS containing myosin-like domains (3, 7, 25, 31, 41). From the homology search, the amino acid sequence of chsVb indicated 71% identity to ChsA from Colletotrichum graminicola (3), 65% to ChsVI from Botrytis cinerea (9), 60% to ChsZ from Aspergillus oryzae (7), 59% to Chs4 from Paracoccidioides brasiliensis (31), 58% to CsmB from Aspergillus nidulans (41), and 58% to CHS7 from Coccidioides posadasii (25).
To establish the correct classification of the new ChsVb, the amino acid sequences of CS2 domains from all the F. oxysporum CHS proteins were aligned (24, 26), together with those representing other filamentous fungal classes. Figure 1 presents the resulting phylogenetic tree, showing the relationship between the CHS. Based on the nomenclature proposed by Niño-Vega et al. (31), ChsVb might be considered a novel member of class VII CHS.
Simultaneous targeting disruption of chsV and chsVb genes.
In order to disrupt both the chsV gene and the chsVb gene, bipartite gene-targeting substrate (13, 30) was used for the cotransformation of F. oxysporum f. sp. lycopersici 4287 protoplasts. We analyzed 40 Hygr transformants by PCR, investigating the loss of the 4.2-kb fragment corresponding to the intergenic region between both genes. One transformant (Tra6) lacked the complete sequence, while another (Tra31) missed a 1.3-kb internal fragment of the chsVb gene (data not shown). Both transformants were further characterized by Southern analysis (data not shown). Tra6 genomic DNA digested with PstI showed the replacement of the 14.2-kb fragment, corresponding to the wild-type chsV and chsVb alleles, by a 1.4-kb fragment when the probe was chsV gene or by a 3.7-kb band when chsVb was used. In contrast, Tra31 maintained the 14.2-kb wild-type band when the probe was the chsV, but the pattern was different in hybridization with the chsVb probe. In addition, analysis of both transformants using the enzyme XhoI and hybridized to the chsV probe, showed the replacement of the 4.4-kb wild-type fragment by a 7.9-kb fragment in Tra6, while Tra31 maintained the wild-type band. The hybridization of transformants Tra6 and Tra31 to chsVb probe showed the loss of the wild-type 3.5-kb fragment, being replaced by two new bands, one of 7.9 kb and a larger one. When DNA from the ectopic transformant (Tra17) was digested with both enzymes and probed with chsV or chsVb, the wild-type pattern was observed. From these results, we conclude that in Tra6, both genes (chsV and chsVb) have been successfully deleted, while only the chsVb gene is disrupted in Tra31.
Expression analysis of F. oxysporum chs genes.
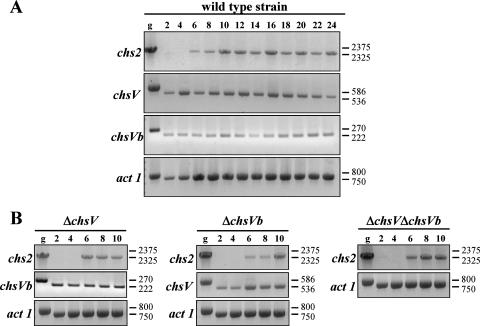
Time course analyses of chs genes during mycelial growth of F. oxysporum wild-type strain and ΔchsV, ΔchsVb, and ΔchsV ΔchsVb mutants were analyzed using RT-PCR. chs1, chs3, chs7 (26), chsV (24), and chsVb showed similar transcription levels during all developmental stages (data not shown and Fig. 3A), while the expression of chs2 was not detected before 6 h of growth on PDB in the wild-type strain (Fig. 3A) as well as in ΔchsV, ΔchsVb, and ΔchsV ΔchsVb mutants (Fig. 3B). These results indicate that the expression of CHS genes during mycelial growth is independent of the stage-specific function, similar to the situation described for C. posadasii CHS genes (25).
FIG. 3.
(A) Time course of chs2, chsV, and chsVb gene transcription during developmental mycelial growth of the F. oxysporum wild-type strain (2 to 24 h). (B) Expression analysis of chs2, chsV, and chsVb genes by RT-PCR during developmental stages of the mycelial growth of the ΔchsV, ΔchsVb, and ΔchsV ΔchsVb mutants for 2 to 10 h. Actin gene transcripts (act 1) were used as controls for cDNA amounts in the different samples. Genomic DNA (g) was used as a control for transcript size. Sizes of bands are in base pairs.
