Abstract
Most eukaryotic C/D small nucleolar RNAs (snoRNAs) guide 2′-O methylation (Nm) on rRNA and are also involved in rRNA processing. The four core proteins that bind C/D snoRNA in Trypanosoma brucei are fibrillarin (NOP1), NOP56, NOP58, and SNU13. Silencing of NOP1 by RNA interference identified rRNA-processing and modification defects that caused lethality. Systematic mapping of 2′-O-methyls on rRNA revealed the existence of hypermethylation at certain positions of the rRNA in the bloodstream form of the parasites, suggesting that this modification may assist the parasites in coping with the major temperature changes during cycling between their insect and mammalian hosts. The rRNA-processing defects of NOP1-depleted cells suggest the involvement of C/D snoRNA in trypanosome-specific rRNA-processing events to generate the small rRNA fragments. MRP RNA, which is involved in rRNA processing, was identified in this study in one of the snoRNA gene clusters, suggesting that trypanosomes utilize a combination of unique C/D snoRNAs and conserved snoRNAs for rRNA processing.
The rRNAs of eukaryotes undergo both processing and extensive covalent modification that involves the function of small nucleolar RNAs (snoRNAs) (2, 8, 12, 22, 42, 63). The modifications include methylation of the 2′-hydroxyl residue (2′-O-methylation [Nm]) and isomerization of the uracil to pseudouridine (11, 22, 23, 53). The snoRNAs guide the modifications upon site-specific base pairing with their targets. The snoRNAs that guide these modifications are designated by their specific sequence motifs: C/D boxed RNA guides Nm, and H/ACA RNA guides pseudouridylation (3, 12, 24, 53). Most relevant to this study are the C/D snoRNA species (1, 2, 8, 22, 26). The box C/D snoRNAs have 10- to 22-nucleotide (nt) sequences of perfect complementarity to the sequences within the mature rRNA. Thus, formation of the snoRNA-rRNA hybrids positions the conserved box D and D′ elements of the snoRNA 5 bp from the nucleotide to be methylated; this is known as the +5 rule. Each C/D snoRNP consists of a specific snoRNA and a set of binding proteins common to either C/D or H/ACA RNAs. The box C/D snoRNPs include four proteins: Nop56p, Nop58p, Snu13p, and Nop1p (26). It has been demonstrated that Snu13p is required in Saccharomyces cerevisiae for the association of other C/D snoRNP-associated proteins (62) and for the stability of all box C/D snoRNAs. The protein interacts with the terminal C/D motif but not with the internal C′/D′ motif (52). The distribution of these associated C/D snoRNPs is asymmetric, with the C′ box contacting Nop56p and fibrillarin (Nop1p), the C box interacting with Nop58p, and the C, D, and D′ boxes contacting Nop1p (5). The association of Nop1p with C/D guide RNAs depends on a bridge formed by the two core proteins, Snu13p and Nop56p/Nop58p (43, 45, 56). Snu13p binds a K-turn motif in the C/D guide RNA, formed by the interaction of box C and D elements. Upon binding of Snu13p, the restructuring of the RNA creates a new binding site that is recognized by the heterodimer Nop56p/Nop58p, which recruits the catalytic protein fibrillarin (Nop1p) to complete the assembly of the functional complex (51).
Fibrillarin (NOP1) is highly conserved throughout evolution from unicellular organisms such as yeast and trypanosomes to humans (19). All these proteins bear both the glycine- and arginine-rich and methyltransferase domains (9, 19). A NOP1 knockout is lethal in S. cerevisiae, and mutants of this gene exhibit defects in pre-rRNA processing, modification, and ribosome assembly (55). Studies of knockout mice suggest that fibrillarin is essential for early development and is required for accumulation of intron-encoded snoRNAs (41). The effects of the other snoRNP protein on snoRNA stability and rRNA were examined, and it was found that Nop56p is stably associated with snoRNAs only in the presence of Nop1p. In contrast, Nop58p and Nop1p associate independently with the snoRNAs (26). Nop58p is closely related to Nop56p, and each bears characteristic KKE/D repeat sequences. Depletion of Nop58p demonstrates that this protein impairs the processing of 35S pre-rRNA at the A0 and A2 cleavage sites (65).
Relatively little is known about snoRNA in trypanosomes. However, recent genome studies with Trypanosoma brucei identified 21 clusters encoding 57 C/D snoRNAs and 34 H/ACA-like RNAs, which have the potential to direct 84 methylations and 32 pseudouridinylations, respectively (35). Previous studies suggest the existence of at least 100 Nms in trypanosomatids (57). The large repertoire of Nm modification and guide RNAs in trypanosomes suggests that these modifications are likely to play a central role in these parasites. We have proposed that since many of the trypanosomatid species undergo temperature changes during their life cycle, from 26°C in the insect host to 37°C in the mammalian host, the hypermodification may be related to the need to preserve ribosomal activity during the organism's cycling between the insect and the mammalian hosts (57).
rRNA processing in trypanosomes seems to differ from maturation in most eukaryotes. The small-subunit (SSU) rRNA in trypanosomes is the largest one known so far, and the large-subunit (LSU) rRNA is processed into six fragments (6, 64). Whereas in most eukaryotes, the first cleavage of LSU takes place in the 5′ external transcribed spacer (5′ ETS), in T. brucei the first cleavage takes place at internal transcribed spacer 1 (ITS1 [position B1]), which separates the pre-SSU from the pre-LSU rRNA (15). Cleavages at the 5′ ETS at sites A′, A0, and A1 generate the 5′ end of the SSU rRNA. The only trypanosome snoRNAs involved in rRNA maturation that have been identified so far are U3 and snR30 (4, 14, 15, 16, 17). Recently, the role of H/ACA RNAs in rRNA processing was examined by RNA interference (RNAi) silencing of the H/ACA RNA protein CBF5. This study identified rRNA-processing defects including the accumulation of pre-rRNA precursors, as well as reductions in the levels of the mature SSU and LSU (4). Many of the snoRNAs that have been shown to function in rRNA processing in other eukaryotes, including U22, U8, U14, and mitochondrial RNA-processing (MRP) RNAs, were not yet identified in trypanosomes. MRP RNA is structurally and functionally related to RNase P, which is involved in the processing of the 5′ end of pre-tRNA (66). MRP has been shown to cleave pre-rRNA within ITS1 (7, 49).
