Abstract
Vegetative hyphal fusion (VHF) is a ubiquitous phenomenon in filamentous fungi whose biological role is poorly understood. In Neurospora crassa, the mitogen-activated protein kinase (MAPK) Mak-2 and the WW domain protein So are required for efficient VHF. A MAPK orthologous to Mak-2, Fmk1, was previously shown to be essential for root penetration and pathogenicity of the vascular wilt fungus Fusarium oxysporum. Here we took a genetic approach to test two hypotheses, that (i) VHF and plant infection have signaling mechanisms in common and (ii) VHF is required for efficient plant infection. F. oxysporum mutants lacking either Fmk1 or Fso1, an orthologue of N. crassa So, were impaired in the fusion of vegetative hyphae and microconidial germ tubes. Δfmk1 Δfso1 double mutants exhibited a more severe fusion phenotype than either single mutant, indicating that the two components function in distinct pathways. Both Δfso1 and Δfmk1 strains were impaired in the formation of hyphal networks on the root surface, a process associated with extensive VHF. The Δfso1 mutants exhibited slightly reduced virulence in tomato fruit infection assays but, in contrast to Δfmk1 strains, were still able to perform functions associated with invasive growth, such as secretion of pectinolytic enzymes or penetration of cellophane sheets, and to infect tomato plants. Thus, although VHF per se is not essential for plant infection, both processes have some signaling components in common, suggesting an evolutionary relationship between the underlying cellular mechanisms.
The ability of filamentous fungi to form multicellular colonies can be regarded as the result of two distinct morphogenetic processes, (i) growth of individual hyphae by tip extension and branching, which requires maintenance of cell polarity and redirection of the growth axis in response to environmental cues (30), and (ii) establishment of an interconnected hyphal network or mycelium through fusion of individual hyphae. The latter process is known as vegetative hyphal fusion (VHF) or anastomosis (2) and appears to be ubiquitous in filamentous fungi, yet its biological function is unknown. Suggested roles which remain to be tested experimentally include general homeostasis, translocation of nutrients and water, and intrahyphal communication within the fungal colony (17, 37).
Evidence from genetic studies in the model ascomycete Neurospora crassa support the notion that VHF is a highly complex and regulated developmental process (14-16). A number of genes required for VHF have recently been identified in N. crassa, including ham-2, which encodes a putative transmembrane protein (47), and so, which encodes a polypeptide with a WW protein-protein interaction domain (12). Besides these genes with hitherto unknown functions, components of a highly conserved mitogen-activated protein kinase (MAPK) cascade, orthologous to the Saccharomyces cerevisiae Fus3 mating pathway, are also required for VHF (16). Mutants of N. crassa lacking the MAPK Mak-2 or the MAPK kinase kinase Nrc-1 (33), as well as Aspergillus nidulans strains affected in the MAPK kinase kinase SteC (46), exhibited severe defects in VHF. The fact that these mutants were also female sterile suggests a possible functional link between signaling mechanisms involved in vegetative and mating cell fusion (33, 38).
Intriguingly, the same conserved MAPK cascade also regulates fungal pathogenicity on plants (22). The essential role of this so-called pathogenicity MAPK (PMK) was first described in the rice BLAST fungus Magnaporthe grisea. Mutants lacking the pmk1 gene were not only defective in the formation of appressoria but also unable to colonize host plants when inoculated through wound sites (48). Since then, PMK was found to be required for virulence in many biologically and taxonomically diverse plant pathogens, suggesting an ancient evolutionary role of this MAPK cascade in fungal pathogenicity on plants (6, 24, 31, 44). While PMK has been shown to control a number of virulence-related functions, the mechanisms through which it regulates plant infection have remained elusive.
In this study, we explored the functional and evolutionary links between VHF and plant pathogenicity in the vascular wilt fungus Fusarium oxysporum. This soil-borne pathogen attacks a wide range of agriculturally important crops (7) and has been reported previously to undergo VHF (19, 34). A genetic approach was used to test two hypotheses, that (i) VHF and plant infection have signaling mechanisms in common and (ii) VHF is required for efficient plant infection. We show that F. oxysporum mutants lacking either the Mak-2 orthologue Fmk1 or the So orthologue Fso1 are severely impaired in VHF. However, while Δfmk1 strains are nonpathogenic on tomato plants, Δfso1 mutants exhibit only slightly reduced virulence. These results demonstrate that (i) VHF and plant infection have some signaling components in common and (ii) VHF is not essential for plant infection.
MATERIALS AND METHODS
Fungal isolates and culture conditions.
For a complete overview of the fungal strains used in this study, see Table 1. F. oxysporum f. sp. lycopersici race 2 wild-type strain 4287 (FGSC 9935) was used in all experiments. Generation and molecular characterization of the F. oxysporum Δfmk1 mutant was described previously (6). All fungal strains were stored as microconidial suspensions at −80°C with 30% glycerol. For extraction of DNA, microconidium production, and fungal development and conidiation studies, cultures were grown for the indicated times in liquid potato dextrose broth (Difco, Detroit, MI) at 28°C with shaking at 170 rpm (8). For determination of heterokaryon formation, non-nitrate-utilizing strains were grown on minimal medium (MM) (34). For phenotypic analysis of colony growth, drops of water containing 2 × 105 microconidia were spotted onto potato dextrose agar plates (Difco, Detroit, MI) and incubated at 28°C for 3 days. Assays for polygalacturonase production on polygalacturonic acid plates were performed as previously described (8). For cellophane invasion assays, autoclaved cellophane sheets were placed on MM plates and the center of each plate was inoculated with a drop of water containing 2 × 105 microconidia. After 4 days at 28°C, the cellophane sheet with the fungal colony was removed carefully. The presence or absence of fungal mycelium on the underlying medium was recorded after incubation of the plates for an additional 24 h at 28°C. For microscopic analysis of VHF, fungal strains were observed in a Leica DMR microscope by the Nomarski technique. Photographs were recorded with a Leica DC 300F digital camera.
TABLE 1.
