Abstract
Systematic disruption of genes encoding kinases and mitogen-activated protein kinases (MAPKs) was performed in Kluyveromyces lactis haploid cells. The mutated strains were assayed by their capacity to mate and to respond to hyperosmotic stress. The K. lactis Ste11p (KlSte11p) MAPK kinase kinase (MAPKKK) was found to act in both mating and osmoresponse pathways while the scaffold KlSte5p and the MAPK KlFus3p appeared to be specific for mating. The p21-activated kinase KlSte20p and the kinase KlSte50p participated in both pathways. Protein association experiments showed interaction of KlSte50p and KlSte20p with Gα and Gβ, respectively, the G protein subunits involved in the mating pathway. Both KlSte50p and KlSte20p also showed interaction with KlSte11p. Disruption mutants of the K. lactis PBS2 (KlPBS2) and KlHOG1 genes of the canonical osmotic response pathway resulted in mutations sensitive to high salt and high sorbitol but dispensable for mating. Mutations that eliminate the MAPKK KlSte7p activity had a strong effect on mating and also showed sensitivity to osmotic stress. Finally, we found evidence of physical interaction between KlSte7p and KlHog1p, in addition to diminished Hog1p phosphorylation after a hyperosmotic shock in cells lacking KlSte7p. This study reveals novel roles for components of transduction systems in yeast.
Cellular signaling transduction networks continuously sense extracellular cues and transduce signals from the cell surface to the interior of the cell. The cell responds to these signals through changes in gene expression and protein activity to yield specific phenotypes.
G-protein signaling pathways in fungi are used to regulate development and virulence and to detect nutrients and other environmental signals. Some of these pathways contain mitogen-activated protein kinase (MAPK) cascades that are highly conserved in metazoans (12, 18). The yeast pheromone signaling, for example, occurs through the action of a G-protein-coupled receptor (GPCR) and the associated G protein in order to activate a MAPK cascade that conducts the signal to the nucleus. Besides G-protein-activated signal transduction pathways, there are at least three more pathways involving MAPK components or their presumed upstream regulators that modulate responses to several stimuli (10).
The best-characterized transduction system in eukaryotic organisms is the Saccharomyces cerevisiae pheromone response pathway. This pathway is initiated by the binding of peptide pheromones to a GPCR (Ste2p in MATa cells and Ste3p in MATα cells) and the dissociation of the heterotrimeric G protein into Gα (Gpa1p) and the stimulatory Gβγ (Ste4p/Ste18p) subunits. The liberated Gβγ dimer directly associates with a scaffold protein, Ste5p, and with a p21-activated kinase (PAK), Ste20p, which is essential for activating the MAPK kinase kinase (MAPKKK) Ste11p. Activation of Ste11p is also promoted by action of the Ste50 protein kinase. Ste11p in turn, activates the MAPKK Ste7p. Downstream from Ste7p, Fus3p and Kss1p, two partially redundant MAPKs, induce the activation of transcription factors, Ste12p among others, that regulate the mating process (2).
Some components of the pheromone response pathway participate in other MAPK cascades. For example, Ste20p, Ste50p, and Ste11p play a role in the high-osmolarity glycerol (HOG) pathway, helping the cell to survive osmotic stress (24), and Ste20p, Ste50p, Ste11p, and Ste7p are used by both the pseudohyphal development pathway and the cell wall integrity pathway (8, 13).
The HOG pathway in S. cerevisiae is regulated by two different inputs that converge to activate the MAPKK Pbs2p. These two branches are composed of membrane-spanning proteins (Sln1p and Sho1p) that respond to variant conditions of external osmolarity.
Under isosmotic conditions, the protein histidine kinase, Sln1p, acting through a phosphorelay reaction with Ypd1p and Ssk1p, keeps the partially redundant MAPKKKs Ssk2p and Ssk22p in an inactive state (27). High external osmolarity blocks Sln1p function, allowing activation of Ssk2p and Ssk22p, which, in turn, phosphorylate Pbs2p. On the other hand, Sho1p uses Ste20p and Ste50p to activate Ste11p (22, 26) in response to high-solute concentrations. Sho1p binds Ste11p and Pbs2p, resulting in activation by phosphorylation of Pbs2p (26, 37). Activation of Pbs2p by the Sln1p and Sho1p branches stimulates phosphorylation and nuclear translocation of Hog1p, triggering expression of a number of genes and production of glycerol to prevent dehydration (13, 33).
While a large amount of information has accumulated on signal transduction cascades in S. cerevisiae and in many other fungi (18), a critical question concerns which features of the transduction systems are generic and which are species specific. For example, in the budding yeast Kluyveromyces lactis, the signal transduction system that mediates the mating response is triggered by both the Gα (K. lactis Gpa1p [KlGpa1p]) (31) and Gβ (KlSte4p) (14) subunits of the heterotrimeric G protein. There are two documented differences between the G protein pheromone response pathways of S. cerevisiae and K. lactis: first, while disruption of GPA1, the gene encoding the G protein α-subunit in S. cerevisiae, leads to permanent growth arrest and therefore to lethality in haploid cells (7), inactivation of Gα (KlGpa1p) in K. lactis does not affect cell viability but produces sterility (31); second, overexpression of the Gβ (Ste4p) subunit induces growth arrest and mating in S. cerevisiae (34) but has no effect in K. lactis (14). Although it has been shown that both Gα and Gβ trigger the mating response by activating the transcription factor KlSte12p (14, 19), nothing is known about the elements that connect the G protein with the transcription factor in K. lactis. We have therefore investigated the phenotypic characteristics of disruption mutants in components of the pheromone response pathway, and in order to elucidate the overlapping of protein function with other transduction systems, we assayed the disruption mutants for their capacity to respond also to osmotic stress.
The budding yeast K. lactis provides an attractive model system for studying cellular differentiation processes in response to environmental cues. It is a unicellular and essentially aerobic and heterothallic organism with a conventionally organized cell cycle. Its entire genome sequence is now available (http://cbi.labri.fr/Genolevures), and it is easily subjected to genetic analysis.
MATERIALS AND METHODS
Strains, media, and genetic techniques.