In this study, we used RNAi silencing of NOP1 to examine the role of C/D snoRNAs in rRNA modification and processing in T. brucei. A machine-learning algorithm was used to predict the level of snoRNA with 85% accuracy. Systematic mapping of Nm sites on rRNA identified an additional 47 Nms beyond the 84 Nms previously identified, suggesting that more C/D snoRNAs than identified in the published repertoire are expected to exist in the genome (31, 35). Mapping of Nms in the two life stages of the parasite, the procyclic and bloodstream forms, revealed hypermethylation in the bloodstream form at certain positions, suggesting that this modification may help the parasite adapt to the higher temperature during cycling from the insect to the mammalian host. Inspection of the rRNA defects in cells in which NOP1 has been silenced suggests the involvement of C/D snoRNAs in trypanosome-specific rRNA-processing events. We also identified a ubiquitous snoRNA involved in rRNA processing, the MRP RNA. This study highlights the role of C/D snoRNAs in trypanosome-specific rRNA-processing events as well as in Nm modification.
MATERIALS AND METHODS
Oligonucleotides.
All the oligonucleotides used are listed in Fig. S-7 in the supplemental material.
Cell growth and transfection.
Procyclic T. brucei strain 29-13 (obtained from the laboratory of Paul England, a gift from the laboratory of George Cross), which carries integrated genes for T7 polymerase and the tetracycline repressor, was grown in SDM-79 medium supplemented with 10% fetal calf serum in the presence of 50 μg/ml hygromycin and 15 μg/ml G418. Cells were transfected as previously described (38). For cloning, the transfected cells were diluted onto microtiter plates in the presence of the parental T. brucei 29-13, which served as feeder cells. The microtiter plate was incubated in a humid chamber at 27°C under a 5% CO2 atmosphere. After 2 to 3 weeks, clonal populations were obtained in the microtiter plates, and the cells were transferred to liquid medium for propagation. Cells from cultures that showed typical growth arrest upon tetracycline induction were grown and frozen. Every 2 weeks, a new culture was started from the original frozen stock.
Northern blot analysis.
Total RNA was prepared with TRIzol reagent (Sigma), and 20 μg/lane was fractionated on a 1.2% agarose-2.2 M formaldehyde gel. The RNA was visualized with ethidium bromide. The NOP1 mRNA and 7SL RNA were detected with randomly labeled probes (Random Primer DNA labeling mix; Biological Industries Co.). rRNA subunits and internal transcribed sequences of pre-rRNA were detected with [γ-32P]ATP-labeled oligonucleotides. For analysis of small rRNAs (srRNAs) and novel C/D snoRNAs, total RNA (10 μg) was fractionated on a 10% polyacrylamide gel containing 7 M urea. The RNA was transferred to a nylon membrane (Hybond; Amersham Biosciences) and probed with [γ-32P]ATP-labeled oligonucleotides.
RT-PCR.
The total RNA was extensively treated with a DNase inactivation reagent (DNA-free; Ambion) to remove DNA contamination. Reverse transcription was performed on the RNA by random priming (Promega). The samples were heated for 5 min at 70°C. After chilling on ice for 2 min, 1 U of superscript III reverse transcriptase (RT) (Invitrogen) and 1 U of RNase inhibitor (Promega) were added, and the reaction mixture was incubated at 55°C for 60 min. The reaction was stopped by heating to 70°C for 15 min, and the reaction mixture was chilled on ice for 2 min. Next, the cDNA was used in PCR amplification as previously described (33).
Primer extension analysis.
RNA was prepared from T. brucei cells using TRIzol reagent (Sigma). Primer extension analysis was performed as described elsewhere (32, 67) using 5′-end-labeled oligonucleotides specific to target RNAs, as indicated in the figure legends. The extension products were analyzed on 6% polyacrylamide-7 M urea gels and visualized by autoradiography.
Mapping of the modified nucleotides.
Nms on rRNA were mapped using primer extension with different levels of deoxynucleoside triphosphates (dNTPs), as described by Xu et al. (67). Primers specific to the relevant region of the rRNA were chosen. In this method, the RT stops 1 nt before the modified base (67). Primer extension products were analyzed on a 6% polyacrylamide-7 M urea gel, alongside results of sequencing reactions performed using the same primer.
TAP tag purification.
The T. brucei 29-13 cell line, coexpressing the Tet repressor and T7 RNA polymerase, was transfected with NotI-linearized plasmid pLew79-snu13pTAP, coding for the Snu13p-TAP-tagged protein under the control of tetracycline and carrying the phleomycin resistance gene (50). Expression of Snu13p-TAP in the transgenic cell line was induced for 72 h with tetracycline (0.1 μg/ml). Cells (1 liter; 107 cells/ml) were harvested, washed with phosphate-buffered saline, and resuspended in IPP150 buffer (10 mM Tris-HCl [pH 8], 150 mM NaCl, 0.1% Nonidet P-40, 1% bovine serum albumin, 5 μg/ml leupeptin). Next, 1% Triton X-100 was added, and the resulting lysate was incubated on ice for 20 min. The lysate was then centrifuged at 10,000 × g for 15 min, and the supernatant was subjected to affinity selection using immunoglobulin G (IgG)-Sepharose beads for 2 h. RNA was extracted from the beads with TRIzol reagent (Sigma) and was subjected to primer extension analysis with specific probes.
Western blot analysis.
Whole-cell extracts (106 cell equivalents per lane) of induced and uninduced cells were separated on a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel, transferred to a Protran membrane (Whatman BioScience), and probed with appropriate antibodies. The anti-fibrillarin antibody (kindly provided by S. J. Baserga, Yale University) and IgG antibodies (Sigma) were diluted 1:1,000 (9) and 1:2,000, respectively. The bound antibodies were detected with goat anti-rabbit IgG coupled to horseradish peroxidase and were visualized by ECL (Amersham Biosciences).
Bioinformatic analysis using SVM.
Support vector machine (SVM) is a machine-learning technique that is used for classification and regression. Recently, it was shown to be useful in bioinformatics studies (68). We applied the SVM algorithm (58) using SVMlight implementation in classification mode (21) to create a model to explain the different expression levels of C/D snoRNA molecules and the different levels of modifications in rRNA sites. Each SVM prediction was evaluated by a five-cross validation test, in which data were randomly divided into five equal parts. Four parts represented the training set for SVM learning, and the final part represented the test set for the SVM model. This procedure was performed 50 times, and the average percentage of successful predictions was calculated.
In vivo labeling of rRNA with [methyl-3H]methionine.
For in vivo labeling of rRNA, NOP1 induced and silenced cells were grown for 2 days. Then cells were washed twice with phosphate-buffered saline and resuspended in 1 ml of methionine-free medium in the presence of 100 μCi of [methyl-3H]methionine (54, 55). Cells were incubated at 27°C for 4 h, and then RNA was extracted from the cells with TRIzol (Sigma). The RNA was separated on a 6% polyacrylamide-7 M urea gel next to a DNA marker (MspI-digested pBR322). Fluorography of the gels was performed using Amplify solution (Amersham).