F. oxysporum strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| 4287 (FGSC 9935) | Wild type | FGSCa |
| Δfmk1 | fmk1::PHLEO | 6 |
| Δfgb1 | fgb1::HYG | 5 |
| Δfmk1 Δfgb1 | fmk1::PHLEO fgb1::HYG | 5 |
| Δfso1 no. 5 | fso1::HYG | This study |
| Δfso1 no. 29 | fso1::HYG | This study |
| Δfso1+fso1 no. 1 | fso1::HYG fso1 PHLEO | This study |
| Δfmk1 Δfso1 no. 2 | fmk1::PHLEO fso1::HYG | This study |
| Δfmk1 Δfso1 no. 4 | fmk1::PHLEO fso1::HYG | This study |
| nit1− | nit1− | This study |
| nitM− | nitM− | This study |
| Δfmk1 nit1− | fmk1::PHLEO nit1− | This study |
| Δfmk1 nitM− | fmk1::PHLEO nitM− | This study |
| Δfgb1 nit1− | fgb1::HYG nit1− | This study |
| Δfso1 no. 5 nit1− | fso1::HYG nit1− | This study |
| Δfmk1 Δfso1 no. 2 nit1− | fmk1::PHLEO fso1::HYG nit1− | This study |
| Δfmk1 Δfso1 no. 2 nitM− | fmk1::PHLEO fso1::HYG nitM− | This study |
FGSC, Fungal Genetics Stock Center.
Nucleic acid manipulations and cloning of the fso1 gene.
Total RNA and genomic DNA were extracted from F. oxysporum mycelium as previously reported (3, 36). Southern and Northern blot analyses and probe labeling were carried out as previously described (8), with the nonisotopic digoxigenin labeling kit (Roche Diagnostics SL, Barcelona, Spain). Other routine DNA procedures were performed as described in standard protocols (42).
Genomic DNA of F. oxysporum strain 4287 was used for PCR amplification on a Perkin-Elmer GeneAmp System 2400 with primers Fso1-1 (5′-GAAGATTGCGACAGCCCGACCG-3′) and Fso1-2 (5′-TCTCCATCATGTCCTCTTTTGTGTC-3′), which were derived from conserved regions of the N. crassa so gene (accession number AB072452) (12). PCRs were routinely performed with the Long Template PCR system (Roche Diagnostics SL). The amplified DNA fragment was cloned into the pGEM-T vector (Promega, Madison, WI) and used to screen a λEMBL3 genomic library of F. oxysporum f. sp. lycopersici isolate 4287. Library screening, subcloning, and other routine procedures were performed as described previously (42). Sequencing of both DNA strands of the clones obtained was performed at the Servicio de Secuenciación Automática de DNA, SCAI (University of Córdoba, Córdoba, Spain), with the DyeDeoxy Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA) on an ABI Prism 377 genetic analyzer (Applied Biosystems). DNA and protein sequence databases were searched with the BLAST algorithm (1).
Construction of plasmid vectors and fungal transformation.
Gene replacement vector pDfso1 was constructed by replacing a 1.1-kb HindIII-XhoI fragment from the fso1 coding region with a 3-kb HindIII-XhoI fragment from plasmid pAN7-1, containing the hygromycin B resistance gene under the control of the A. nidulans gpdA promoter (35). A linear fragment containing the interrupted fso1 allele was generated by amplifying the entire construct with primers Fso1-9 (CGGGCGCTTGCATCGAGGC) and Fso1-10 (CCATGCGTATCCTCAAGGAACT) and used to transform protoplasts of the F. oxysporum wild-type strain or the Δfmk1 mutant to hygromycin resistance as previously described (8). For complementation experiments, an 8.3-kb DNA fragment encompassing the entire fso1 gene was amplified by PCR from F. oxysporum genomic DNA with primers Fso1-13 (CTTGTGCTCGGCTTTGCTTCT) and Fso1-14 (CGTCAAGCGCGGCAACTTCA) and introduced into protoplasts of Δfso1 strain 29 by cotransformation with the phleomycin resistance cassette amplified from plasmid pAN8-1 (27). Hygromycin- and/or phleomycin-resistant transformants were selected and purified by monoconidial isolation as previously described (6). The presence of the wild-type fso1 allele in the complemented transformants was confirmed by PCR on genomic DNA with primers Fso1-3 (GGTACGCTCGGATCGGCAG) and Fso1-5 (CGCCTCCCCTCCCGAAAC).
Heterokaryon tests.
To determine heterokaryon formation between two strains carrying different nitrate auxotrophic markers, drops of microconidial suspensions or small pieces of mycelia from the respective strains were inoculated on MM at a distance of 1 cm (34). Heterokaryons exhibiting vigorous wild-type growth on nitrate were observed in the colony contact zone after approximately 4 days of incubation at 28°C.
To measure the abilities of the different strains to undergo VHF, quantitative heterokaryon tests were performed (47). Conidial suspensions (106 conidia) of a nitM or a nit1 tester strain in a wild-type background were mixed with 106, 105, 104, 103, or 102 conidia of nit1 or nitM mutants, respectively, obtained in the different genetic backgrounds (Table 1). The concentration of conidial suspensions was determined by counting with a hemacytometer. Conidial mixtures were plated on MM with nitrate, and the number of heterokaryotic colonies per plate was determined after 3 days of incubation at 28°C. Conidial suspensions of the different mutants, in the absence of the tester strain, were used as controls to check for revertants. Viability of conidia was determined by plating 50 and 500 conidia from each strain individually on MM with ammonium. Treatments were done in triplicate, and experiments were performed at least twice with similar results. For statistical analysis of results, Scheffe's test for pairwise comparisons was used (49).
Virulence and root adhesion assays.
Tomato plant inoculation assays in a growth chamber were performed as previously described (8). At different times after inoculation, severity of disease symptoms was recorded with indices ranging from 1 (healthy plant) to 5 (dead plant). Twenty plants were used for each treatment. Assays for root adhesion and invasive growth on tomato fruits (cultivar Daniela) were carried out as previously described, with three replicates (6), except that potato dextrose broth diluted 1:50 with water and supplemented with 20 mM glutamic acid was used for incubation of roots with fungal spores. All virulence and root adhesion experiments were performed three times with similar results.