The yeast strains used in this work were K. lactis 155 (MATa ade2 his3 ura3) and 12/8 (MATα lysA argA ura3). S. cerevisiae strain EGY48 (MATα his3 trp1 ura3-52 leu2::pLeu-LexAop6) was used for two-hybrid assays (23). Escherichia coli strains DH5α and Gm33 (for preparation of unmethylated DNA) were used to propagate plasmids. YPD medium consisted of 1% yeast extract, 2% Bacto-peptone, and 2% glucose. Synthetic dextrose (SD) minimal medium consisted of 0.67% yeast nitrogen base without amino acids (Difco) and 2% glucose. SD medium was supplemented with the required amino acids and nitrogen bases (50 μg/ml). SGal medium was the same as SD medium except for the substitution of glucose for galactose. Selective SD medium containing 10 mM KH2PO4 was used for induction of pPHO constructs. LB medium plus ampicillin (100 μg/ml) was used to propagate recombinant bacteria. Standard protocols were followed for Southern blot analysis, recombinant DNA technology, and yeast manipulation.
Gene identification and cloning.
A BLAST search of the K. lactis genome database, Genolevures project II (http://cbi.labri.fr/Genolevures/elt/KLLA), allowed the identification of putative open reading frames (ORFs) encoding the orthologues of S. cerevisiae protein kinases. Pairs a and b of the oligodeoxynucleotides listed in Table 1 were used for PCR amplification. Chromosomal DNA from K. lactis strain 155 was used as a template. Standard PCRs were carried out to amplify the desired gene fragments. The PCR products were cloned into the pGEMTeasy vector (Promega) and sequenced in full. Plasmid pPHO (containing the PHO5 promoter from S. cerevisiae and a K. lactis replication origin) was obtained from Hiroshi Fukuhara. A six-histidine tag was fused in frame to the C terminus of KlSte7p [KlSte7(His6)] by PCR-mediated amplification, employing primers Ste7c and Ste7d (Table 1). An XhoI-SmaI fragment of the KlSte7(His6) construct was Klenow filled and subcloned into pPHO previously digested and filled in at EcoRI. Recombinant plasmid containing the KlSte7(His6) gene under the control of the PHO5 promoter was identified by restriction analysis.
TABLE 1.
Primers used for PCR amplification
| Primer name | Sequencea |
|---|---|
| Ste50a | (+88) CTAGATAGAAccCGgGAGAACAAGATA (+114) |
| SmaI | |
| Ste50b | (+741) TTGATCTCCAAAGCtTATCACTAAAAC (+715) |
| HindIII | |
| Ste50c | (−19) ATAGAAACAAAAACtcgagATGAGCGGTAC(+11) |
| XhoI | |
| Ste50d | (+929) AAGTGTTTTTTCtcGagGATGAAGGAGGAG (+899) |
| XhoI | |
| Ste20a | (−14) TTTGTCTCCtcgagATGACGGATACTGGAT (+16) |
| XhoI | |
| Ste20b | (+3002) TAAATTCATGTAcTGCaGTAAAACAGAAAT (+2972) |
| PstI | |
| Ste5a | (+1) ATGTCTAGAGGTAATAC (+17) |
| Ste5b | (+2300) GTTTAGAGAGAGAACGG (+2284) |
| Ste11a | (+360) GCAAAGAAAGcTtACGTTAACGGGT (+385) |
| HindIII | |
| Ste11b | (+1210) TGGATTCTGAGGTACCAG (+1192) |
| Asp718 | |
| Ste11c | (−19) TTTTTAAGATCTctcgAgATGAGCAGTGAC (+12) |
| XhoI | |
| Ste11d | (+2222) AGTAACAATACCTCGAGACTGAAAACGGC (+2194) |
| XhoI | |
| Ste7a | (+232) GAGAAGCTTCCGGTCC (+247) |
| HindIII | |
| Ste7b | (+1303) CTTTCAAAGAgGATCcTCCTCTCTCCTC (+1275) |
| BamHI | |
| Ste7c | (−18) AGATTGACTTAActcgAgATGATTACACGT (+12) |
| XhoI | |
| Ste7d | (+1543) CTAATATAGTGCtcgagCTTAATTTATTTC (+1514) |
| XhoI | |
| Ste7e | (+1513) cccgggTCAgtggtggtggtggtggtgTCGCCTTGTACT (+1501) |
| SmaI | |
| Fus3a | (+498) ACAGATGGCGgTaCCCATGTTGACG (+523) |
| Asp718 | |
| Fus3b | (+1171) TGATATAATAaGCTtGTGGTTACTTAG (+1144) |
| HindIII | |
| Pbs2a | (−21) GGCGATTAAGTGgtaCCAATTATGAGT (+6) |
| Asp718 | |
| Pbs2b | (+2293) GGCGAGGCAAAGgaTccCTTACAATCCGCC (+2264) |
| BamHI | |
| Hog1a | (+490) CTTGCTAGGATcCAAGATCCTCAAATG (+517) |
| BamHI | |
| Hog1b | (+1366) CTTCTGTCAAGCttATTTCATTACATTG (+1339) |
| HindIII | |
| Hog1c | (−18) TTTTCCAATTGAGAATTCATGTCGAATGAG (+12) |
| EcoRI | |
| Hog1d | (+1397) TATACTTCTGTTCCTCGAGATTTCATTAC (+1343) |
| XhoI |
Numbers in parentheses correspond to coordinates considering A in the translation start codon as the +1 position. Nucleotide changes introduced to generate restriction sites or tag sequences are indicated by lowercase letters.
Gene disruptions.