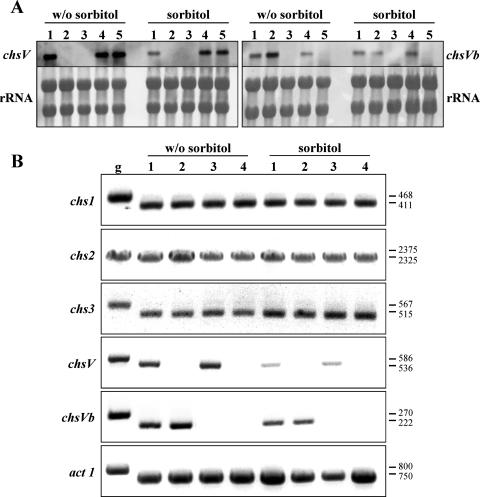
To determine whether the expression of chsV and chsVb genes is regulated by osmotic stress, total RNA from the wild-type strain and ΔchsV, ΔchsVb, and ΔchsV ΔchsVb mutants grown on SM with or without 1.2 M sorbitol was analyzed by northern blotting and RT-PCR. The transcription level of chsV gene is strongly reduced in the presence of sorbitol, both in wild-type and ΔchsVb strains (Fig. 4). On the other hand, the expression of the chsVb gene in all the strains was lower in comparison to that of chsV and was unaffected by the presence of sorbitol. Additionally, the transcription of the chsVb gene was increased in the ΔchsV mutant background, although this result was suppressed by the addition of sorbitol (Fig. 4A and B). These results indicate a compensatory mechanism by the overexpression of chsVb in the absence of the functional chsV gene but not vice versa. Therefore, our results are only partially in agreement with those reported by Takeshita et al. for A. nidulans (41), where the deletion of either csmA or csmB increased the expression levels of its counterpart.
FIG. 4.
(A) Northern analysis of chsV and chsVb genes from F. oxysporum grown on SM with or without (w/o) 1.2 M sorbitol. Lanes: 1, wild-type strain; 2, ΔchsV; 3, ΔchsV ΔchsVb; 4, ectopic transformant; 5, ΔchsVb. (B) RT-PCR analysis of the indicated F. oxysporum chs genes. Lanes: 1, wild-type strain; 2, ΔchsV; 3, ΔchsVb; 4, ΔchsV ΔchsVb. Growth conditions were the same as those described for panel A. Size bands are in base pairs. g, genomic DNA.
As expected, of the ΔchsV, ΔchsVb, and ΔchsV ΔchsVb mutants, none showed any expression of the corresponding disrupted genes.
In order to assess whether the compensatory mechanism in the defective chsV and/or chsVb mutants was extensive to other available structural chs genes, the transcription levels of chs1, chs2, and chs3 was determined in strains grown under low and high osmotic conditions. Considering the low expression levels detected in the genes under study (26), a RT-PCR approach was performed using RNA from mycelia obtained from wild-type, ΔchsV, ΔchsVb, and ΔchsV ΔchsVb strains grown with or without sorbitol. The amplified cDNA bands observed for all genes suggested a similar expression pattern for the three chs genes (Fig. 4B).
Phenotypes of ΔchsVb and ΔchsV ΔchsVb mutants.