RESULTS
Cloning and targeted knockout of the F. oxysporum fso1 gene.
The so gene was previously reported to encode a protein with a WW domain required for VHF in N. crassa (12). A TBLASTN search of the F. graminearum complete genome sequence available at http://www.broad.mit.edu/annotation/genome/fusarium_graminearum/Home.html revealed the presence of two DNA regions located at the ends of contigs 298 and 299, corresponding to the 5′ and 3′ regions, respectively, of a putative orthologue of N. crassa so. The complete fso1 gene of F. oxysporum f. sp. lycopersici strain 4287 was cloned from a λEMBL3 genomic library using as a probe a 2.1-kb PCR fragment amplified from genomic DNA with primers Fso1-1 and Fso1-2 derived from conserved regions of the N. crassa so gene (see Materials and Methods for details). Sequencing of a hybridizing λ genomic clone identified an open reading frame of 3,591 bp, which encodes a putative 1,197-amino-acid protein with a predicted molecular mass of 129.5 kDa and a pI of 6.92. The sequence of the F. oxysporum fso1 gene has been deposited in GenBank under accession number EF593098. Alignment of the nucleotide sequences of the fso1 cDNA obtained by reverse transcription-PCR with gene-specific primers, and the genomic clone showed the presence of two introns of 49 and 55 bp, respectively. Southern analysis of total genomic DNA treated with different restriction enzymes suggested that fso1 is present in F. oxysporum as a single copy (data not shown). In agreement with this result, a BLASTP search of the complete genome database of F. oxysporum (http://www.broad.mit.edu/annotation/genome/fusarium_group.1/MultiHome.html) detected one single significant match (FOXG_01690.2). Alignment of the amino acid sequence of the predicted F. oxysporum Fso1 protein with sequences in the databases revealed 61% identity with the N. crassa So protein. Based on these data, we conclude that F. oxysporum fso1 is the structural orthologue of N. crassa so.
The biological role of fso1 in F. oxysporum was explored by generating a Δfso1 loss-of-function allele by replacement of a 1.1-kb HindIII-XhoI fragment in the open reading frame with the hygromycin resistance cassette (see Fig. S1A in the supplemental material; see Materials and Methods for details). The knockout construct was introduced into both the wild-type strain and the Δfmk1 mutant to study possible epistatic relationships between the two genes. Thirty-five and 12 hygromycin-resistant transformants, respectively, were analyzed by PCR with different combinations of gene-specific primers. Four and three transformants, respectively, produced amplification products that were indicative of homologous integration-mediated gene replacement (data not shown). Southern blot analysis confirmed the replacement of a 4.1-kb PstI fragment corresponding to the wild-type fso1 allele, with a fragment of 5.5 kb (see Fig. S1B in the supplemental material), demonstrating that these transformants, which were named Δfso1 and Δfmk1 Δfso1, respectively, carry a disrupted copy of the fso1 gene.
To confirm that the phenotype of the Δfso1 mutants was indeed caused by loss of fso1 function, an 8.3-kb DNA fragment encompassing the complete F. oxysporum fso1 gene was introduced into Δfso1 strain 29 by cotransformation with the phleomycin resistance marker. Twenty-five phleomycin-resistant transformants were analyzed for the presence of a functional fso1 allele by PCR with gene-specific primers Fso1-3 and Fso1-5. A 2.1-kb amplification product identical to that obtained from the wild type was detected in several phleomycin-resistant transformants but not in the Δfso1 strain (see Fig. S1C in the supplemental material). We concluded that these transformants, designated Δfso1+fso1, had integrated an intact copy of the fso1 gene into the genome.
Fmk1 and Fso1 are required for efficient fusion of vegetative hyphae and conidial anastomosis tubes in F. oxysporum.
To test the role of the MAPK Fmk1 and the putative WW domain protein Fso1 in VHF of F. oxysporum, qualitative and quantitative heterokaryon tests were performed. Non-nitrate-utilizing mutants were first obtained from the different genetic backgrounds (wild type, Δfmk1, Δfso1, and Δfmk1 Δfso1) by selection on chlorate (34). Chlorate-resistant strains were classified into nit1 (deficient in nitrate reductase) and nitM (deficient in molybdenum cofactor) according to their growth on different nitrogen sources (34). When conidial suspensions from non-nitrate-utilizing mutants obtained in the wild-type background were inoculated 1 cm apart onto MM with nitrate as the sole nitrogen source, sparsely growing colonies emerged. After a few days, vigorous hyphal growth was observed in the region of mycelial contact between nit1 and nitM strains but not between two nit1 or two nitM strains, indicating vegetative complementation by heterokaryon formation (Fig. 1). A nit1 strain obtained from an ectopic transformant with the fso1 knockout construct also showed heterokaryon formation. By contrast, no complementation was detected in nit1-nitM pairings in which at least one of the strains lacked a functional allele of either fmk1 or fso1, suggesting that both genes are required for efficient VHF.
FIG. 1.
Fmk1 and Fso1 are required for vegetative complementation of nitrate utilization deficiency in F. oxysporum. Non-nitrate-utilizing nit1 mutants were obtained from different genetic backgrounds (wild type, Δfmk1, Δfso1, and an ectopic transformant with the fso1 knockout construct) and inoculated, 1 cm from a nitM mutant, onto MM with nitrate as the sole nitrogen source. Vigorous hyphal growth in the region of contact between colonies denotes the presence of vegetative complementation through heterokaryon formation.
To quantify the hyphal fusion defect in the mutants, a conidial suspension of a nit1 or nitM tester strain (106 cells per plate) was mixed with serial dilutions of conidia from the wild type or the mutant strains with complementing auxotrophic markers and then plated onto MM before determining the number of heterokaryotic colonies after 3 days. Conidial suspensions of the mutants in the absence of the tester strain were used to determine the frequency of reversion, which was extremely low (approximately 10−6). The frequency of heterokaryon formation between the Δfso1 nit1 mutant and a nitM strain was reduced roughly 1,000-fold compared to that between nit1 and nitM strains (Table 2). Frequencies of heterokaryon formation in the Δfmk1 and Δfmk1 Δfso1 mutants were 2,000- and 10,000-fold lower, respectively, than those of the wild type. We also tested heterokaryon frequency in a mutant lacking the G-protein β subunit Fgb1, which was previously shown to signal via a cyclic-AMP-dependent pathway distinct from the Fmk1 cascade (5). VHF frequency in the Δfgb1 strain was increased 50% compared to that of the wild type (Table 2). These results indicate that fmk1 and fso1, but not fgb1, are required for efficient heterokaryon formation in F. oxysporum.