Gene disruptions were achieved by homologous recombination using the integrative vector YIp352 (11). Disruption mutants were selected by their capacity to grow in Ura-deficient medium. All gene fragments were obtained from the pGEMTeasy clones. For K. lactis STE50 (KlSTE50), a SmaI-HindIII 634-bp fragment was introduced into the integrative vector YIp352 previously digested with EcoRI (Klenow filled) and HindIII. The resulting plasmid was digested with EcoRI and SacI (natural restriction sites of KlSTE50 ORF located at positions 298 and 438, respectively). This yields a linear YIp352 plasmid flanked by 298 bp and 196 bp of KlSTE50 recombinant ends. For KlSTE20, a 1,046-bp fragment obtained by EcoRI and SalI digestion was subcloned into YIp352 digested with the same enzymes. The resulting integrative plasmid was opened with BglII, which cuts the KlSTE20 ORF at position 268. The resulting construct contains recombinant ends of 267 bp and 779 bp. KlSTE5 was obtained as a 1,266-bp EcoRI fragment and ligated into YIp352 opened with the same enzyme. The resulting construct was digested with HpaI and SpeI, yielding an integrating molecule with 573 bp and 140 bp of recombinant ends. A KlSTE11 850-bp fragment obtained with HindIII-Asp718 was introduced into YIp352 digested with the same enzymes. This plasmid was opened with SpeI, yielding an integrating fragment containing 190 bp and 275 bp of recombinant ends. For KlSTE7, a 1,071-bp fragment was obtained by HindIII-BamHI digestion and ligated into YIp352 digested with the same enzymes. This construct was digested with XbaI and BglII, leaving recombinant ends of 333 bp and 338 bp. For KlFUS3, a 670-bp EcoRI fragment was subcloned into YIp352 vector prepared with the same enzyme. A linear molecule was generated by digesting the resulting plasmid with SmaI and PstI. This yields an integrating molecule containing 227 bp and 277 bp of recombinant ends. A 967-bp KpnI-PstI fragment of KlPBS2 was subcloned into YIp352 opened with the same enzymes. The YIp352-KlPBS2 construct was digested with XbaI and BstEII, yielding a linear molecule containing 431 bp and 313 bp of recombinant ends. For KlHOG1, a BamHI-HindIII 876-bp fragment was subcloned into YIp352 digested with the same enzymes. BglII digestion of YIp352-KlHOG1 yielded a linear molecule flanked by recombinant ends of 331 bp and 350 bp. When required, YPD medium containing 1 mg/ml of 5-fluoroorotic acid (5-FOA) was used for negative selection of the URA3 cassette.
Protein interactions.
Assays of physical interaction were done with a LexA-B42 two-hybrid system as described previously (14). The KlGPA1 and KlSTE4 genes were subcloned into pEG202 as previously reported (14, 31). KlGPA1 was also subcloned into pJG4-5 as reported previously (14). Genes of interest (with the exception of KlSTE20 and KlPBS2) were amplified by PCR employing specific c and d primers (Table 1) and ligated in frame into two-hybrid plasmids. The full KlSTE50 gene was ligated into pJG4-5 as an XhoI fragment. KlSTE11 was subcloned into pEG202 as a 2,215-bp XhoI fragment. The KlSTE7 gene was obtained as a 1,536-bp XhoI fragment and ligated into pJG4-5. The gene encoding KlPbs2p was subcloned into pEG202 as a 2,285-bp Asp718-BamHI fragment. KlHOG1 was subcloned in both pEG202 and pJG4-5 as a 1,364-bp EcoRI-XhoI fragment. The KlSTE20 gene was obtained as a 2,987-bp fragment from the pGEMTeasy clone by XhoI digestion and subcloned into pJG4-5 digested with the same enzyme. S. cerevisiae endochitinase gene (CTS1) cloned into either plasmid pEG202 or pJG4-5 (14) was used as a negative interaction control for each interacting couple. All recombinant genes were sequenced in full. S. cerevisiae strain EGY48 was transfected with two-hybrid plasmids and grown overnight in SD medium at 30°C. Induction of genes subcloned into pJG4-5 was done by shifting cells to SGal medium 4 h prior to harvesting. Protein interactions were determined by the ability of hybrid proteins to induce blue colonies by expression of the LACZ reporter gene located in the pSH18-34 plasmid and by reversion of the leu2 deficiency.
Mating assays.
A patch of cells of the strain to be tested was grown on a plate of selective medium for 24 h. The tester strain was grown as a lawn on a YPD plate for 24 h. Both strains were replica plated onto a YPD plate and incubated for 8 h at 30°C, allowing cells to mate. Diploids were selected on SD medium by replica plating.
Osmotic stress assays.
Strains to be tested were grown overnight on SD medium. Cells were washed, suspended in YPD medium at an optical density at 600 nm of 0.1, and grown to mid-exponential phase. Serial dilutions of cells were spotted on YPD medium containing 0.5 M KCl or 1 M d-sorbitol and incubated at 30°C. Plates were photographed 48 h later.
Phosphorylation assays.
To estimate the cellular content of both total and phosphorylated KlHog1p in wild-type, ΔKlpbs2, and ΔKlste7 strains, exponentially growing cells in 5 ml of YPD medium were pelleted and suspended in YPD medium or YPD medium plus 0.5 M KCl for different times. Cells were concentrated by centrifugation, and the supernatant was removed by aspiration. Cells were resuspended in loading buffer (5% sodium dodecyl sulfate [SDS], 0.1 M Tris-HCl [pH 7.5], 5% glycerol, 0.07 M 2-mercaptoethanol, 0.02 mM bromophenol blue) and boiled for 5 min. Samples were centrifuged, and the supernatants were transferred to a clean tube. An aliquot of the samples was subjected to SDS-polyacrylamide gel electrophoresis on an 10% gel and transferred to polyvinylidene difluoride membrane (Millipore Corporation).
Immunoblotting.
Blots were incubated with blocking buffer (1× phosphate-buffered saline, 1% albumin, 0.05% Tween 20) for 1 h at 37°C. Blots were incubated in the same buffer containing rabbit polyclonal anti-Hog1p (Santa Cruz Biotechnology) or rabbit monoclonal anti-phospho-p38 antibody (Cell Signaling Technology) for 1 h at room temperature. Filter-bound antibodies were detected with horseradish peroxidase-conjugated secondary goat anti-rabbit immunoglobulin G antibody (Zymed) and visualized with chemiluminescent horseradish peroxidase substrate (Millipore Corporation).
Immunoprecipitation assays.
Cells bearing pPHO or pPHO-KlSte7(His6) were grown at 30°C to mid-log phase in 50 ml of selective medium. At mid-log phase, KH2PO4 was added at a final concentration of 10 mM, and cells were incubated for another 2 h at 30°C. Half of the culture was treated with 0.5 M KCl for 10 min. Cells were collected by centrifugation and resuspended in 300 μl of TEA buffer containing 10 mM Tris (pH 7.4), 1 mM EDTA, 100 mM phenylmethylsulfonyl fluoride, 20 mM NaN3, and 1× complete protease inhibitor cocktail (Roche). Sterile acid-washed glass beads were added, and cell disruption was performed by vortexing for 5 min at 4°C. Lysates were cleared in a microcentrifuge (3 min at 2,000 rpm), and the supernatant was adjusted to an 800-μl final volume with TEA buffer. Ten microliters of anti-His6-peroxidase antibody (Roche) was added and incubated at 4°C overnight with gentle shaking. Sixty microliters of protein G-agarose beads (Upstate) was added, and incubation continued for 2 h. The beads were washed three times with 1.0 ml of cold TNTE buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Triton X-100, 1 mM EDTA). Thirty microliters of loading buffer was added, followed by 5 min of boiling. Samples were chilled on ice and centrifuged briefly. Twenty microliters of each sample was used for immunoblotting. Blots were probed with anti-Hog1p as described above. Reciprocal immunoprecipitation was done in the same way except that 5 μl of anti-Hog1p (Santa Cruz Biotechnology) antibodies was used for precipitation, and blots were probed with anti-His-peroxidase antibodies.