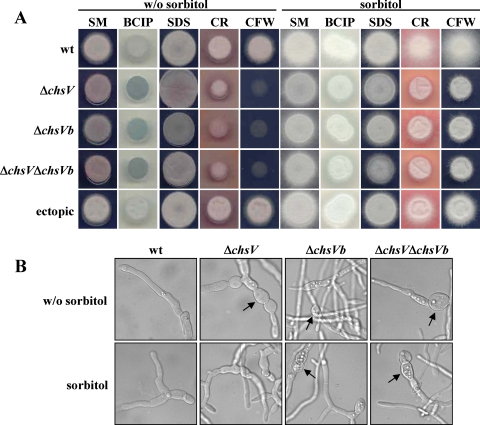
To investigate the physiological function of ChsVb, the phenotypes of single- and double-null mutants, ΔchsVb and ΔchsV ΔchsVb, respectively, were compared to those of the wild-type strain and ΔchsV mutant. The colony sizes of the defective mutants grown on SM plates were slightly smaller than those of the wild-type strain or the ectopic transformant (Fig. 5A). Furthermore, staining of the chsV and/or chsVb mutant colonies grown on plates containing the vital stain BCIP (60 μg ml−1) produced a blue zone around the colony, indicating that hyphal lysis occurred at their margins. This effect was suppressed by the presence of 1.2 M sorbitol (Fig. 5A). No such blue zones were observed for the wild-type or the ectopic transformant. Colonial growth in the presence of Congo red (50 μg ml−1), a compound interfering with fungal cell wall assembly, was strongly inhibited in the ΔchsV, ΔchsVb, and ΔchsV ΔchsVb mutants. Nevertheless, no inhibitory effect was observed in the presence of the detergent SDS (0.015%), which affects membrane integrity (Fig. 5A), or with the oxidative stress compound menadione (10 μg ml−1) (data not shown). Increased sensitivity to CFW (50 μg ml−1), a chitin binding dye, was observed in the defective mutants chsV and/or chsVb by the strong inhibition of their colonial growth (Fig. 5A).
FIG. 5.
(A) Phenotypes of colonies from F. oxysporum wild-type and mutant strains, showing alterations in cell wall or membrane integrity. (B) Germlings from the wild-type (wt) strain and ΔchsV, ΔchsVb, and ΔchsV ΔchsVb mutants grown on SM with or without (w/o) 1.2 M sorbitol (magnification, ×50). Arrows indicate swollen ballon-like structures. BCIP, 5-bromo-4-chloro-3-indolylphosphate; SDS, sodium dodecyl sulfate; CR, Congo red; CFW, calcofluor white.
Similarly to the phenotype previously described for ΔchsV (24), the chsVb mutants formed thick walls and swollen balloon-like structures in subapical regions of the hyphae (Fig. 5B) as well as lemon-like shape microconidia (data not shown). These aberrant structures are never observed in the wild-type strain. Madrid et al. (24) stated that aberrant structures are alleviated when the ΔchsV mutant was grown in the presence of 1.2 M sorbitol. In contrast, the abnormally shaped hyphae caused by the nonfunctionality of the chsVb gene newly described in this work were not restored by the addition of the osmotic stabilizer sorbitol (Fig. 5B).
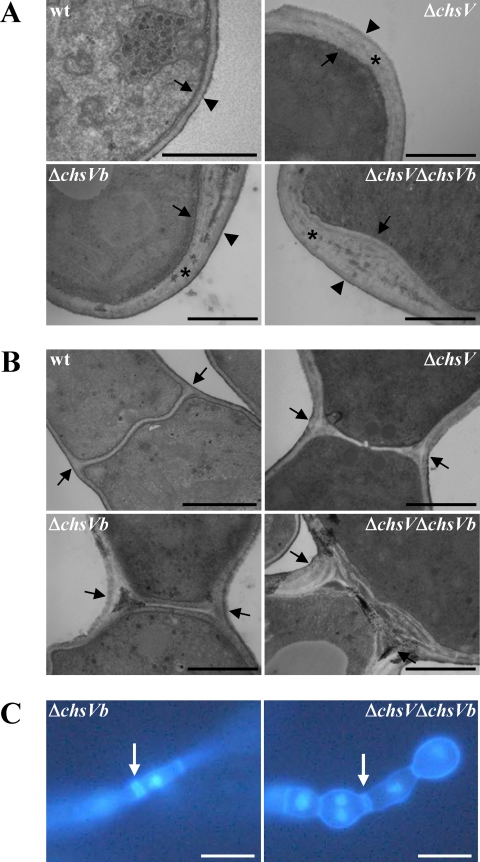
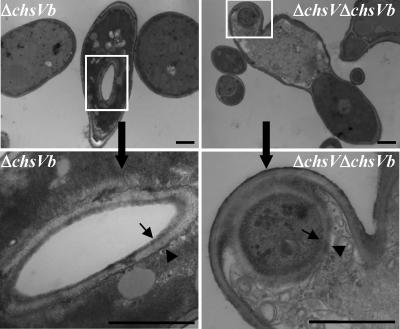
Previous studies have shown that the F. oxysporum cell wall is composed of three layers: the inner electron-dense layer, corresponding to the plasma membrane and adjacent periplasmic proteins, the electron-transparent layer enriched in carbohydrate polymers representing the skeletal layer, and the outer electron-dense layer enriched in proteins associated with the underlying skeletal layer (37). Electron microscopy observations of the cell wall from the wild-type strain showed that the skeletal and the outer layers were approximately 70-nm wide. In contrast, the CHS null mutants ΔchsV, ΔchsVb, and ΔchsV ΔchsVb showed an electron-transparent layer threefold thicker than that of the wild-type strain (Fig. 6A). In addition, aberrant septa were occasionally observed by TEM in hyphal subapical regions of ΔchsVb and ΔchsV ΔchsVb mutants (Fig. 6B). This fact was further investigated by simultaneous DAPI and CFW staining of germlings from both strains (Fig. 6C). Furthermore, a three-layer-like structure very similar to typical cell walls was detected by TEM analysis inside the hyphae from ΔchsVb and ΔchsV ΔchsVb mutants (Fig. 7) resembling the intrahyphal hyphae described for csmA and csmB null mutants from A. nidulans (20, 41).