TABLE 2.
Frequencies of heterokaryon formation
| Strain | Tester strain | Frequencya |
|---|---|---|
| nit1− | nitM− | 1† |
| nitM− | nit1− | 1† |
| Δfgb1 nit1− | nitM− | 1.52‡ |
| Δfso1 nit1− | nitM− | 0.0017§ |
| Δfmk1 nit1− | nitM− | 0.00057¶ |
| Δfmk1 nitM− | nit1− | 0.00067¶ |
| Δfmk1Δfso1 nit1− | nitM− | 0.00016‖ |
| Δfmk1Δfso1 nitM− | nit1− | 0.00018‖ |
Values followed by the same symbol do not differ significantly (P < 0.01) according to Scheffe's test (49).
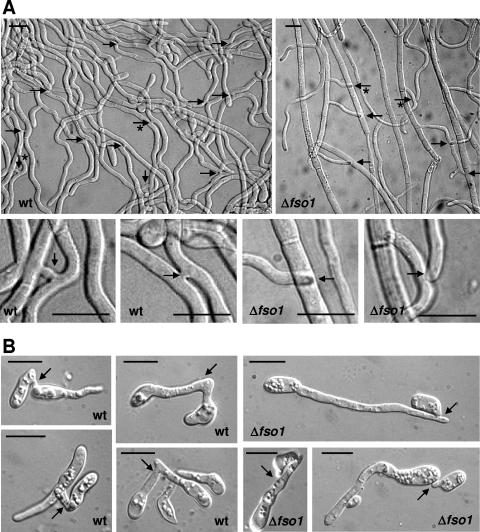
Microscopic examination of wild-type colonies revealed the presence of a large number of VHF events, mainly in the form of short fusion bridges between parallelly growing hyphae (Fig. 2A). By contrast, hyphal fusion bridges were not detected in colonies of the Δfmk1 and Δfso1 mutants after extensive microscopic analyses, even though hyphae frequently made physical contact with each other.
FIG. 2.
Vegetative fusion between hyphae and microconidial germ tubes of F. oxysporum. Hyphae of the indicated strains from subperipheral regions of colonies grown on solid MM (A) or microconidia germinated in liquid MM (B) were imaged by the Nomarski technique. Fusion events in the wild type (wt) and points of contact without fusion in the Δfso1 strain are indicated by arrows. In panel A, details of the regions marked by asterisks are shown below the general view. Bar, 10 μm.
In N. crassa, germinating conidia fuse during early stages of colony establishment by means of conidial anastomosis tubes (39, 40). We found that microconidial germ tubes of the F. oxysporum wild-type strain fused approximately 6 to 8 h after initiating germination on solid medium or in liquid medium (Fig. 2). Similar to N. crassa (40), the germ tubes of F. oxysporum frequently changed the original axis of growth before fusing with each other. Conidia from Δfmk1 and Δfso1 strains germinated within the same time frame as wild-type conidia, but fused germ tubes exhibiting cytoplasmic continuity with each other were never observed under the microscope (Fig. 2). In the complemented Δfso1+fso1 strain, both hyphal fusion bridges and germ tube fusion events were observed at similar frequencies as in the wild type (data not shown). We conclude that both the MAPK Fmk1 and the WW domain protein Fso1 are essential for efficient VHF and that this essential role is conserved between F. oxysporum and N. crassa.
VHF affects development and conidiation during late stages of submerged culture.
To test whether VHF affects fungal development, we first determined the radial growth of the strains on different media. No differences were detected between the wild type and the Δfso1 mutants, whereas the Δfmk1 and Δfmk1 Δfso1 strains had a significantly reduced growth rate, particularly on MM (see Fig. S2 in the supplemental material). These results argue that growth reduction in these mutants is a consequence of fmk1 deletion rather than impairment of VHF. Both Δfso1 and Δfmk1 colonies produced slightly fewer aerial hyphae than those of the wild type (results not shown).
When grown in submerged culture, F. oxysporum produced abundant hyphae bearing phialides with microconidia. These microconidia detached and germinated, giving rise to more hyphae and microconidia. After a number developmental cycles, the newly produced microconidia failed to germinate and the culture became stationary (approximately 5 × 107 microconidia ml−1). During late stages of submerged culture, large mycelial aggregates formed and the number of microconidia in the culture decreased approximately 1,000-fold in the wild-type strain (Fig. 3). By contrast, no hyphal aggregates were observed in the Δfmk1 and Δfso1 strains, suggesting that VHF is required for the formation of these structures (Fig. 3A). Moreover, the number of microconidia per milliliter at late stages of culture was four- to sixfold higher in the cultures of VHF-impaired mutants (Δfmk1, Δfso1, and Δfmk1 Δfso1) compared to the wild type and the complemented Δfso1+fso1 strain (Fig. 3B). These results suggest that VHF plays a role in fungal development during late stages of submerged culture.
FIG. 3.
VHF affects fungal development and conidiation. The indicated strains were grown for 35 days on potato dextrose broth in submerged culture with shaking. (A) The entire culture was transferred to a petri dish and photographed. Note the presence of large mycelial aggregates in the wild-type (wt) and Δfso1+fso1 strains. (B) Concentrations of microconidia in the culture supernatants of the different strains. Error bars indicate standard deviations from three samples. The experiment was performed twice with similar results.
VHF contributes to the formation of hyphal networks on tomato roots.