RESULTS
In previous work, we have demonstrated that both the Gα (KlGpa1p) and Gβ (KlSte4p) subunits of the heterotrimeric G protein trigger the mating pheromone response pathway in K. lactis (6, 14, 31). Additionally, we have found that this activation requires the transcription factor KlSte12p (14, 19). In order to elucidate further the pathway that promotes mating in K. lactis, we constructed disruption mutants of genes encoding putative intermediates between the G protein and the KlSte12p transcription factor. We therefore inactivated the genes encoding components of the MAPK cascade, and we also inactivated the genes for KlSte50p and KlSte20p protein kinases, two putative inputs of the MAPK module. To investigate the overlapping of protein function with other transduction systems, we assayed the disruption mutants for their capacity to respond to osmotic stress, and for this purpose we added to this screening mutants of genes encoding KlPbs2p and KlHog1p, which have been shown to be key elements in the osmotic adaptation response in other yeast species (10, 15). Table 2 shows relevant features of the studied genes and their products as well as the degrees of homology with their S. cerevisiae counterparts.
TABLE 2.
Proven components of K. lactis mating and osmoresponse pathways
| Gene name | Mutant phenotypea
|
S. cerevisiae homologueb
|
Reference or source | ||
|---|---|---|---|---|---|
| Mating | High osmolarity | % Identity | E value | ||
| KlSTE2 | Sterile | ND | 48 | 5e-92 | 32 |
| KlSTE3 | Sterile | ND | 51 | 1e-131 | 32 |
| KlGPA1 | Sterile | ND | 62 | 8e-154 | 31 |
| KlSTE4 | Sterile | ND | 52 | 1e-119 | 14 |
| KlSTE18 | Fertile | ND | 53 | 3e-24 | 6 |
| KlSTE20 | Sterile | Sensitive | 50 | 0.0 | This work |
| KlSTE50 | Sterile | Sensitive | 41 | 9e-60 | This work |
| KlSTE5 | Sterile | Resistant | 27 | 3e-56 | This work |
| KlSTE11 | Sterile | Sensitive | 58 | 1e-60 | This work |
| KlSTE7 | Sterile | Sensitive | 52 | 1e-130 | This work |
| KlFUS3 | Sterile | Resistant | 69 | 1e-150 | This work |
| KlSTE12 | Sterile | ND | 35 | 1e-107 | 19 |
| KlPBS2 | Fertile | Sensitive | 57 | 0.0 | This work |
| KlHOG1 | Fertile | Sensitive | 80 | 0.0 | This work |
ND, not determined.
Identity values were determined by BLAST using the BLOSUM62 matrix.
Because each particular protein may modulate responses in different degrees and its inactivation may have several effects, detailed studies on the structure and the role that each protein may have in different cell functions will be described elsewhere.
All disruption mutants were constructed by integrating a URA3 cassette in haploid cells of both mating types; therefore, transformants were selected by their capacity to grow in Ura-deficient medium, and the disrupted alleles were confirmed by Southern blot. In all cases, plasmidic gene copies reversed the phenotypic defects of 5-FOA-resistant strains to almost wild-type levels (data not shown). Since our results suggest that gene disruptions seem to have the same phenotypic effects in MATa and MATα cells, results described in this work refer solely to MATα strains.
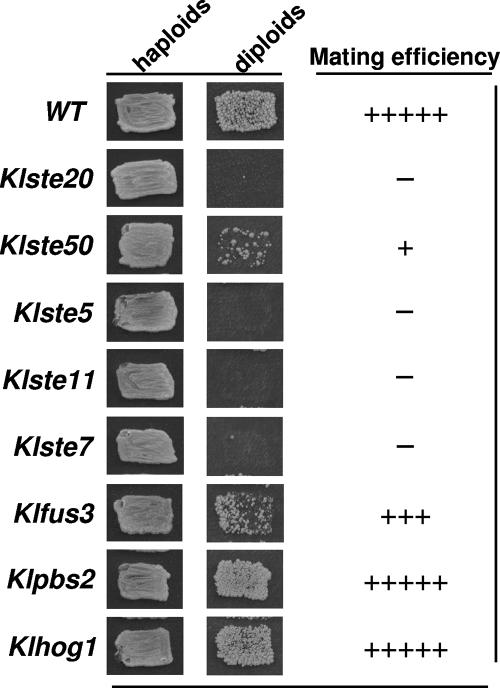
All mutant strains were viable, and they exhibited normal growth; however, when assayed in sexual crosses with a tester strain, they showed (with the exception of Klhog1 and Klpbs2 mutants) mating defects (Fig. 1). The hierarchy of defects in mating was determined as follows: Klste5 = Klste20 = Klste11 = Klste7 > Klste50 > Klfus3 (Fig. 1). The strongest effect on mating was observed when the scaffold protein KlSte5p and the KlSte20p, KlSte11p, and KlSte7p kinases were inactivated, indicating that all these proteins play essential roles in haploid mating. KlSte5p shows a moderate degree of identity with its counterpart in S. cerevisiae (27% identity) (Table 2). S. cerevisiae Ste5p (ScSte5p) has no catalytic activity but acts as a scaffold, organizing and forming a complex with the MAPK module composed by ScSte11p, ScSte7p, and ScFus3p (4, 35). The KlSte20p is a member of the PAK family (17), which is 50% identical to ScSte20p (Table 2) and shows high degree of conservation in the CRIB domain, needed for Cdc42p binding (a small Rho-like G protein) (1), and in the Gβ and kinase domains (16). KlSte11p is 58% identical to ScSte11p (Table 2), which in S. cerevisiae also binds to Gβ and is phosphorylated and activated by the ScSte20p kinase (28). ScSte11p has an essential role in mating since it appears to be an essential bridge between proteins acting as inputs of the MAPK module and the G protein (10). KlSte7p is 52% identical to ScSte7p (Table 2). In the S. cerevisiae pheromone response pathway, Ste7p is required for efficient activation of the MAPK Fus3p via its recruitment to the scaffold protein Ste5 and its activation by Ste11p (10, 20). Disruption of the KlSTE50 gene affected mating with moderate strength. Comparison of the primary structure of KlSte50p with its S. cerevisiae homologue showed overall 41% identity (Table 2). The SAM (sterile alpha motif) domain, involved in Ste11p binding, and the Ras association domain (30) are the most conserved regions within the primary structure of Ste50 proteins. Inactivation of ScSte50p leads to attenuation of pheromone signaling and to sterility in S. cerevisiae (36). Finally, inactivation of KlFus3p yielded the weakest sterile phenotype. The K. lactis and S. cerevisiae Fus3 proteins share a relatively high degree of identity (69%) (Table 2). In S. cerevisiae inactivation of Fus3p leads to partial sterility (21) due to the fact that it can be replaced by ScKss1p, the MAPK involved in pseudohyphal development (5). The K. lactis genome contains an ORF (KLLA0A02497) that encodes a putative KlKss1p. Although we have not investigated the role of this kinase, we assume, by analogy to S. cerevisiae, that KlKss1p can replace KlFus3p in the mating pathway, leading to the observed weak sterile phenotype of the ΔKlfus3 mutant.