FIG. 6.
Ultrastructural characteristics of cell walls (A) and septa (B) of the wild-type (wt) strain and the ΔchsV, ΔchsVb, and ΔchsV ΔchsVb mutants. Mutant cell walls are significantly larger than those of the wild-type strain. Arrows, plasma membrane; asterisks, skeletal layer; arrowheads, outer layers. In panel B, septa are denoted by arrows. (C) Septa from ΔchsVb and ΔchsV ΔchsVb were observed using fluorescence microscopy after staining with DAPI and CFW. Scale bars, 1 μm (A and B) and 100 μm (C).
FIG. 7.
Intrahyphal hypha-like structures detected in ΔchsVb and ΔchsV ΔchsVb mutants observed under TEM. Lower panels correspond to magnified areas boxed in the upper panels. Three layer-like structures can be observed: the plasma membrane (arrow), the outer layer (arrowhead), and the skeletal layer in between. Bars, 1 μm.
Role of ChsVb in pathogenicity.
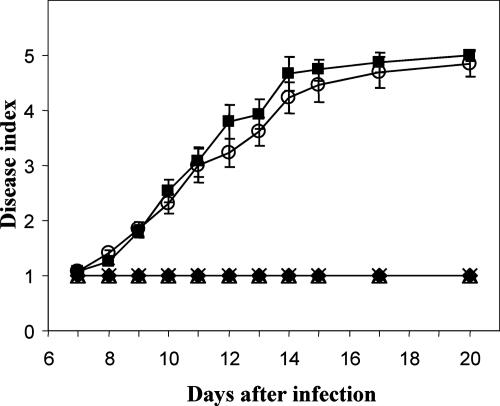
To determine the pathotypic behavior of the chsVb null mutants, root infection assays with tomato plants were performed by immersing roots of 2-week-old tomato plants in a microconidial suspension of the wild-type strain, the ectopic transformant or the ΔchsVb and ΔchsV ΔchsVb mutants. Plants were scored for vascular wilt symptoms at different time intervals (12). The development of disease is shown in Fig. 8. Plants inoculated with the wild-type strain showed characteristic wilt symptoms starting 7 days after inoculation. Disease severity increased steadily throughout the experiment, and all the plants were dead 20 days after inoculation. Plants inoculated with the ectopic transformant showed a very similar disease profile. In contrast, plants inoculated with single- and double-chsVb null mutants failed to show any visible disease symptoms and remained as healthy as the water controls throughout the experiment.
FIG. 8.
Virulence of F. oxysporum f. sp. lycopersici chs-deficient mutants on tomato plants (cv. Monika). Severity of disease symptoms was recorded at different times after inoculation by using an index ranging from 1 (healthy plant) to 5 (dead plant). Symbols refer to plants inoculated with the wild-type strain 4287 (▪), ΔchsVb (×), ΔchsV ΔchsVb (▵), the ectopic transformant (○), and the noninoculated control (⧫). Error bars indicate the standard errors from 15 plants for each treatment.
To study the effect of the chsVb mutation on the capacity to proliferate on living host tissue, tomato fruits were injected with a microconidial suspension of the different strains. Figure 9 shows the tomato fruits inoculated with the wild-type strain, the ectopic transformant, and the chsVb single- and double-null mutants after 24 h of incubation at 100% relative humidity. The wild-type strain and the ectopic transformant colonized and macerated the fruit tissue surrounding the site of inoculation, forming a dense mycelium on the surface of the fruit. Single- or double-chsVb mutants did not show the capacity for invading the fruit tissue; however, the ΔchsVb mutant could macerate the tissue in contrast to the ΔchsV ΔchsVb mutant and the previously described ΔchsV (24).