Virulence-related phenotypes reported in the Δfmk1 mutants include the inability to adhere to and colonize tomato roots (6). We tested the hypothesis that VHF contributes to efficient root colonization by allowing the formation of large hyphal networks on the root surface. Roots of tomato seedlings were immersed for 24 h in microconidial suspensions of the wild-type and mutant strains and then washed by vigorous shaking in water and observed in a stereomicroscope. After this time period, the wild type and the complemented Δfso1+fso1 strain had efficiently adhered to the lateral roots and formed a dense hyphal network covering the root surface whereas the Δfmk1 strain failed to adhere to and colonize the roots (Fig. 4A and B). The Δfso1 strains showed significantly reduced root colonization ability but were still able to colonize the roots to some extent. Microscopic observation of the colonized root surface revealed the presence of frequent hyphal fusion events between germ tubes and hyphae of F. oxysporum (Fig. 4B). Thus, VHF not only takes place during the initial stages of fungal growth on the root but also appears to contribute to efficient colonization of the root surface.
FIG. 4.
(A) VHF is required for efficient adhesion and root colonization. Roots of tomato seedlings were immersed for 24 h in microconidial suspensions of the indicated strains, washed by vigorous shaking in water, and observed in a stereomicroscope. Adhering fungal mycelium is visible as a white mass covering lateral roots. wt, wild type. (B) VHF during colonization of tomato roots by F. oxysporum wild-type strain 4287. Upper left, hyphal aggregates on the root surface (indicated by arrows). Right, F. oxysporum germlings undergoing anastomosis on a tomato root surface. An example of a hyphal fusion bridge is indicated by an arrow. Lower left, detail of the fusion bridge.
Fso1 is not essential for pathogenicity on tomato.
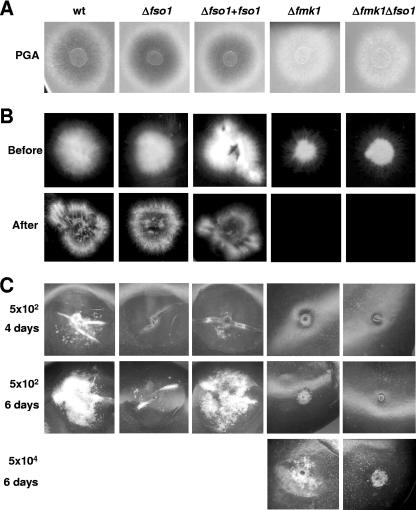
Fmk1 is essential for infection of tomato plants by F. oxysporum (6). Because Fmk1 is also required for efficient VHF, we forwarded two contrasting working hypotheses, i.e., that (i) VHF and pathogenicity are controlled by shared signaling components but are functionally unrelated and (ii) VHF is required for fungal pathogenicity on plants. If the second hypothesis were true, the inability of the Δfmk1 strain to undergo VHF would account, at least in part, for its nonpathogenic phenotype. To explore this question, we asked whether the Δfso1 strains, which are impaired in VHF, are also affected in virulence-related functions such as production of pectinolytic enzymes, penetration of cellophane sheets, and invasive growth on tomato fruits. The Δfmk1 mutant showed strongly reduced clear halo production on polygalacturonic acid, was unable to penetrate cellophane sheets, and failed to invade tomato fruit tissue (Fig. 5). By contrast, the Δfso1 strains performed as efficiently as the wild type in these tests, except for a minor but consistently reproducible delay in fruit tissue invasion (Fig. 5C). The Δfmk1 Δfso1 double mutants had a phenotype similar to that of the Δfmk1 strain, but fruit tissue invasion was even more reduced than in the Δfmk1 single mutant.
FIG. 5.
Fmk1 and Fso1 contribute differentially to invasive growth. Microconidial suspensions of the indicated strains were used for the different phenotypic assays. (A) Clear halo production on polygalacturonic acid (PGA)-containing plates for visualization of extracellular polygalacturonase activity (visible as a dark halo underneath the fungal colony). (B) Penetration of cellophane sheets. Colonies were grown for 4 days on a plate with MM covered by a cellophane sheet (before), and then the cellophane with the colony was removed and the plate was incubated for 1 additional day (after). (C) Invasive growth on tomato fruits inoculated with the indicated concentrations of microconidia and incubated for 4 or 6 days at 28°C. wt, wild type.
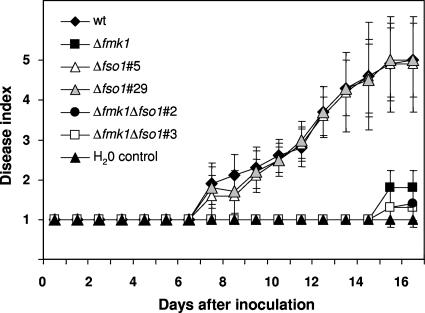
Infection of tomato plants was performed by inoculating the roots of seedlings with microconidial suspensions. Disease symptoms in plants inoculated with the wild-type strain increased steadily throughout the experiment, and most of the plants were dead 16 days after inoculation (Fig. 6). As reported previously (6), plants inoculated with the Δfmk1 strain showed extremely low disease ratings. By contrast, the Δfso1 mutant did not differ significantly from the wild-type strain in the pattern of disease development, whereas the Δfmk1 Δfso1 double mutant behaved similarly to the Δfmk1 single mutant. We conclude that, in contrast to Fmk1, Fso1 does not control functions related to invasive growth and is not essential for the pathogenicity of F. oxysporum on tomato plants.
FIG. 6.
Fso1 is not required for virulence of F. oxysporum on tomato plants. The graph shows the incidence of Fusarium wilt on tomato plants (cultivar Monica) inoculated with the indicated strains. Severity of disease symptoms was recorded at different times after inoculation, with indices ranging from 1 (healthy plant) to 5 (dead plant). Error bars represent the standard deviations calculated from 20 plants. wt, wild type.
DISCUSSION
In this study, we explored the functional relationship between cellular mechanisms that regulate VHF and virulence in the plant pathogen F. oxysporum. VHF occurs at crucial stages during the life cycle of filamentous fungi and therefore is generally assumed to serve important functions in intrahyphal communication, nutrient transport, and colony homeostasis. So far, however, none of these predicted functions has been analyzed experimentally. Although mutants of N. crassa lacking genes required for VHF show pleiotropic phenotypes such as slow vegetative growth or altered conidiation (12, 16, 33, 47), the causal relationship between these phenotypes and lack of VHF remains to be elucidated. Thus, in spite of the emerging knowledge of the underlying molecular mechanisms, the biological role of VHF is still poorly understood.