FIG. 1.
Mating of MATα disruption mutants. Mating was done by replica plating. Patches of strains to be tested were streaked on selective medium and replicated onto YPD plates containing a lawn of the MATa wild-type tester strain, followed by incubation overnight at 30°C. Diploid selection was done by replica plating onto SD medium. Plates were photographed 48 h later. WT, wild type.
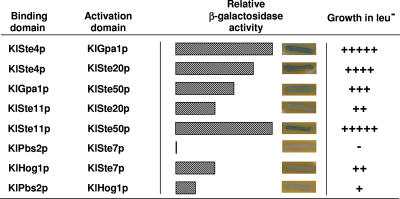
To determine the connection between the heterotrimeric G protein and the MAPK module, we assayed protein-protein interactions of KlSte50p and KlSte20p with the MAPKKK KlSte11p and with the Gα (KlGpa1p) and Gβ (KlSte4p) subunits of the heterotrimeric G protein. Although it should be confirmed that protein interactions have a functional meaning, as a first approach we chose the two-hybrid system to measure interactions because its reliability has been proved in a number of cases involving components of the mating pathway. We used the LexA-B42-based two-hybrid system in which protein association leads to both activation of the reporter LACZ gene located in plasmid pSH18-34 and activation of the chromosomal LEU2 gene, controlled by several lexA-op sites, in the host strain (23). The KlSte50p and KlSte20p kinases were the focus of this analysis because we have observed that they are implicated in the pheromone response pathway, possibly acting as effectors of the G protein (data not shown). The two-hybrid assay detected that KlSte50p makes a moderate interaction with KlGpa1p, the Gα subunit of the heterotrimeric G protein, while the KlSte20p kinase interacts strongly with the Gβ subunit KlSte4p (Fig. 2). These assays also revealed that both the KlSte50p and the KlSte20p kinases associate with the MAPKKK KlSte11p. The KlSte50p-KlSte11p interaction is stronger than that of KlSte20p with KlSte11p (Fig. 2), most probably due to the presence of the strongly interacting SAM domains in KlSte50p and KlSte11p (30). These findings suggest that KlSte50p and KlSte20p may serve as links between the G protein and the MAPK module in the transmission of the pheromone stimulus in K. lactis.
FIG. 2.
Protein interactions determined by the two-hybrid system. The binding domain corresponds to LexA-fused proteins cloned into pEG202. The activation domain corresponds to B42-fused proteins cloned into pJG4-5. Two-hybrid plasmids were introduced into strain EGY48. β-Galactosidase activity was determined by the relative intensity of blue staining in cells growing on SGal plates containing 1 mg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), pH 7.0. Growth in SD medium without leucine (leu−) was determined by relative size of the colony after a 24-h incubation at 30°C. The S. cerevisiae endochitinase gene (CTS1) cloned either into pEG202 or pJG4-5 was used as negative interaction control for each interacting couple.
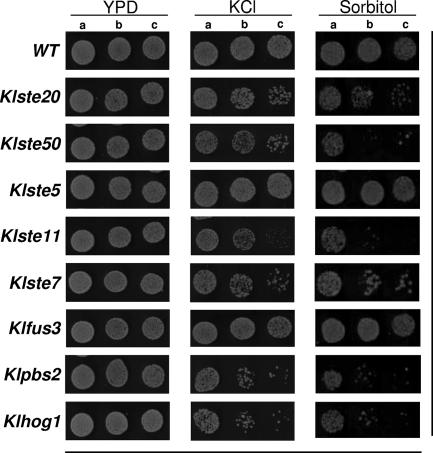
Several elements of the yeast pheromone response pathway play a role in other signal transduction cascades (10, 13, 20). Concerning the high-osmolarity response pathway in S. cerevisiae, at least three elements of the pheromone pathway are needed to regulate the osmoadaptation program: Ste50p, Ste20p, and Ste11p, which function as upstream elements of the scaffold and kinase protein Pbs2p and the MAPK Hog1p in one of the branches dedicated to adaptation of yeast to high osmolarity (13). All these proteins are components of the so-called HOG pathway. We assayed the K. lactis disrupted strains under conditions that produced osmotic (high K+ and high sorbitol) stress (Fig. 3). In this set of experiments we found increased sensitivity to sorbitol and KCl of mutant strains ΔKlPbs2 and ΔKlHog1 compared to the wild-type strain, which is in agreement with their expected roles in osmoadaptation. The strong sensitivity of mutant ΔKlSte11 to high osmolarity was also evident, indicating some parallelism of KlSte11p function with Ste11p from S. cerevisiae (26). Surprisingly, in this set of experiments we observed that the ΔKlSte7 mutant showed sensitivity to stress conditions, as indicated by its marked growth delay in 0.5 M KCl and 1 M sorbitol compared to the growth of wild-type cells (Fig. 3). This finding is striking and suggests that the MAPKK KlSte7p is involved in a signal transduction cascade related to the osmotic response. Inactivation of the KlSte50p and KlSte20p kinases produced severe sensitivity to high osmotic stress, confirming that these kinases play overlapping roles in the propagation of different signals (Fig. 3). As expected, the ΔKlste5 and ΔKlfus3 mutants were insensitive to hyperosmotic stress (Fig. 3).
FIG. 3.