Determination of 2′-O-methylated nucleotides using alkaline hydrolysis.
For partial alkaline hydrolysis, 50 μg of total RNA was resuspended in 10 mM NaOH and 0.2 mM EDTA, and the samples were boiled for either 30 s or 1, 2, 5, 10, or 30 min. The RNA samples were mixed and precipitated with ethanol. One-tenth of the RNA sample was subjected to primer extension as previously described (27).
RESULTS
Silencing of fibrillarin (NOP1) by RNAi demonstrates that it is essential for life.
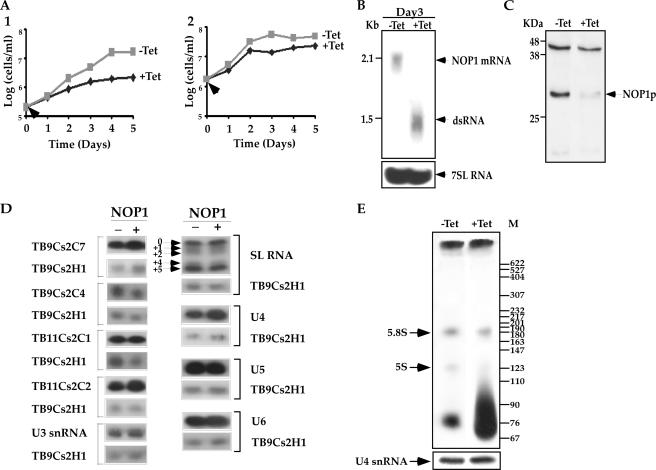
The fibrillarin (NOP1) gene (Tb10.6k15.3160) was previously cloned and analyzed, and antibodies were used to immunoprecipitate and characterize the first T. brucei snoRNP protein (9). To further elucidate the function of the C/D pathway for rRNA processing and modification, the NOP1 gene was silenced by RNAi. A 511-bp fragment carrying the coding information was used to prepare a stem-loop silencing construct. The construct was linearized for integration into the nontranscribed rRNA spacer, and a clonal population of cells was prepared, as previously described (38). A typical growth curve of the silenced cells at different temperatures is presented in Fig. 1A. At the normal growth temperature, the silenced cells stopped growing after 3 days, and growth remained arrested (Fig. 1A1). If the silenced cells grew at 37°C, growth arrest occurred earlier, after 2 days (Fig. 1A2). These results suggest that the NOP1 gene is essential and that its absence causes temperature sensitivity. A similar phenotype was observed in yeast, suggesting that fibrillarin is an essential gene (19). To demonstrate that the growth arrest stems from the silencing of the gene, the levels of fibrillarin mRNA and protein were examined 3 days after silencing. The results, shown in Fig. 1B, demonstrate down-regulation of the fibrillarin mRNA. The level of fibrillarin protein was also reduced by 75%, as presented in Fig. 1C. We next examined the effects of fibrillarin silencing on the levels of C/D snoRNAs and Nm on rRNA. The results presented in Fig. 1D suggest that silencing of fibrillarin did not affect the levels of C/D snoRNAs. These results resemble the observation that in yeast, C/D snoRNAs, which are independently transcribed, are not affected by fibrillarin depletion (54). The levels of H/ACA RNAs, snRNAs, and spliced leader (SL) RNA also were not altered by silencing of the C/D pathway (Fig. 1D). The effect of C/D silencing on the SL RNA level suggests that Nm modification is not essential for trans-splicing. This is especially interesting, because RNAi silencing of the H/ACA core protein CBF5 results in splicing defects, which are manifested by the accumulation of SL RNA and reductions in the levels of the Y structure intermediate and mature mRNA (4).
FIG. 1.
NOP1 is essential for life. (A) Growth curves of cells carrying the NOP1 RNAi construct. The growth of uninduced cells (−Tet [gray]) was compared with that of cells in which dsRNA production was induced (+Tet [black]). Arrows indicate the times when tetracycline was added. (Panel 1) Cell growth at 27°C; (panel 2) cell growth at 37°C. (B) Northern blot analysis of NOP1 mRNA upon RNAi silencing. RNA was prepared from cells carrying the RNAi construct, either uninduced (−Tet) or induced for 3 days (+Tet). Total RNA (30 μg) was separated on a 1.2% agarose-2.2 M formaldehyde gel. The RNA was blotted and hybridized with a randomly labeled probe specific for the NOP1 gene (positions 389 to 899 of the coding sequence). The level of 7SL RNA (used as a control for equal loading) was determined using a randomly labeled probe specific for 7SL RNA as described in Fig. S-7 in the supplemental material. Positions of markers are indicated on the left. (C) Western blot analysis of Nop1p. Whole-cell extracts (106 cells per lane) were prepared from cells carrying the RNAi construct, either uninduced (−Tet) or after 3 days of induction (+Tet). The extracts were separated on a 12% SDS-polyacrylamide gel and subjected to Western blot analysis with the anti-fibrillarin antibody (9). (D) Effects of NOP1 silencing on the levels of C/D snoRNAs and snRNAs. Total RNA was prepared from cells carrying the RNAi construct either without induction (−) or after 3 days of induction (+). Total RNA (10 μg) was subjected to primer extension with radiolabeled oligonucleotides complementary to each of the C/D snoRNAs and to SL RNA and snRNAs. The oligonucleotides and their sequences are summarized in Fig. S-7 in the supplemental material. cDNA was separated on a 6% sequencing gel. To control for the level of RNA in each sample, primer extension was performed with primer TBsno-H-1, specific to TB9Cs2H1 H/ACA snoRNA. (E) In vivo labeling of rRNA under NOP1 depletion. Uninduced cells and cells induced for 2 days were labeled with 100 μCi of [methyl-3H]methionine as described in Materials and Methods. RNA was separated on a 6% polyacrylamide-7 M urea gel next to a DNA marker (MspI-digested pBR322). The levels of RNA in the samples were determined by analyzing the U4 snRNA by primer extension.