FIG. 9.
Invasive growth of F. oxysporum strains on tomato fruits at the inoculation site after 24 h of incubation at 100% relative humidity.
DISCUSSION
F. oxysporum f. sp. lycopersici contains two genes encoding myosin motor-like CHS, chsVb and the previously identified chsV (24). Three additional CHS genes and a chaperone-like protein related to the class IV CHS activity of S. cerevisiae (43) had previously been isolated (26). In spite of the targeted inactivation of chs1, chs2, chsV, and chs7 genes, their specific functions in chitin biosynthesis have scarcely been revealed. Expression analyses of these genes during the developmental process of F. oxysporum showed that chs2 transcription appears to be inhibited at the onset of germination (2 and 4 h), while no significant differences were detected with the rest of the genes.
ChsVb shows high similarity (71 to 58%) to other class VII CHS. Like other fungal CHS belonging to this class, ChsVb lacks the characteristic myosin motor signatures, such as the P-loop or switch I and switch II, present in class V CHS (3, 7, 25, 31, 41). The highest identity was with ChsA from C. graminicola, which has been classified as class V, subgroup B, or class VI or class VII CHS, depending on the authors (3, 4, 7, 9, 25, 27, 31). An analysis of phylogenetic relationships among fungal CHS revealed that ChsVb clustered into class VII CHS according to the nomenclature proposed by Niño-Vega et al. (31), allowing us to propose ChsVb as a new member of the class VII CHS.
F. oxysporum ChsVb and ChsV perform different roles in growth and morphogenesis.
The biological function of ChsVb in F. oxysporum has been demonstrated in the present study by targeted inactivation of the gene. Phenotypes similar to those of the chsV null mutant have been observed: lemon-like shaped conidia, swelling hyphae, hyphal lysis, and sensitivity to CFW and Congo red that could be ameliorated by the addition of osmotic stabilizers (24). However, unlike in the chsV null mutants, hyphal morphological abnormalities of chsVb null mutants could not be restored by supplementing the growth medium with osmotic stabilizers, suggesting that ChsVb functions not only in the maintenance of cell wall integrity under different osmotic conditions but also in polarized cell wall synthesis. Takeshita et al. (40, 41) demonstrated the increased transcription levels of csmA and csmB genes when A. nidulans was grown under low osmotic conditions, thus indicating an important role of these genes in growth and morphogenesis. Our studies on the two F. oxysporum orthologue genes (chsV and chsVb) found similar expression levels for chsVb, both under low or high osmotic conditions, while chsV seems to be down regulated under high osmotic conditions. These differential gene expression profiles are consistent with the opposite phenotypes observed between both types of mutants on medium supplemented with sorbitol: the swelling hyphae of ΔchsV mutant are restored in the presence of 1.2 M sorbitol. Meanwhile, these aberrant structures persist in chsVb mutants under similar growth conditions. Thus, we speculate that ChsVb functions both under low or high osmotic conditions, in contrast to a specific ChsV role, being produced mainly in the absence of osmotic stabilizers. Furthermore, overexpression of the chsVb gene in the ΔchsV mutant under low osmotic conditions did not suppress the defects caused by the deletion of chsV. The existence of different gene expression patterns, in addition to the functional differences shown by chsV and chsVb null mutants, support the idea that ChsV and ChsVb play different roles in growth and morphogenesis as well as during the infection process in tomato plants. Class V CHS of A. nidulans (CsmA) has been reported to play an important role in the maintenance of cell wall integrity under low osmotic conditions. Its localization at the hyphal tips and septation sites by binding its MMD to the actin cytoskeleton suggests its involvement in determination of hyphal polarity (39-41). A similar function was described for CsmB, the second CHS with MMD from A. nidulans (41). Conservation of CHS from classes V and VII among filamentous fungi might imply that their functions in characteristic polarized growth are also conserved. However, further analyses must be performed for F. oxysporum in order to determine the localization and precise functions of ChsV and ChsVb CHS.