Effect of high osmolarity on the growth properties of disruption mutants. Tenfold serial dilutions of the indicated strains were spotted onto plates containing YPD medium alone, YPD medium with 0.5 M KCl, and YPD medium with 1.0 M sorbitol and incubated at 30°C for 48 h. Lanes a, 106 cells; lanes b, 105 cells; lanes c, 104 cells. WT, wild type.
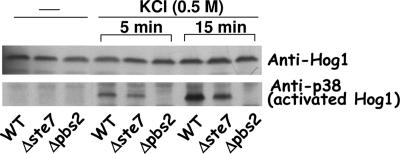
In S. cerevisiae, Pbs2p acts as a MAPKK in phosphorylating Hog1p, and in addition to its catalytic activity, it also serves as a scaffold, binding Sho1p, Ste11p, and Hog1p (9). Comparison of the primary sequences of ScPbs2p and KlPbs2p indicates that KlPbs2p conserves the kinase domain which in S. cerevisiae has been shown to phosphorylate ScHog1p (3), suggesting that KlPbs2p may acts as a MAPKK over KlHog1p. Since the sensitivity to high osmolarity of the ΔKlste7 mutant is not as strong as the one of ΔKlpbs2 (Fig. 3), we hypothesize that either KlPbs2p lacks a kinase activity and KlSte7p may replace it or that the propagation of the osmoresponse requires both proteins acting in parallel. In order to gain some light on the role that KlSte7p plays in the osmotolerance process, we determined the KlHog1p phosphorylation levels in wild-type, ΔKlste7, and ΔKlpbs2 backgrounds (Fig. 4). Dual phosphorylation of Hog1p (Thr173 and Tyr175 in K. lactis) can be detected by immunoblotting with an antibody raised against activated mammalian p38, as previously reported (24). Upon exposure for 5 min of wild-type cells to 0.5 M KCl, KlHog1p was phosphorylated. By 15 min, the amount of activated KlHog1p increased 1.4-fold (Fig. 4). KlHog1p phosphorylation levels were 30% (5 min) and 60% (15 min) lower in the ΔKlste7 mutant than in the wild-type strain even though the amount of total KlHog1p did not show significant variation, as determined by an anti-Hog1 antibody (Fig. 4). No detectable phosphorylation of KlHog1p was observed in the ΔKlpbs2 mutant, even at 15 min of exposure to KCl. These results suggest that KlSte7p may partially activate KlHog1p in response to hyperosmotic stress and that this activation is totally dependent on KlPbs2p.
FIG. 4.
Phosphorylation of KlHog1p in wild-type (WT), ΔKlste7, and ΔKlpbs2 strains. Cells were grown to mid-exponential phase in YPD medium and not treated or treated with 0.5 M KCl. After the indicated times, the cells were lysed as described in Materials and Methods. Samples of the resulting extracts were analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting with anti-Hog1 or anti-p38.
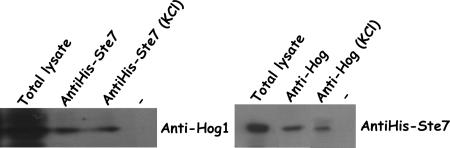
The above observations suggest that KlSte7p and KlHog1p are able to physically associate with each other. By means of the two-hybrid assay, we observed that KlSte7p made a significant interaction with KlHog1p but failed to associate with KlPbs2p (Fig. 2). KlPbs2p and KlHog1p showed very weak interaction in the two-hybrid assay. Confirmatory data were obtained by immunoprecipitation assays. We transformed a 5-FOA-resistant ΔKlste7 strain with the pPHO (phosphate-dependent promoter) plasmid carrying the gene encoding KlSte7p tagged with six His residues at its C terminus. This plasmid reverses the sterile phenotype of the 5-FOA-resistant mutant. Under phosphate induction conditions, KlHog1p was immunoprecipitated by anti-His antibodies in the presence of KlSte7(His6)p (Fig. 5) and, vice-versa, the KlSte7(His6) protein was immunoprecipitated by anti-Hog1p antibodies in the presence of KlHog1p (Fig. 5). Coimmunoprecipitation was independent of hyperosmotic shock induced with 0.5 M KCl. Anti-Hog1p antibodies were able to precipitate KlSte7(His6)p at different phosphate concentrations which induce variable expression levels of KlSte7(His6)p (data not shown). These findings suggest that KlSte7p makes physical contact with both activated and inactivated KlHog1p and suggest a model whereby KlSte7p is able to phosphorylate KlHog1p during the response to high-osmolarity conditions only when KlHog1p has been recruited by KlPbs2p.
FIG. 5.
Physical interaction between KlSte7p and KlHog1p. KlSte7p and KlHog1p coimmunoprecipitation from cells extracts of the ΔKlste7 strain carrying the pPHO-KlSte7(His6) plasmid. Cells were grown to mid-exponential phase in selective medium and not treated or treated with 0.5 M KCl for 10 min. Cells were lysed as described in Materials and Methods. KlSte7(His6)p was precipitated with anti-His antibodies, and the recruited KlHog1p was detected by immunoblotting using an anti-Hog1 antibody. KlHog1p was precipitated with anti-Hog1, and the recruited KlSte7p was detected by immunoblotting using an anti-His antibody.
DISCUSSION
In S. cerevisiae, the Gβγ complex of the heterotrimeric G protein transmits the pheromone signal to different effectors, two of which are the Ste20p kinase and the Ste5p/Ste11p complex. The PAK kinase Ste20p is essential for Ste11p activation (2), and Ste50p has been implicated as an adaptor protein linking the G-protein-associated Cdc42p-Ste20p kinase complex to Ste11p via its SAM domain, thus modulating the pheromone response pathway (30). In K. lactis disruption of KlSTE20 produced complete sterility while disruption of KlSTE50 reduced but did not eliminate mating. Our experiments reveal that the MAPK cascade that drives mating in K. lactis is composed of the proteins KlSte5p, KlSte11p, KlSte7p, and KlFus3p and suggest that this MAPK module is triggered by KlSte50p and KlSte20p kinases in response to activation of the heterotrimeric G protein. It is reasonable to assume that the K. lactis MAPK module is formed by KlSte11p (MAPKKK), KlSte7p (MAPKK), and KlFus3p (MAPK), anchored by KlSte5p (scaffold). However, detailed epistasis experiments should be performed to confirm this arrangement. Our preliminary studies indicate that both KlSte50p and KlSte20p interact with KlSte11p, thus activating the MAPK cascade that elicits mating in K. lactis. The actual picture of the pheromone response pathway in K. lactis points to a model where activation of G protein by binding of pheromone to GPCR (32) triggers two branches that converge in the MAPKKK KlSte11p; one is essential for mating and is formed by Gβ-KlSte20p, and the second is dispensable and is formed by Gα-KlSte50p. Additional support for this model is given by the disruption phenotypes of mutants that inactivate the G protein subunits. While inactivation of KlSte4p (Gβ) leads to complete incapacity to mate (14), inactivation of KlGpa1p (Gα) only reduces mating but does not eliminate it (31). The physical association between Gα and KlSte50p opens a new window for the study of new functional relationships of G protein signaling components.