We next examined how the depletion of NOP1 affects Nms. Cells (untreated) in which NOP1 was depleted (on the second day following induction) were labeled in vivo with [methyl-3H]methionine for 4 h, and the incorporation of methyl groups into the rRNA was examined after separation of the RNA on a denaturing gel and fluorography. The results, presented in Fig. 1E, indicate reductions in the incorporation of methyl groups into rRNA (5.8S and 5S rRNA) in NOP1-depleted cells. Note that the level of incorporation of methyls into tRNA, which is usually mediated by specific methyltransferases (40), was even somewhat increased in the silenced cells. These results indicate an overall effect of NOP1 depletion on the level of methylation on rRNA. To locate specific defects in Nm under conditions of NOP1 depletion, systematic mapping was performed covering 67% of the entire LSU and SSU rRNAs. Mapping was performed using the dNTP starvation approach. In this reaction, at low concentrations of dNTPs, the RT pauses 1 nt before the Nm (24). In Fig. 2a, we present one example of such mapping data; three additional examples are presented in Fig. S-1A to S-1C in the supplemental material. The secondary structure of the relevant domain whose Nms were mapped is shown in Fig. 2b. Note that the RT stops not only due to Nm modifications but also in regions of complex secondary structure. However, such structural stops are insensitive to changes in dNTP concentrations and thus appear at the same intensity in reactions conducted with either low or high dNTP levels. To control for the amount of RNA that was used in the samples, primer extension was also performed with an antisense primer for U4 snRNA (Fig. 2a). The percentage of reduction in the stop due to Nm modification in RNA from the silenced cells is indicated.
FIG. 2.
Mapping of the modified nucleotides in SSU by a dNTP starvation assay and alkaline hydrolysis. (a) Total RNA was prepared from NOP1-silenced cells without induction (−Tet) or after 3 days of induction (+Tet). Total RNA (8 μg) was subjected to primer extension with radiolabeled oligonucleotide 1923, specified in Fig. S-7 in the supplemental material. Primer extension was performed using different concentrations of dNTPs (0.05, 0.5, and 5 mM) to detect the methylations. Extension products were analyzed on a 6% polyacrylamide-7 M urea gel, and DNA sequencing was performed with the same oligonucleotides used in the primer extension assay. Partial sequences are given; open arrows indicate structural stops. To control for the level of RNA in each sample, primer extension was performed with primers specific to U4 snRNA. The percentages of reduction in the levels of the stops and the standard deviations based on three independent experiments are given on the right. (b) Locations of the Nms on rRNA. Nm modifications in T. brucei (T), S. cerevisiae (Y), the plant A. thaliana (P), and humans (H) are indicated. (c) Mapping of the Nms by alkaline hydrolysis. RNA (50 μg) was subjected to alkaline hydrolysis. The same primer used for panel a was used for primer extension of the partially hydrolyzed RNA (L). The sequence of the DNA using the same primer is given, and mapping with different dNTP levels, as in panel a, is shown in the four right lanes. Arrows mark the positions of the methylated nucleotides, and their identities are given.
The dNTP starvation approach, presented in Fig. 2 and in Fig. S-1A to S-1C in the supplemental material, is commonly used to map DNA modifications. However, we wanted to confirm the mapping data using an alternative method in order to verify the starvation protocol for large-scale mapping. For this purpose, we used the partial alkaline hydrolysis technique, which is based on the principle that a phosphodiester bond adjacent to an Nm modification is completely resistant to hydrolysis under basic conditions (37). This method requires a greater amount of rRNA than does the low-dNTP method, as well as an additional step of alkaline hydrolysis, and therefore is not used as often as the dNTP starvation assay for high-throughput experiments. We compared the mapping of Nms of the same domain by the two methods. The results, presented in the leftmost lane of Fig. 2c, show the alkaline hydrolysis ladder generated by primer extension. The presence of an Nm modification is indicated by a gap in the ladder, since the phosphodiester bond adjacent to Nm modification is resistant to alkaline hydrolysis. The right lanes show the results of primer extension under low dNTP concentrations, analyzed on the same gel. The modifications on positions G1900, G1895, U1875, and C1868 were confirmed by the two methods.
In all the mapping experiments (Fig. 2a to c; see also Fig. S-1A to S-1C in the supplemental material) using the dNTP starvation assay, we noticed major differences in the intensity of primer extension stops and observed weaker stops in between the strong stops. Multiple factors can affect the level of modification and the stops due to the presence of Nm on a particular site. Factors that may affect the level of Nm modifications are the abundance of the snoRNA guiding the modification and the strength of the interaction between the snoRNA and its target site. In addition, the stops on the Nm may be affected by the location of Nm on the rRNA secondary structure. In the alkaline hydrolysis method as well, we observed differences between the intensity of the extension products within the gap, indicating the lack of modification on this site. However, we could not find a correlation between the intensity of the stop in the dNTP experiment and the degree of alkaline cleavage, suggesting that comparison between the two methods can be used to confirm the presence of modification but not to determine the intensity of modification, especially when different sites are compared.
Effect of NOP1 depletion on rRNA processing.