Previous studies have established that MAPK orthologues of MAK-2 are essential for virulence in many plant-pathogenic fungi (6, 22, 24, 31, 44, 48). Because MAK-2 is required for VHF in N. crassa (33), it was predicted that MAPK-deficient mutants of these pathogenic species would also be unable perform VHF and to form an interconnected mycelial network and that this aspect could be important for pathogenesis (14, 16, 40). Indeed, a different MAPK cascade that controls cell wall integrity was previously shown to be required for full virulence and heterokaryon formation in the cereal pathogen F. graminearum (18).
To explore the functional links between VHF and pathogenicity, we put forward two hypotheses, i.e., that (i) VHF and plant infection have signaling mechanisms in common and (ii) VHF is essential for virulence on plants. We chose F. oxysporum as a model to test our hypotheses since this plant pathogen has been known for many years to undergo VHF (21, 29). Indeed, the ability to form heterokaryons is used routinely to classify F. oxysporum field isolates into vegetative complementation groups (19, 23, 34). Moreover, a number of molecular tools and well-characterized virulence mutants are available in F. oxysporum, including strains affected in different signaling pathways (5-7). Thus, F. oxysporum provides a suitable experimental model to dissect VHF and pathogenicity within a single organism.
Characteristics of VHF in F. oxysporum.
Microscopic analysis of tomato-pathogenic isolate 4287 revealed the presence of frequent hyphal fusion events, in agreement with early reports reporting the occurrence of VHF in this species (21, 29). The hallmarks of VHF in F. oxysporum were similar to those reported in N. crassa (16). Fusion occurred mainly at the interior of mature colonies and between germinating conidia. Short fusion bridges between parallelly growing hyphae were very frequent under the conditions studied, but longer hyphal connections were also observed. The presence of positive tropic responses, previously reported in several fungal species (2, 16, 20, 40), was also evident in F. oxysporum. These responses include reorientation of the hyphal growth axis toward a neighboring hypha or germ tube, as well as the emergence of fusion pegs in receptive hyphae. The nature of the chemoattractant compounds involved in hyphal or germling fusion, as well as their specificity across fungal species, is unknown (14, 38).
Fmk1 and Fso1 control VHF through distinct pathways.
Here we focused on two genes known to play a key role in VHF of N. crassa, MAK-2 and SO (12, 33). Strains of F. oxysporum lacking the orthologue fmk1 or fso1 were severely affected in the fusion of vegetative hyphae and microconidial germ tubes, suggesting that the essential role in anastomosis of the MAPK and WW domain protein is conserved between Neurospora and Fusarium. In our study, deletion of fmk1 had a slightly but reproducibly more severe effect on the frequency of heterokaryon formation than inactivation of fso1. Interestingly, the Δfmk1 Δfso1 double mutants had a clearly lower heterokaryon frequency than either of the single mutants. The simplest interpretation of this result is that mutations in fmk1 and fso1 are not epistatic because the two genes function in distinct pathways. While the PMK cascade has been studied in more detail (22), the biological role of Fso1 remains largely unknown. In N. crassa, the SO protein was shown to localize to septal plugs, but this localization appeared to be independent of the WW protein interaction domain which, in turn, was required for VHF (11). The SO orthologue of Sordaria macrospora, PRO40, which is essential for fruiting-body development, also accumulated in septal plugs of injured hyphae and colocalized with the Woronin body-specific protein HEX-1 (10). It is not clear how the subcellular localization of the SO protein relates to its role in vegetative and sexual hyphal fusion.
Besides their severe impairment in VHF, F. oxysporum Δfso1 mutants displayed a clear phenotype in aging submerged cultures. In contrast to the wild type, they failed to produce large hyphal aggregates and maintained higher levels of microconidia at late stages of culture. Both phenotypes were also observed in the Δfmk1 and Δfmk1 Δfso1 strains, suggesting that they may be related to the absence of anastomosis in these mutants. The requirement of VHF for the formation of hyphal aggregates points to a role of VHF in the formation of large multicellular structures, although the biological significance of these structures is unknown.
Link between VHF and pathogenicity.
The finding that the MAPK Fmk1 controls pathogenicity and anastomosis in a single organism, F. oxysporum, validates the hypothesis that the two processes have some signaling mechanisms in common. The abilities of this conserved MAPK cascade to integrate a variety of signals and to produce distinct developmental outputs were first reported in Saccharomyces cerevisiae, where it controls mating, haploid invasion, and pseudohyphal development (25, 26). Subsequently, it was found to regulate both virulence and sexual development in fungal plant pathogens (24, 31, 48) and, more recently, anastomosis and sexual development in A. nidulans and N. crassa (33, 46). The exact mechanisms through which a single MAPK cascade imparts signaling specificity to these different developmental outputs remain to be elucidated. Mating, anastomosis, and plant infection clearly have characteristics in common, such as activation by diffusible substances, redirection of growth, attachment to the partner or host, differentiation of specialized structures, and deployment of cell wall-degrading enzymes to the site of attachment (14, 38). While the activation of sexual development by pheromones and G-protein-coupled receptors has been studied in detail (9), the molecular nature of the signals and receptors triggering VHF and plant infection remains to be elucidated.
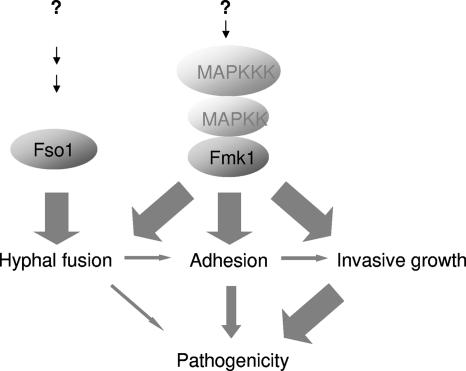
In contrast to Δfmk1 mutants, which were impaired in both VHF and pathogenicity, Δfso1 mutants were only affected in anastomosis but retained full virulence on tomato plants. Thus, Fso1 must function either in an anastomosis-specific branch downstream of the Fmk1 cascade or in a distinct pathway. Based on genetic evidence, we favor the second hypothesis because Δfmk1 Δfso1 double mutants display a more severe phenotype than each of the single mutants. The Δfmk1 Δfso1 strains also consistently showed slightly further reduced virulence compared to the already very weak virulence of the Δfmk1 strain. Thus, although Fso1 contributes only marginally to virulence, its contribution appears to come through a pathway distinct from that of Fmk1 (Fig. 7).