In the dimorphic fungus Ustilago maydis, simultaneous inactivation of mcs1 and chs6 genes resulted in increased swollen yeast-like cells (44). On the other hand, double-null mutants in the filamentous fungus A. nidulans for both CsmA and CsmB CHS with MMDs were not viable, suggesting that they play compensatory roles essential for hyphal growth (41). In contrast, in F. oxysporum, viable double-chsV chsVb mutants were obtained, showing hyphal structure defects similar to those of single chsV or chsVb mutant strains, suggesting that both enzymes may play different noncompensatory roles in development. Furthermore, single chsVb and double chsV chsVb mutants, but not the single chsV mutant, showed intrahyphal hyphae as well as morphological abnormalities in septum formation and distribution, the latter being more profound in the double-chsV chsVb mutant, denoting that ChsV and ChsVb do perform additive, but not redundant, functions in septum formation. These phenotypes are similar to those of A. nidulans csmA and csmB single mutants (20, 41) and chsA chsC double mutants (21). The formation of intrahyphal hyphae in csmA mutants seemed to be caused by septa in the hyphal compartments of the old regions due to the intracellular conditions of old mycelia. Thus, CsmA might function in chitin metabolism in the septated hyphal compartments of the old compartments (20). TEM images of F. oxysporum ΔchsVb and ΔchsV ΔchsVb mutants denoted intrahyphal hyphae whose origin remains unknown.
The characteristic head-to-head configuration of genes chsV and chsVb from the F. oxysporum genome is conserved among orthologous genes in other ascomycete filamentous fungi, such as A. nidulans (41) or C. posadasii (25). In A. nidulans, this configuration implies a common transcriptional regulation for csmA and csmB genes, the start translation codons are 2.4 kb apart and mutational deletion of either csmA or csmB increases the expression levels of each counterpart (41). Nevertheless, in F. oxysporum the larger distance between chsV and chsVb (4.2 kb), together with the results obtained by northern analysis, suggests independent transcriptional regulation for both genes, as it was speculated for A. oryzae chsY and chsZ genes (7).
No striking changes in transcription levels of the chs1, chs2, and chs3 structural CHS genes were detected in ΔchsV, ΔchsVb, or ΔchsV ΔchsVb mutants. Thus, no compensatory mechanisms occur at the transcriptional level in response to defective chsV and/or chsVb genes.
The class VII CHS is essential for pathogenesis of F. oxysporum.
The inability of chsVb mutants to cause disease in tomato plants might be due to cell wall structure defects. Schoffelmeer et al. (37) described the F. oxysporum cell wall as composed of three layers. The cell wall in chsVb null mutants has a skeletal layer three times thicker than that of the wild-type strain. This engrossed layer indicates the disorganization of structural components such as chitin. In addition, a compensatory mechanism of chitin and glucan polysaccharides synthesis might exist, producing larger amounts of these polymers. All these changes could alter the normal cross-linking process of components in the cell wall space (6). Thus, differences in permeability could cause higher sensitivity to plant defense compounds in the class V and VII null mutants, resulting in the nonpathogenic behavior of these strains. Alternatively, wall defects could lead to partial or total destruction of the pathogen by the host, as a result of a differential exposure of avirulence factors that may be recognized by the plant.
Recently, studies of the phytopathogenic basidiomycete U. maydis have shown that mcs1 and chs6 genes are essential for pathogenicity (14, 44). In contrast to the F. oxysporum chsV mutant, the absence of virulence of U. maydis mcs1 mutant was the result of its defects in invading deeper layers of the host tissue, while the Δchs6 mutant failed to colonize the plant tissues, suggesting the inability to overcome the host plant defense mechanisms (14).
In F. oxysporum, the chsVb gene is likely to function in polarized growth during the infection process of the host plant, supporting the critical importance of the cell wall integrity in pathogenicity and the complexity of the infection process.
Supplementary Material
Acknowledgments
We gratefully acknowledge Nicolas Rispail for valuable help in phylogenetic analysis, Antonio Di Pietro for helpful discussions, and Esther Martínez for technical assistance (all from the University of Córdoba).
This research was supported by the Ministerio de Educación y Ciencia of Spain (BIO2004-0276) and Junta de Andalucía (CVI-138). M.M.U. was supported by a Ph.D. fellowship from Junta de Andalucía.