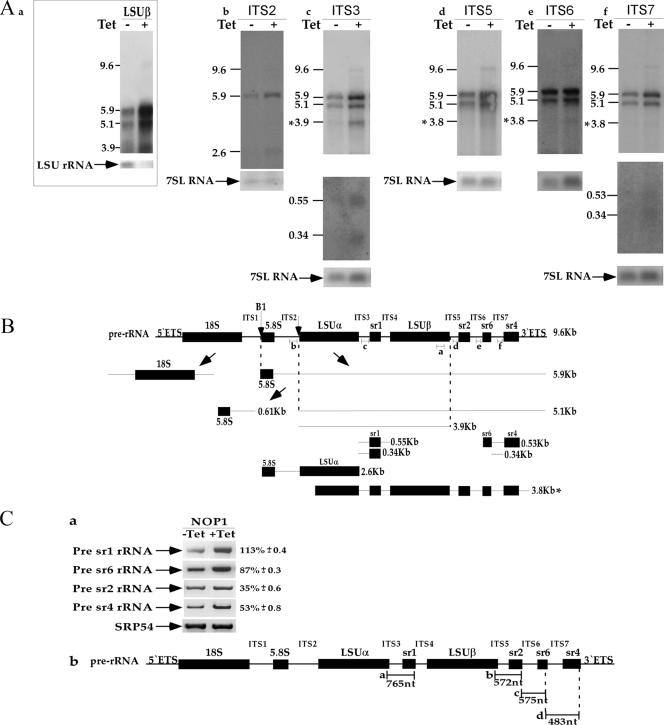
We next used the NOP1-silenced cells as a tool to decipher the complex process of rRNA processing, especially that of the LSU, aiming to identify rRNA defects that are informative as to the possible involvement of C/D snoRNA in trypanosome-specific rRNA events. Trypanosomes, as opposed to other eukaryotes, have additional rRNA-processing events that separate the LSUα and LSUβ as well as releasing the srRNA fragments (6, 64). If C/D snoRNAs are involved in these specific cleavages, we would expect to observe a decrease in the production of srRNAs and an accumulation of precursors flanking the RNAs. To identify such defects, two approaches were taken. First, the accumulation of precursors and reductions in the levels of mature RNAs were examined by Northern blot analysis (Fig. 3). RNA was extracted from cells before (−Tet) and after (+Tet) induction of silencing and was subjected to Northern blot analysis with probes specific to the SSU. The results are presented in Fig. 3Aa, and a schematic presentation of the rRNA domain and its cleavage sites is given in Fig. 3Ac. Northern blotting detected three rRNA precursors of 3.7, 3.3, and 2.6 kb. The 3.7-kb precursor results from cleavage at B1, releasing the entire pre-SSU portion of the precursor that was not cleaved at site A0, A1, or A2. The 3.3-kb species is derived by further cleavage of the 3.7-kb precursor at A2 but not at B1. The 2.6-kb species is cleaved at the A0 site but not at the A2 site. The identities of these precursors were confirmed by hybridization with ETS (data not shown) and ITS1 (Fig. 3Ab) probes. The most abundant precursor is the 3.7-kb species, which results from a major defect in cleavage at the A0, A1, or A2 sites. To precisely evaluate the rRNA defects, we mapped the precursors and the cleavages in the pre-SSU region. Primer extension was performed with oligonucleotides situated in regions downstream from the A′, A0, A1, and A2 sites (indicated in Fig. 3Ba). Increases in the levels of primer extension products, obtained by using primers situated downstream of A0 (Fig. 3Bb2) and A2 (Fig. 3Bb4), suggest the accumulation of the pre-rRNA harboring these sites, since these cleavages were inhibited by silencing. The accumulation of RNA carrying the A′ site (Fig. 3Bb1) suggests additional defects in removal of the ETS, which may stem from defects in cleavage at A0 and A1. Indeed, cleavage at A1 was inhibited, as can be seen in Fig. 3Bb3, demonstrating a reduction in the stop at the A1 position. The rRNA defects observed suggest the inhibition of cleavages at positions A1 and A2. The defect in rRNA processing at sites A1 and A0 may be attributed to defects in U3 function. However, other C/D snoRNAs should exist in trypanosomes to guide these cleavages. In yeast and mammals, a similar cleavage is mediated by U14 (28, 30, 39). So far, we have failed to identify a U14 homologue. However, TB11Cs2C1 might functionally replace U14 in trypanosomes (see Discussion). The most abundant precursor that accumulates in NOP1-silenced cells is the 3.7-kb transcript, which results from cleavage at the B1 site, situated near the cleavage site of MRP RNA (see below).
FIG. 3.
Effect of NOP1 silencing on SSU rRNA maturation. (A) (a and b) Northern blot analysis of SSU rRNA and ITS1 in a NOP1-silenced cell line. Total RNA was extracted from cells carrying the NOP1 RNAi construct without induction (−Tet) or after 3 days of induction (+Tet) and was separated on a 1.2% agarose gel containing 2.2 M formaldehyde. The RNA was blotted and hybridized with probes for SSU rRNA (a) (SSU1923; probe h in panel Ac) and ITS1 rRNA (b) (probe g in panel Ac). Long and short exposures are presented. The 7SL RNA probe was used to control for the level of RNA in each sample. Positions of markers, in kilobases, are given on the left. (c) Schematic representation of the rRNA-processing sites (indicated by arrows) on SSU rRNA. The sizes of the fragments are given on the left, and the positions of the probes used for hybridization (g, h, and i) are indicated. (B) (a) Schematic representation of the pre-rRNA SSU. The positions of oligonucleotides 1 to 4, used for primer extension, are indicated. Processing sites are indicated by arrows. (b) Primer extension mapping of rRNA precursors. Total RNA was extracted from cells carrying the NOP1 RNAi construct without induction (−Tet) or after 3 days of induction (+Tet). The RNA was analyzed by primer extension with a radiolabeled oligonucleotide complementary to each of the sites: oligonucleotide A′-2730 (oligonucleotide 1 in panel Ba), oligonucleotide A0-5470 (oligonucleotide 2 in panel Ba), oligonucleotide A1-2729 (oligonucleotide 3 in panel Ba), and oligonucleotide A2-5472 (oligonucleotide 4 in panel Ba). A DNA sequence ladder for each of the primers is shown on the left. The position on the rRNA is indicated by the corresponding number. Cleavage sites are marked next to the sequence with asterisks. U3 snoRNA was used as a control for the amount of RNA in each sample.
To examine possible defects in LSU rRNA, the Northern blots were probed with primers specific to LSU-β, ITS2, ITS3, and ITS5 to -7 (Fig. 4Aa to f). A schematic representation of the cleavages that take place to generate LSU and srRNA molecules, along with the predicted precursors that accumulate during the knockdown, is given in Fig. 4B. The results in Fig. 4Aa to f demonstrate the accumulation of the 9.6- and 5.9-kb precursors. The 9.6-kb precursor is the full-length pre-rRNA transcript. The 5.9-kb transcript emerges from cleavage of the 9.6-kb precursor at the B1 site, which is most probably mediated by MRP RNA. The 5.1-kb precursor emerges from cleavage at the ITS2 site to separate the 5.8S precursor from the remaining genes, suggesting that this cleavage is not mediated by C/D snoRNA (Fig. 4Ac to f). The 3.9-kb precursor emerges from cleavage at the ITS5 site, which separates the two large structural RNAs (LSUα and LSUβ) from the three distal small RNAs (sr2, sr6, and sr4) at the 3′ end, suggesting that cleavage at ITS5 is not affected in NOP1-depleted cells (Fig. 4Aa and c). In addition, the 2.6-kb precursor was detected at low levels with an ITS2-specific probe. This intermediate spans the 5.8S precursor through LSUα and did not hybridize to the ITS3 or the ITS1 probe (Fig. 4Ab). It may represent an alternative processing intermediate whose generation does not depend on C/D snoRNA, again suggesting that cleavage at the 5′ end of srRNA-2 is not mediated by C/D snoRNA. In addition, we identified two precursors that hybridized only to ITS3 (0.55 and 0.34 kb) (Fig. 4Ac), suggesting that separation of srRNA-1 from LSUα depends on C/D snoRNA. Two precursors that hybridized only to ITS7 (0.53 and 0.34 kb) (Fig. 4Af) were identified, suggesting that cleavage at srRNA-6 depends on C/D snoRNA. Probes corresponding to ITS5 to -7 identified a 3.8-kb precursor, which most probably represents an aberrant processing intermediate resulting from cleavage upstream of LSUα rRNA. The defects in rRNA processing, especially in the processing of the LSU, suggest that additional C/D snoRNAs should exist in trypanosomes to mediate the cleavages generating srRNAs 1, 4, and 6.