Participation of Ste50p, Ste20p, and Ste11p in the Sho1p branch of the HOG pathway has been extensively documented in S. cerevisiae (12, 15). In a brief view of the pathway, the osmosensor Sho1p recruits Pbs2p to the membrane during signaling. Both Sho1p and Pbs2p can bind Ste11p. Ste11p is activated by phosphorylation, which is mediated by the Ste20p and Cla4p kinases and requires Ste50p (30). Ste11p activates Pbs2p, which in turn activates Hog1p (15). The high-osmolarity response pathway in S. cerevisiae is also regulated by the Sln1p branch, which consists of the Sln1p-Ypd1p-Ssk1p phosphorelay system. Sln1p is an osmosensor histidine kinase, Ypd1p is a phosphotransfer protein, and Ssk1p is a response regulator (25). Hyperosmotic shock deactivates Sln1p, leading to activation of Ssk2p and Ssk22p (two redundant kinases) via the Ssk1p response regulator. Ssk2p and Ssk22p activate the MAPKK Pbs2p, which in turn activates, by phosphorylation, the MAPK Hog1p (22). The two branches for Hog1p activation are not redundant, since the Sln1p-Ssk1p branch has a more prominent role than the Sho1p-Ste11p branch in intermediate osmolarity (0.1 M KCl), but both branches function at high osmolarity (24). At present, it is not known if these two branches are active in K. lactis; however, the phenotype of strong sensitivity to high sorbitol (1.0 M) and high salt (0.5 M KCl) displayed by mutants ΔKlSte50, ΔKlSte20, and ΔKlste11, strongly suggests that the Sho1p pathway has remarkable participation in osmoadaptation to high-stress conditions. Moreover, disruption of KlSTE11 induces severe growing defects under high osmotic stress in K. lactis, while in S. cerevisiae cells, inactivation of Ste11p induces sensitivity to hyperosmotic conditions only in Δssk1 or Δssk2 Δssk22 backgrounds (22, 24). In addition, it is worth noting that the Sln1p branch of the HOG pathway in K. lactis lacks the homologue Ssk22p (15) although we actually do not know the functional meaning for the absence of this MAPKKK.
In addition to the interaction detected between components of the MAPK module and scaffold proteins, two-hybrid studies indicate that some of the protein kinases associate with each other independently of their association with the scaffold protein. In particular, both ScSte7p and ScSte11p interact with the MAPKs ScFus3p and ScKss1p, independently of Ste5p (4, 29). However, due to the sensitivity to conditions of high osmotic stress and to the lower KlHog1p phosphorylation level displayed by the ΔKlste7 strain, we believe that the interaction observed between KlSte7p and KlHog1p is significant and might play an important role in osmoadaptation. In S. cerevisiae, the Ste7p kinase has been shown to participate not only in mating but also in filamentous and invasive growth (20); however, a mutant strain lacking Ste7p shows normal osmotolerance, which indicates that this kinase does not participate in the HOG response pathway in this yeast species. We were unable to find physical interaction between KlSte7p and KlPbs2p, and the interaction between KlPbs2p and KlHog1p was rather weak. It is well known that failure to find an association by the two-hybrid system can occur for many reasons; therefore, association between these proteins should be tested by different techniques.
Sensitivity of the ΔKlste7 mutant to high osmotic stress and association of KlSte7p with KlHog1p in K. lactis are, at least, intriguing observations. Either KlPbs2p lacks a kinase activity and KlSte7p is able to replace it, or KlHog1p can be phosphorylated by redundant MAPKK activities in the HOG pathway. Our results discarded the first hypothesis since KlHog1p phosphorylation is still observed in ΔKlste7 mutant cells. Although we have no direct evidence that KlSte7p phosphorylates KlHog1p, we found that the amount of phosphorylated KlHog1p diminished in cells lacking the KlSte7p kinase. This may affect the growing rate of the Klste7 mutant under hyperosmotic conditions. Additionally, our results showed that KlHog1p phosphorylation is completely dependent on KlPbs2p, suggesting that KlHog1p needs to be recruited by the scaffold activity of KlPbs2p in order to be phosphorylated by KlSte7p; i.e., KlSte7p fails to phosphorylate soluble KlHog1p.
We have provided evidence that in addition to KlSte50p, KlSte20p, and KlSte11p, KlSte7p also participates in both the pheromone response pathway and in the high-osmolarity response pathway in K. lactis. Despite the common use of protein kinases in both pathways, our observations also indicate that there is no cross talk between these two signaling systems; i.e., sexual pheromone does not induce a high-osmolarity response and conditions of high osmotic stress do not induce mating (data not shown).
The composition of the G protein signaling pathway for mating, the participation of the KlSte7p in the HOG pathway, and the strong sensitivity phenotype of the ΔKlste11 mutant to hyperosmotic stress indicate that signaling systems in K. lactis follow, at least in part, mechanisms different from those of S. cerevisiae. The perspective ahead is to understand further the organization and functional interactions of the elements belonging to the intracellular MAPK signaling pathways in K. lactis. The results will be of interest for the understanding of related problems in other organisms.
Acknowledgments
We thank Guadalupe Codiz and Minerva Mora (Molecular Biology Unit) for technical assistance and the staff of the Computer Facility at Instituto de Fisiología Celular, Universidad Nacional Autónoma de México. We also thank Marisela Bolaños for technical assistance.
This work was partially funded by grants 44178 from CONACyT, México, and IN211906 from PAPIIT, DGAPA, Universidad Nacional Autónoma de México.
Footnotes
Published ahead of print on 16 November 2007.