FIG. 7.
VHF and pathogenicity have some signaling mechanisms in common. The Fmk1 MAPK cascade regulates both VHF and virulence-related functions such as adhesion and invasive growth. By contrast, Fso1 functions in a distinct pathway which is essential for VHF but contributes only marginally to virulence, indicating that VHF is not essential for pathogenicity on plants.
The finding that Δfso1 mutants are still virulent on tomato plants rules out the direct implication of anastomosis in plant infection. However, hyphal fusion still may play subtle yet significant roles in fungal pathogenicity. The establishment of hyphal networks may help in optimizing virulence-related functions such as adhesion to host surfaces or exploitation of the limited nutrient resources encountered during infection. Conidial anastomosis tubes were previously reported in leaf pathogens such as Colletotrichum spp. (41). Here we observed frequent anastomosis events during the growth of F. oxysporum on tomato roots and found that strains impaired in VHF were reduced in root adhesion and colonization efficiency. Both pathogenic and nonpathogenic F. oxysporum strains extensively colonize the entire root surface of host plants, with the exception of the apical zone (32). Under natural conditions, the formation of hyphal networks during root colonization may be important for efficient adhesion, nutrient utilization, and competition with other soil microorganisms. In bacterial pathogens, the formation of biofilms on the root surface has been implicated in virulence (45).
A second possible role of anastomosis in pathogenicity is the generation of variability through the transfer of genetic material among different strains or even species (39, 43). This mechanism could be particularly important in pathogens lacking a known sexual cycle such as F. oxysporum. The occurrence of horizontal gene transfer between F. oxysporum and other fusaria has been suggested based on the species distribution of the transposon Fot1 (4). Although fusion among incompatible strains usually results in the death of the fused cells (15), certain DNA regions may escape degradation and become integrated in the genome of the fusion partner (28, 43). Evidence for horizontal transfer of a critical virulence factor, the toxA gene, between two fungal pathogen species was reported recently (13).
In summary, the present study shows that hyphal fusion in F. oxysporum is regulated by distinct cellular pathways. While some of these signaling components are also required for virulence, others are not. These results indicate that VHF is not essential for plant infection.
Supplementary Material
Acknowledgments
We gratefully acknowledge Andre Fleissner, University of California, Berkeley, for providing the Neurospora so sequence before publication.
This research was supported by grant BIO-2004-01240 from the Ministerio de Ciencia y Tecnología to A.D.P. R.P.R. has a Ph.D. fellowship from the Ministerio de Educación y Ciencia. A.D.P. was the recipient of a Ramón y Cajal grant from the Ministerio de Ciencia y Tecnología.
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, C. W. Myers, and D. L. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Buller, A. 1933. Researches on fungi, vol. 5. Longman, London, United Kingdom.
- 3.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 4.Daboussi, M.-J., J.-M. Davière, S. Graziani, and T. Langin. 2002. Evolution of the Fot1 transposons in the genus Fusarium: discontinuous distribution and epigenetic inactivation. Mol. Biol. Evol. 19510-520. [DOI] [PubMed] [Google Scholar]
- 5.Delgado-Jarana, J., A. L. Martinez-Rocha, R. Roldan-Rodriguez, M. I. Roncero, and A. Di Pietro. 2005. Fusarium oxysporum G-protein β subunit Fgb1 regulates hyphal growth, development, and virulence through multiple signalling pathways. Fungal Genet. Biol. 4261-72. [DOI] [PubMed] [Google Scholar]
- 6.Di Pietro, A., F. I. Garcia-Maceira, E. Méglecz, and M. I. Roncero. 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 391140-1152. [PubMed] [Google Scholar]
- 7.Di Pietro, A., M. P. Madrid, Z. Caracuel, J. Delgado-Jarana, and M. I. G. Roncero. 2003. Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 4315-326. [DOI] [PubMed] [Google Scholar]
- 8.Di Pietro, A., and M. I. Roncero. 1998. Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant-Microbe Interact. 1191-98. [DOI] [PubMed] [Google Scholar]
- 9.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70703-754. [DOI] [PubMed] [Google Scholar]
- 10.Engh, I., C. Würtz, K. Witzel-Schlömp, H. Y. Zhang, B. Hoff, M. Nowrousian, H. Rottensteiner, and U. Kuck. 2007. The WW domain protein PRO40 is required for fungal fertility and associates with woronin bodies. Eukaryot. Cell 6831-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleissner, A., and N. L. Glass. 2007. SO, a protein involved in hyphal fusion in Neurospora crassa, localizes to septal plugs. Eukaryot. Cell 684-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleissner, A., S. Sarkar, D. J. Jacobson, M. G. Roca, N. D. Read, and N. L. Glass. 2005. The so locus is required for vegetative cell fusion and postfertilization events in Neurospora crassa. Eukaryot. Cell 4920-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friesen, T. L., E. H. Stukenbrock, Z. Liu, S. Meinhardt, H. Ling, J. D. Faris, J. B. Rasmussen, P. S. Solomon, B. A. McDonald, and R. P. Oliver. 2006. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38953-956. [DOI] [PubMed] [Google Scholar]
- 14.Glass, N. L., and A. Fleissner. 2006. Re-wiring the network: understanding the mechanism and function of anastomosis in filamentous ascomycete fungi, p. 123-139. In U. Kües and R. Fischer (ed.), The Mycota. I. Growth, differentiation and sexuality. Springer-Verlag, Berlin, Germany.