FIG. 4.
Effect of NOP1 silencing on LSU rRNA maturation. (A) Northern blot analysis of LSU rRNA in a NOP1-silenced cell line. Total RNA was extracted from cells carrying the NOP1 RNAi construct without induction (−Tet) or after 3 days of induction (+Tet) and was separated on a 1.2% agarose gel containing 2.2 M formaldehyde. The RNA was blotted and hybridized with probes for LSUβ (a), ITS2 (b), ITS3 (c), ITS5 (d), ITS6 (e), and ITS7 (f) rRNA. The lower panels for the ITS3 and ITS7 rRNA probes represent the results of long exposure of the small transcripts. The 7SL RNA probe was used to control for the level of RNA in each sample. Positions of markers (in kilobases) are given on the left. (B) Scheme of pre-rRNA processing of LSU rRNA. Processing sites are indicated by arrows. The size of each fragment, corresponding to the sizes in panel A, is given on the right. Probes a to f are each represented by a short line and are complementary to LSU-β, ITS2, ITS3, ITS5, ITS6, and ITS7 rRNA, respectively. Aberrant processing intermediates are indicated by an asterisk. (C) (a) Levels of the srRNA precursors upon NOP1 silencing. RT-PCR was performed on total RNA (DNA free) prepared from NOP1 uninduced cells (−Tet) or induced cells 3 days after induction (+Tet). The levels of increase in pre-rRNA accumulation ± standard deviation based on three independent experiments are presented. The level of SRP54 mRNA served as a control for the amount of cDNA used in the experiments. (b) Schematic representation of the srRNA genes demonstrating the positions of the primers used for amplifying the precursors. The expected sizes of the products and the oligonucleotides used for the RT-PCR are shown.
To further examine the role of C/D snoRNA in the processing of srRNAs, and to fine map the defects in generating srRNA, the level of ITS flanking srRNA was examined using a sensitive RT-PCR assay. RNA was prepared from cells before silencing and 2 days after induction, and cDNA was prepared using random primers. The level of the RNA in each sample was calibrated based on the level of SRP54 mRNA. The cDNA was amplified with primers such that the reverse primer was situated in the coding sequence of srRNA and the forward primer in the upstream ITS. The results indicate differential effects on the levels of precursors. The level of the precursor situated between srRNA-1 and LSUα was increased by 113% (Fig. 4Ca). These data are in agreement with the data presented in Fig. 4Ac, where two precursors in this area were detected, suggesting that C/D snoRNAs are likely to be involved in the processing of srRNA-1, most probably by mediating cleavages at both the 5′ and 3′ ends of the molecule. The other precursors whose levels were elevated are the ones situated between srRNA-2 and srRNA-6 as well as between srRNA-6 and srRNA-4 (Fig. 4Ca), suggesting that cleavages that release srRNA-6 are also dependent on C/D snoRNAs. These data are also in agreement with the detection of 0.53- and 0.43-kb precursors covering both srRNA-4 and srRNA-6 (Fig. 4Af). These results are also consistent with the rRNA precursors recently described by Jensen et al. (20). Interestingly, the levels of all the precursors were not affected to the same extent, but it was possible to detect specific defects greater than the general elevation in the level of the pre-rRNA precursor (∼35%). The general increase of 30 to 40% in each of the precursors reflects the accumulation in the silenced cells of the 9.6-kb precursor, which covers the entire rRNA repeat unit.
MRP RNA exists within a snoRNA cluster.
Cleavage at the B1 site was not affected by silencing of either the C/D or the H/ACA pathway (4), suggesting that, as in other eukaryotes, cleavage in the vicinity of this site is most probably mediated by MRP RNA which, in yeast, cleaves at the A3 site, situated just upstream (59). Since snR30 was recently identified in a cluster carrying C/D and H/ACA RNA, we searched the snoRNA clusters for RNAs longer than the average size of snoRNAs (70 to 90 nt). Such an RNA was identified in cluster 10 as TB10Cs1. The RNA is 521 nt long. Multiple alignments of the RNAs from different species such as humans, S. cerevisiae, and D. melanogaster are presented in Fig. S-6 in the supplemental material. Primer extension was used to map the 5′ end of the RNA (Fig. 5B). The secondary structure of the T. brucei RNA is presented next to the secondary structure of the S. cerevisiae RNA (Fig. 5A). The data presented in Fig. 5 suggest that TB10Cs1 is most probably the T. brucei MRP homologue. Like all MRP RNAs in nature, the trypanosome homologue is divided into two domains (Fig. 5A). Whereas subdomains P1, P2, P3, and P4 within domain 1 are highly conserved, the entire domain 2 is less conserved throughout evolution (29, 44). Interestingly, the study that identified various MRP RNAs from multiple species failed to find the trypanosomatid RNA (44). The identification of TB10Cs1 as MRP RNA is based on several criteria. CR-I (conserved region I) (Fig. 5C1) obeys the consensus sequence of MRP CR-1 but not that of RNase P; all nine of the most conserved nucleotides exist in the T. brucei sequence. The sequence of CR-V also resembles the consensus sequence and is G rich, but CR-V is less conserved than CR-I, and only 6 out of 8 nt present are conserved in the T. brucei sequence (Fig. 5C2). The sequence of CR-IV contains the three conserved nucleotides found in all MRP RNAs (Fig. 5A).
FIG. 5.
Identification of the MRP RNA in the T. brucei genome. (A) (Left) Predicted secondary structure of the T. brucei MRP RNA. The conserved nucleotides of the conserved regions CR-I, CR-IV, and CR-V are circled. The nomenclature for the helices P1, P2, P3, P4, P5, P6, CR-IV, mCR-I, and mCR-V is based on the nomenclature of the S. cerevisiae MRP homologue in the work of Piccinelli et al. (44). (Right) Predicted secondary structure of S. cerevisiae MRP RNA, based on the method of Piccinelli et al. (44). (B) Total RNA (20 μg) was prepared from procyclic T. brucei and subjected to primer extension with a radiolabeled oligonucleotide (designated MRP50). The extension product was analyzed on a 6% polyacrylamide-7 M urea gel, and DNA sequencing was performed with the same oligonucleotides used in the primer extension assay. Partial sequences are given, and the MRP 5′ end is marked by an arrow. (C) Sequence logos of CR-I (panel 1) and CR-V (panel 2) of MRP RNA based on the method of Piccinelli et al. (44). The sequence of the T. brucei domain is given below each logo.