Footnotes
Published ahead of print on 9 November 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aljanabi, S. M., and I. Martinez. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 254692-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amnuaykanjanasin, A., and L. Epstein. 2003. A class V chitin synthase gene, chsA is essential for conidial and hyphal wall strength in the fungus Colletotrichum graminicola (Glomerella graminicola). Fungal Genet. Biol. 38272-285. [DOI] [PubMed] [Google Scholar]
- 4.Amnuaykanjanasin, A., and L. Epstein. 2006. A class Vb chitin synthase in Colletotrichum graminicola is localized in the growing tips of multiple cell types, in nascent septa, and during septum conversion to an end wall after hyphal breakage. Protoplasma 227155-164. [DOI] [PubMed] [Google Scholar]
- 5.Bartnicki-Garcia, S. 1968. Cell wall chemistry, morphogenesis, and taxonomy of fungi. Annu. Rev. Microbiol. 2287-108. [DOI] [PubMed] [Google Scholar]
- 6.Bowman, S. M., and S. J. Free. 2006. The structure and synthesis of the fungal cell wall. Bioessays 28799-808. [DOI] [PubMed] [Google Scholar]
- 7.Chigira, Y., K. Abe, K. Gomi, and T. Nakajima. 2002. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr. Genet. 41261-267. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 9.Choquer, M., M. Boccara, I. R. Goncalves, M. C. Soulie, and A. Vidal-Cros. 2004. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur. J. Biochem. 2712153-2164. [DOI] [PubMed] [Google Scholar]
- 10.Dimmic, M. W., J. S. Rest, D. P. Mindell, and R. A. Goldstein. 2002. rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J. Mol. Evol. 5565-73. [DOI] [PubMed] [Google Scholar]
- 11.Di Pietro, A., F. I. García-Maceira, E. Méglecz, and M. I. G. Roncero. 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 391140-1152. [PubMed] [Google Scholar]
- 12.Di Pietro, A., and M. I. G. Roncero. 1998. Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant-Microbe Interact. 1191-98. [DOI] [PubMed] [Google Scholar]
- 13.Erdeniz, N., U. H. Mortensen, and R. Rothstein. 1997. Cloning-free PCR-based allele replacement methods. Genome Res. 71174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcerá-Teruel, A., B. Xoconostle-Cázares, R. Rosas-Quijano, L. Ortiz, C. León-Ramírez, C. A. Specht, R. Sentandreu, and J. Ruiz-Herrera. 2004. Loss of virulence in Ustilago maydis by Umchs6 gene disruption. Res. Microbiol. 15587-97. [DOI] [PubMed] [Google Scholar]
- 15.Gow, N. A. R., T. H. Perera, J. Sherwood-Higham, G. W. Gooday, D. W. Gregory, and D. Marshall. 1994. Investigation of touch-sensitive responses by hyphae of the human pathogenic fungus Candida albicans. Scanning Microsc. 8705-710. [PubMed] [Google Scholar]
- 16.Gow, N. A. R., A. J. P. Brown, and F. C. Odds. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5366-371. [DOI] [PubMed] [Google Scholar]
- 17.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52696-704. [DOI] [PubMed] [Google Scholar]
- 18.Hardham, A. R. 2001. Cell biology of fungal infection of plants, p. 91-123. In N. A. R. Gow and R. J. Howard (ed.), The mycota VIII. Springer-Verlag, Berlin, Germany.