REFERENCES
- 1.Ash, J., C. Wu, R. Larocque, M. Jamal, W. Stevens, and M. Osborne. 2003. Genetic analysis of the interface between Cdc42p and the CRIB domain of Ste20p in Saccharomyces cerevisiae. Genetics 1639-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell, L. 2004. A walk-through of the yeast mating pheromone response pathway. Peptides 251465-1476. [DOI] [PubMed] [Google Scholar]
- 3.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 2591760-1763. [DOI] [PubMed] [Google Scholar]
- 4.Choi, K.-Y., B. Satterberg, D. M. Lyons, and E. A. Elion. 1994. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78499-512. [DOI] [PubMed] [Google Scholar]
- 5.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signaling pathway. Nature 39085-88. [DOI] [PubMed] [Google Scholar]
- 6.Coria, R., L. Kawasaki, F. Torres-Quiroz, L. Ongay-Larios, E. Sánchez-Paredes, N. Velázquez-Zavala, R. Navarro-Olmos, M. Rodríguez-González, R. Aguilar-Corachán, and G. Coello. 2006. The pheromone response pathway of Kluyveromyces lactis. FEMS Yeast Res. 6336-344. [DOI] [PubMed] [Google Scholar]
- 7.Dietzel, C., and J. Kurjan. 1987. The yeast Scg1 gene: a Gα-like protein implicated in the a- and α-factor response pathway. Cell 501001-1010. [DOI] [PubMed] [Google Scholar]
- 8.Elion, E. A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3573-581. [DOI] [PubMed] [Google Scholar]
- 9.Garrington, T. P., and G. L. Johnson. 1999. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr. Opin. Cell Biol. 11211-218. [DOI] [PubMed] [Google Scholar]
- 10.Herskowitz, I. 1995. MAP kinase pathways in yeast: for mating and more. Cell 80187-197. [DOI] [PubMed] [Google Scholar]
- 11.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2163-167. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman, C. S. 2005. Except in every detail: comparing and contrasting G-protein signaling in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Eukaryot. Cell 4495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeast. Microbiol. Mol. Biol. Rev. 66300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawasaki, L., A. L. Saviñón-Tejeda, L. Ongay-Larios, J. Ramírez, and R. Coria. 2005. The Gβ(KlSte4p) subunit of the heterotrimeric G protein has a positive and essential role in the induction of mating in the yeast Kluyveromyces lactis. Yeast 22947-956. [DOI] [PubMed] [Google Scholar]
- 15.Krantz, M., E. Becit, and S. Hohmann. 2006. Comparative genomics of the HOG-signalling system in fungi. Curr. Genet. 49137-151. [DOI] [PubMed] [Google Scholar]
- 16.Lamson, R. E., M. J. Winters, and P. M. Pryciak. 2002. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 222939-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leberer, E., D. Dignard, D. Harcus, D. Y. Thomas, and M. Whiteway. 1992. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 114815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W-C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloret, A., A. Saviñón-Tejeda, L. Ongay-Larios, E. P. Tenorio, and R. Coria. 2003. The KlFUS1 gene is required for proper haploid mating and its expression is enhanced by the active form of the Gα (Gpa1) subunit involved in the pheromone response pathway of the yeast Kluyveromyces lactis. FEMS Microbiol. Lett. 219105-113. [DOI] [PubMed] [Google Scholar]
- 20.Madhani, H. D., and G. R. Fink. 1998. The riddle of MAP kinase signaling specificity. Trends Genet. 14151-155. [DOI] [PubMed] [Google Scholar]
- 21.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91673-684. [DOI] [PubMed] [Google Scholar]
- 22.Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269554-558. [DOI] [PubMed] [Google Scholar]
- 23.Mendelsohn, A. R., and R. Brent. 1994. Applications of interaction traps/two-hybrid systems to biotechnology research. Curr. Opin. Biotechnol. 5482-486. [DOI] [PubMed] [Google Scholar]
- 24.O'Rourke, S. M., and I. Herskowitz. 1998. The HOG1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 122874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD-SSK1 “two-component” osmosensor. Cell 86865-875. [DOI] [PubMed] [Google Scholar]
- 26.Posas, F., and H. Saito. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 2761702-1705. [DOI] [PubMed] [Google Scholar]
- 27.Posas, F., and H. Saito. 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 171385-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Priciak, P. M., and F. A. Huntress. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 122684-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Printen, J. A., and G. F. Sprague, Jr. 1994. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138609-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramezani-Rad, M. 2003. The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast. Curr. Genet. 43161-170. [DOI] [PubMed] [Google Scholar]
- 31.Saviñón-Tejeda, A., L. Ongay-Larios, J. Valdés-Rodríguez, and R. Coria. 2001. The KlGpa1 gene encodes a G-protein α subunit that is a positive control element in the mating pathway of the budding yeast Kluyveromyces lactis. J. Bacteriol. 183229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres-Quiroz, F., L. Kawasaki, M. Rodríguez-González, A. Patrón-Soberano, and R. Coria. 2007. The KlSTE2 and KlSTE3 genes encode MATα and MATa specific G-protein-coupled receptors, respectively, which are required for mating of Kluyveromyces lactis haploid cells. Yeast 2417-25. [DOI] [PubMed] [Google Scholar]
- 33.Westfall, P. J., D. R. Ballon, and J. Thorner. 2004. When the stress of your environment makes you go HOG wild. Science 3061511-1512. [DOI] [PubMed] [Google Scholar]
- 34.Whiteway, M., L. Hougan, D. Dignard, D. Y. Thomas, L. Bell, G. C. Saari, F. J. Grant, P. O'Hara, and V. L. MacKay. 1989. The Ste4 and Ste18 genes of yeast encode potential β and γ subunits of the mating factor receptor-coupled G protein. Cell 56467-477. [DOI] [PubMed] [Google Scholar]
- 35.Winters, M. J., R. E. Lamson, H. Nakanishi, A. M. Neiman, and P. M. Pryciak. 2005. A membrane binding domain in the Ste5 scaffold synergizes with Gβγ binding to control localization and signaling in pheromone response. Mol. Cell 2021-32. [DOI] [PubMed] [Google Scholar]
- 36.Xu, G., G. Jansen, D. Y. Thomas, C. P. Hollenberg, and M. Ramezani-Rad. 1996. Ste50p sustains mating pheromone-induced signal transduction in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 20773-783. [DOI] [PubMed] [Google Scholar]
- 37.Zarrinpar, A., R. P. Bhattacharyya, M. P. Nittler, and W. A. Lim. 2004. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high-osmolarity MAP kinase pathway. Mol. Cell 14825-832. [DOI] [PubMed] [Google Scholar]