Domain 2 of MRP RNA is less conserved among all eukaryotes. This domain is composed of several stem-loop structures that emerge from a single-stranded region. The number of stem-loops and the size of the stems differ in different organisms. For instance, in yeast, three stem-loops (ymP5, ymP6, and ymP7) exist (29), and in humans, these stem-loops are designated P8, P9, and P12 (60). Inspection of the trypanosome sequence identified five stem-loop structures in domain 2, whereas in other organisms, such as the yeast S. cerevisiae, Chlamydomonas spp., and Drosophila, only three stem-loops exist (44). The first two stem-loops in the trypanosome MRP were designated P5 and P6, as in yeast, but P7 and P8 seem to be specific to trypanosomes, whereas P9 seems to be related to human P9 in its length, though it lacks the second stem-loop that carries the K-turn motif in the mammalian RNA. Such long stem-loops in similar positions are also found in the structures of the Chlamydomonas and Drosophila homologues (44). Additional stem-loops, which we termed P10, exist in the trypanosome RNA and might be equivalent to eP15 in the yeast molecule. Two major differences exist between trypanosome MRP and its homologues in other eukaryotes: trypanosome MRP lacks eP19, which is normally found between P4 and P2, but carries an extended stem-loop at the 5′ end, which we termed Pext, that seems to be trypanosome specific. The major deviation in domains 1 and 2, summarized above, and the sequence divergence in CR-V most probably explain why this RNA was not identified by the systematic search of MRP RNA in the recent study by Piccinelli et al., which failed to identify MRP and RNase P homologues in any trypanosomatid species (44).
Prediction of the cellular level of snoRNA using the SVM algorithm.
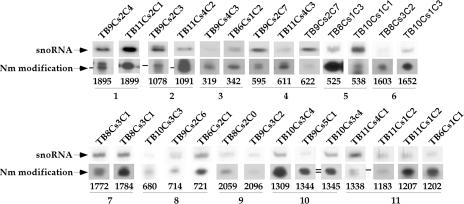
This study identified two observations that require explanation: the variability in the expression level of each C/D molecule and the variability in the modification level of each Nm site, as described above and as can be clearly seen in Fig. 6 (determined by the dNTP starvation assay). Since the level of stops might be affected by the local structure and environment of the rRNA, we assigned the modifications to 11 groups, each of which represents modifications situated at a single region. Within each group, the level of each modification and the level of the snoRNA were examined by primer extension and were normalized to the level of U4 snRNA in each sample. This analysis was performed for 27 sites (Fig. 6; see also Fig. S-8 in the supplemental material). In addition, we collected other parameters for Nm modifications and the snoRNAs that guide these modifications, including the location of the modification on the secondary structure, the conservation of the modification, and the conservation of the snoRNA and its boxes (see Fig. S-8 in the supplemental material).
FIG. 6.
Levels of modifications and their corresponding guiding snoRNAs. The levels of modifications and the levels of the snoRNAs that guide these modifications were determined for 11 different groups representing different regions on the rRNA. The level of the snoRNA guiding the modification was determined relative to the level of U4 snRNA in each sample. Quantitations of the levels of modification and snoRNAs are given in Fig. S-8 in the supplemental material. Numbers below the gels indicate the Nm positions on the rRNA.
Manual inspection of the level of modification guided by different snoRNA species suggested that there is no obvious correlation between the snoRNA level and the extent of modification. As presented in Fig. 2c, primer extension by dNTP starvation can faithfully identify the Nm, but it is difficult, based on this assay, to learn about the relative abundance of the modification. The differential strength of stops on certain positions is very reproducible and has been described previously in other systems (37). The basis for this phenomenon is currently unknown, but it remains difficult to use the differential level of the stops to deduce the actual level of modification, which may differ along the rRNA.
However, several of the parameters described in Fig. S-8 in the supplemental material could explain the differential abundance of the snoRNAs guiding the different modifications. We decided to use a sophisticated machine-learning algorithm that can search for a combination of parameters (21, 68) to help us predict the abundance of snoRNA. To predict the abundances of C/D molecules, the SVM approach was used in the following manner. For each C/D molecule, 10 parameters that may be related to its expression level or to the function of these molecules were compiled (see Fig. S-8 in the supplemental material). For each pair of C/D molecules that guide adjacent modifications and whose expression level could be compared, we created a vector describing the differences between the 10 parameters associated with each molecule involved in these modifications. The algorithm was trained to choose the C/D molecule with the higher expression level. Clearly, a random prediction would yield a 50% success rate, and a significant deviation from that value would indicate that the algorithm was able to capture a combination of parameters that affects the expression level. When the SVM was trained on the set of parameters, the average percentage of successful predictions for the level of snoRNA was 65.5%, suggesting that indeed several or all of the parameters used can explain the differential level of snoRNA abundance. To further examine which factors were the most relevant to the prediction, we removed one parameter at a time from the list and ran the SVM with the remaining variables. When a significant deterioration in the performance of the model occurred due to the removal of a single parameter, we concluded that this variable was crucial for the system. Using this approach, we identified four features that best predicted the snoRNA level: conservation of the C box, the number of modifications guided by the snoRNA, location of the modification on a stem or loop, and the level of conservation of the Nm modification among eukaryotes. Interestingly, although the level of conservation of Nm modifications among eukaryotes has no statistically significant correlation to the abundance of the corresponding C/D molecule, somehow it is essential for the prediction model. The new SVM model was created based only on these four parameters, and the average percentage of successful prediction was 85%. The significant improvement in the prediction success rate with only 4 parameters over that for the initial set with 10 parameters is probably due to the decrease in the noise level introduced by the irrelevant parameters.
The level of modifications is increased at certain positions in bloodstream-form trypanosomes.