- 19.Harris, S. D., J. L. Morrell, and J. E. Hamer. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136517-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horiuchi, H., M. Fujiwara, S. Yamashita, A. Ohta, and M. Takagi. 1999. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 1813721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichinomiya, M., E. Yamada, S. Yamashita, A. Ohta, and H. Horiuchi. 2005. Class I and class II chitin synthases are involved in septum formation in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell 41125-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keane, T. M., C. J. Creevey, M. M. Pentony, T. J. Naughton, and J. O. McLnerney. 2006. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, H. B., S. Kauffman, J. M. Becker, and P. J. Szaniszlo. 2004. Wangiella (Exophiala) dermatitidis WdChs5p, a class V chitin synthase, is essential for sustained cell growth at temperature of infection. Eukaryot. Cell 340-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madrid, M. P., A. Di Pietro, and M. I. G. Roncero. 2003. Class V chitin synthase determines pathogenesis in the vascular wilt fungus Fusarium oxysporum and mediates resistance to plant defence compounds. Mol. Microbiol. 47257-266. [DOI] [PubMed] [Google Scholar]
- 25.Mandel, M. A., J. N. Galgiani, S. Kroken, and M. J. Orbach. 2006. Coccidioides posadasii contains single chitin synthase genes corresponding to classes I to VII. Fungal Genet. Biol. 43775-788. [DOI] [PubMed] [Google Scholar]
- 26.Martín-Udíroz, M., M. P. Madrid, and M. I. G. Roncero. 2004. Role of chitin synthase genes in Fusarium oxysporum. Microbiology 1503175-3187. [DOI] [PubMed] [Google Scholar]
- 27.Mellado, E., A. Aufauvre-Brown, N. A. R. Gow, and D. W. Holden. 1996. The Aspergillus fumigatus chsC and chsG genes encode class III chitin synthases with different functions. Mol. Microbiol. 20667-679. [DOI] [PubMed] [Google Scholar]
- 28.Money, N. P. 2001. Biomechanics of invasive hyphal growth, p. 3-17. In N. A. Gow and R. J. Howard (ed.), The mycota VIII. Springer-Verlag, Berlin, Germany.
- 29.Munro, C. A., and N. A. R. Gow. 2001. Chitin synthesis in human pathogenic fungi. Med. Mycol. 3941-53. [PubMed] [Google Scholar]
- 30.Nielsen, M. L., L. Albertsen, G. Lettier, J. B. Nielsen, and U. H. Mortensen. 2006. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 4354-64. [DOI] [PubMed] [Google Scholar]
- 31.Niño-Vega, G. A., L. Carrero, and G. San-Blas. 2004. Isolation of the CHS4 gene of Paracoccidioides brasiliensis and its accommodation in a new class of chitin synthases. Med. Mycol. 4251-57. [DOI] [PubMed] [Google Scholar]
- 32.Ortoneda, M., J. Guarro, M. P. Madrid, Z. Caracuel, M. I. G. Roncero, E. Mayayo, and A. Di Pietro. 2004. Fusarium oxysporum as a multihost model for the genetic dissection of fungal virulence in plants and mammals. Infect. Immun. 721760-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12357-358. [DOI] [PubMed] [Google Scholar]
- 34.Roncero, C. 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41367-378. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Herrera, J., J. Manuel González-Prieto, and R. Ruiz-Medrano. 2002. Evolution and phylogenetic relationships of chitin synthases from yeasts and fungi. FEMS Yeast Res. 1247-256. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3th ed. Cold Spring Harbor Laboratory, New York, NY.
- 37.Schoffelmeer, E. A. M., F. M. Klis, J. H. Sietsma, and B. J. C. Cornelissen. 1999. The cell wall of Fusarium oxysporum. Fungal Genet. Biol. 27275-282. [DOI] [PubMed] [Google Scholar]
- 38.Specht, C. A., Y. L. Liu, P. W. Robbins, C. E. Bulawa, N. Iartchouk, K. R. Winter, P. J. Riggle, J. C. Rhodes, C. L. Dodge, D. W. Culp, and P. T. Borgia. 1996. The chsD and chsE genes of Aspergillus nidulans and their roles in chitin synthesis. Fungal Genet. Biol. 20153-167. [DOI] [PubMed] [Google Scholar]
- 39.Takeshita, N., A. Ohta, and H. Horiuchi. 2005. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans. Mol. Biol. Cell 161961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeshita, N., A. Ohta, and H. Horiuchi. 2002. csmA, a gene encoding a class V chitin synthase with a myosin motor-like domain of Aspergillus nidulans, is translated as a single polypeptide and regulated in response to osmotic conditions. Biochem. Biophys. Res. Commun. 298103-109. [DOI] [PubMed] [Google Scholar]
- 41.Takeshita, N., S. Yamashita, A. Ohta, and H. Horiuchi. 2006. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 591380-1394. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trilla, J. A., A. Durán, and C. Roncero. 1999. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin. J. Cell Biol. 146265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber, I., D. Assmann, E. Thines, and G. Steinberg. 2006. Polar localizing class V myosin chitin synthases are essential during early plant infection in the plant pathogenic fungus Ustilago maydis. Plant Cell 18225-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamada, E., M. Ichinomiya, A. Ohta, and H. Horiuchi. 2005. The class V chitin synthase gene csmA is crucial for the growth of the chsA chsC double mutant in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 6987-97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.