- 15.Glass, N. L., D. J. Jacobson, and P. K. Shiu. 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 34165-186. [DOI] [PubMed] [Google Scholar]
- 16.Glass, N. L., C. Rasmussen, M. G. Roca, and N. D. Read. 2004. Hyphal homing, fusion and mycelial interconnectedness. Trends Microbiol. 12135-141. [DOI] [PubMed] [Google Scholar]
- 17.Gregory, P. H. 1984. The fungal mycelium: a historical perspective. Trans. Br. Mycol. Soc. 821-11. [Google Scholar]
- 18.Hou, Z., C. Xue, Y. Peng, T. Katan, H. C. Kistler, and J.-R. Xu. 2002. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant-Microbe Interact. 151119-1127. [DOI] [PubMed] [Google Scholar]
- 19.Jacobson, D. J., and T. R. Gordon. 1988. Vegetative compatibility and self-incompatibility within Fusarium oxysporum f. sp. melonis. Phytopathology 78668-672. [Google Scholar]
- 20.Köhler, E. 1929. Beiträge zur Kenntnis der vegetativen Anastomosen der Pilze. I. Planta 8140-153. [Google Scholar]
- 21.Köhler, E. 1930. Zur Kenntnis der vegetativen Anastomosen der Pilze. II. Mitteilung. Planta 10495-522. [Google Scholar]
- 22.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leslie, J. 1993. Fungal vegetative compatibility. Annu. Rev. Phytopathol. 31127-150. [DOI] [PubMed] [Google Scholar]
- 24.Lev, S., A. Sharon, R. Hadar, H. Ma, and B. A. Horwitz. 1999. A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA 9613542-13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, H., C. A. Styles, and G. R. Fink. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 2621741-1744. [DOI] [PubMed] [Google Scholar]
- 26.Madhani, H. D., and G. R. Fink. 1998. The riddle of MAP kinase signaling specificity. Trends Genet. 14151-155. [DOI] [PubMed] [Google Scholar]
- 27.Mattern, I. E., P. Punt, and C. A. M. J. J. Van den Hondel. 1988. A vector for Aspergillus transformation conferring phleomycin resistance. Fungal Genet. Newsl. 3525. [Google Scholar]
- 28.McGuire, I. C., J. E. Davis, M. L. Double, W. L. MacDonald, J. T. Rauscher, S. McCawley, and M. G. Milgroom. 2005. Heterokaryon formation and parasexual recombination between vegetatively incompatible lineages in a population of the chestnut blight fungus, Cryphonectria parasitica. Mol. Ecol. 143657-3669. [DOI] [PubMed] [Google Scholar]
- 29.Mesterhazy, A. 1973. The morphology of an undescribed form of anastomosis in Fusarium. Mycologia 65916-919. [PubMed] [Google Scholar]
- 30.Momany, M. 2002. Polarity in filamentous fungi: establishment, maintenance and new axes. Curr. Opin. Microbiol. 5580-585. [DOI] [PubMed] [Google Scholar]
- 31.Müller, P., C. Aichinger, M. Feldbrügge, and R. Kahmann. 1999. The MAP kinase Kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 341007-1017. [DOI] [PubMed] [Google Scholar]
- 32.Olivain, C., C. Humbert, J. Nahalkova, J. Fatehi, F. L'Haridon, and C. Alabouvette. 2006. Colonization of tomato root by pathogenic and nonpathogenic Fusarium oxysporum strains inoculated together and separately into the soil. Appl. Environ. Microbiol. 721523-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey, A., M. G. Roca, N. D. Read, and N. L. Glass. 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puhalla, J. E. 1985. Classification of strains of Fusarium oxysporum on the basis of vegetative incompatibility. Can. J. Bot. 63179-183. [Google Scholar]
- 35.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56117-124. [DOI] [PubMed] [Google Scholar]
- 36.Raeder, U., and P. Broda. 1985. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 117-20. [Google Scholar]
- 37.Rayner, A. D. M. 1996. Interconnectedness and individualism in fungal mycelia, p. 193-232. In B. C. Sutton (ed.), A century of mycology. Cambridge University Press, Cambridge, United Kingdom.
- 38.Read, N. D., and M. G. Roca. 2006. Vegetative hyphal fusion in filamentous fungi, p. 87-98. In F. Baluska, D. Volkmann, and P. W. Barlow (ed.), Cell-cell channels. Landes Bioscience, Georgetown, TX.
- 39.Roca, G. M., N. D. Read, and A. E. Wheals. 2005. Conidial anastomosis tubes in filamentous fungi. FEMS Microbiol. Lett. 249191-198. [DOI] [PubMed] [Google Scholar]
- 40.Roca, M. G., J. Arlt, C. E. Jeffree, and N. D. Read. 2005. Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot. Cell 4911-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roca, M. G., L. C. Davide, M. C. Mendes-Costa, and A. Wheals. 2003. Conidial anastomosis tubes in Colletotrichum. Fungal Genet. Biol. 40138-145. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Sanders, I. R. 2006. Rapid disease emergence through horizontal gene transfer between eukaryotes. Trends Ecol. Evol. 21656-658. [DOI] [PubMed] [Google Scholar]
- 44.Takano, Y., T. Kikuchi, Y. Kubo, J. E. Hamer, K. Mise, and I. Furusawa. 2000. The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13374-383. [DOI] [PubMed] [Google Scholar]
- 45.Walker, T. S., H. P. Bais, E. Déziel, H. P. Schweizer, L. G. Rahme, R. Fall, and J. M. Vivanco. 2004. Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 134320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei, H., N. Requena, and R. Fischer. 2003. The MAPKK kinase SteC regulates conidiophore morphology and is essential for heterokaryon formation and sexual development in the homothallic fungus Aspergillus nidulans. Mol. Microbiol. 471577-1588. [DOI] [PubMed] [Google Scholar]
- 47.Xiang, Q., C. Rasmussen, and N. L. Glass. 2002. The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, J.-R., and J. E. Hamer. 1996. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 102696-2706. [DOI] [PubMed] [Google Scholar]
- 49.Zar, J. H. 1996. Biostatistical analysis, 3rd ed. Prentice-Hall International, London, United Kingdom.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.