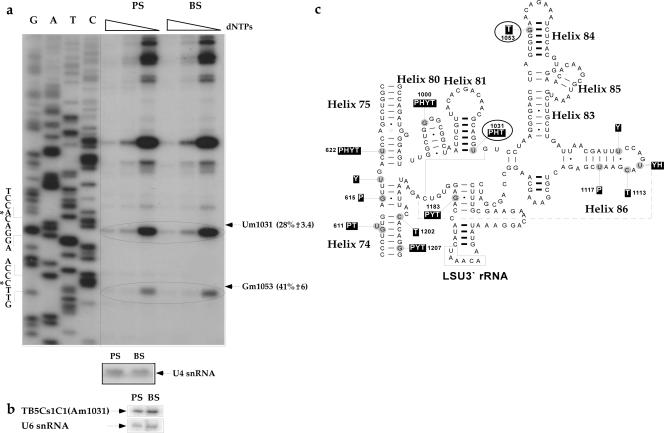
Based on the large amount of C/D snoRNAs and the high level of Nm modifications in trypanosomes, we previously proposed that these modifications may assist the trypanosomes in coping with the temperature shift during their cycling between the insect and mammalian hosts (57). It is well documented that in hyperthermophilic archaea, a large number of Nms stabilizes the ribosomes at high temperatures (57). We therefore systematically mapped the modifications during the two life stages of the parasites. To this end, total RNA was prepared from procyclic and bloodstream parasites and was subjected to mapping that covered ∼21% of the rRNA. In each experiment, the level of RNA in each sample was calibrated by primer extension using an antisense U4 snRNA probe. The results are presented in Fig. 7 and Fig. S-2A to S-2C in the supplemental material. The increase in the level of stops in RNA from bloodstream-form parasites over that for procyclic-form parasites ranged from 59 to 500%. These experiments represent only a small portion of many examples observed. However, the extent of modification did not increase for all positions, and only about 20% of the modified nucleotides seemed to increase in the bloodstream form.
FIG. 7.
Comparison of the levels of Nm modifications in bloodstream-stage (BS) versus procyclic-stage (PS) trypanosomes. Total RNA (8 μg) was prepared from T. brucei procyclic and bloodstream forms and was subjected to primer extension with radiolabeled oligonucleotides, performed as described for Fig. 2. Partial sequences are given, and the modified nucleotides are marked by solid arrows and circles. To control for the level of RNA in each sample, primer extension was performed with a primer specific to U4 snRNA. (a) Mapping of Nms on LSU rRNA (3′ half) using primer 2 LSU-AS 1098-1117. The modified nucleotides that are developmentally regulated are circled. (b) The level of TB5Cs1C1 snoRNA, which directs the Am1031 modification, was analyzed by primer extension using a radiolabeled oligonucleotide against TB5Cs1C1 snoRNA. U6 snRNA served as a control for the level of RNA in each sample. (c) Positions of the modified nucleotides on the secondary structure of T. brucei LSU 3′ rRNA. Nm modifications in T. brucei (T), S. cerevisiae (Y), the plant A. thaliana (P), and humans (H) are indicated.
Although we reached the conclusion that we cannot deduce the level of modification based on the extent of stops at specific Nm sites, the situation is different when the intensity of stops at a single Nm site is studied under different conditions. It is therefore possible to compare the level of stops at the same site but under different conditions, such as at two developmental stages of the parasite. In this case, the extent of stops should be proportional to the abundance of the modification. Interestingly, we did not observe any bloodstream-specific modification that does not exist in the procyclic form. However, we cannot exclude the possibility that such modifications may exist. We noticed that the majority of modifications whose levels are increased in the bloodstream form either exist in important functional domains or are conserved in evolution. For instance, Cm1207 is conserved in plants, yeast, and trypanosomes and is located in the peptidyltransferase center domain, and Gm1895 is also conserved in plants, humans, yeast, and trypanosomes. The mechanism that leads to the developmental regulation of these modifications is currently unknown. One mechanism to increase the modification might be an increase in the expression of the snoRNA that guides this particular modification. Indeed, the results in Fig. 7b suggest that this is the case for TB5Cs1C1, which guides the modification on Am1031, since the snoRNA level was higher in bloodstream-form than in procyclic-form parasites.
Silencing of NOP58 and TAP tagging of Snu13p homologues serve as tools to identify novel C/D snoRNAs.
We previously estimated that the collection of C/D snoRNAs that had been identified covered only 70% of the expected repertoire (35). To test this prediction, we systematically mapped ∼67% of the entire LSU and SSU for Nm modifications. The results, presented in Fig. S-9 in the supplemental material, summarize modifications identified based on the primer extension assay. The levels of all modifications listed in Fig. S-9 in the supplemental material were reduced in NOP1-silenced cells, indicating that these modifications are likely to be guided by C/D snoRNAs. Note that these modifications were not predicted previously on the basis of the snoRNAs identified in the T. brucei genome (35). Interestingly, most of these modifications (60%) are trypanosome specific; 30% are present only in trypanosomes and plants; and only three were found in humans, yeast, trypanosomes, and plants. These data suggest that trypanosome rRNA carries at least 131 Nms, the sum of the 84 modifications listed by Liang et al. in 2005 (35) and the 47 new modifications mapped in this study. However, the true number might be even larger, since our mapping did not cover the entire rRNA but only the domains that are also rich in modifications in other eukaryotes. These data further suggest that additional snoRNAs may exist in trypanosomes to carry out the modifications identified in this study (see below).
Since NOP1 silencing did not affect the levels of snoRNAs, we wanted to generate a tool that would enable us unequivocally identify RNA molecules as C/D snoRNAs. We therefore searched for other snoRNP proteins in the T. brucei genome. NOP58 depletion destabilizes snoRNA in yeast (25); we therefore sought to identify its trypanosome homologue. The T. brucei genome was searched with the human homologue, and a sequence alignment of T. brucei NOP58 with those of other eukaryotes is presented in Fig. S-3 in the supplemental material. The results indicate 53% identity to human, 52% identity to S. cerevisiae, and 55% identity to Drosophila NOP58. Our search also revealed a homologue to NOP56, and its sequence alignment is presented in Fig. S-4 in the supplemental material. The results indicate 49.5% identity to human, 48% identity to S. cerevisiae, and 49% identity to Drosophila NOP56. As in other eukaryotes, NOP56 and NOP58 are closely related. Like the yeast proteins, both proteins carry a KKE/D motif at the carboxy terminus. Interestingly, whereas depletion of NOP58 resulted in destabilization of snoRNAs (25), depletion of NOP56 did not affect snoRNA levels (26). To confirm that we had identified the homologue of NOP58, the expression of the putative homologue was down-regulated by RNAi. To silence the gene, T7 opposing promoters were used to produce the gene-specific double-stranded RNA (dsRNA) under tetracycline induction (61). Parasites stopped growing 3 days after dsRNA induction, suggesting the essential nature of this gene. We next examined whether silencing affects the levels of snoRNAs. RNA was extracted from cells before and 3 days after induction and was subjected to primer extension to determine the levels of snoRNAs. The results, presented in Fig. 8A, indicate specific reductions of all C/D snoRNAs by 50 to 90% in the silenced cells. These reductions are specific, since no effect on the levels of snRNA or H/ACA molecules was observed. Of special interest are two C/D snoRNAs, TB11Cs2C1 and TB11Cs2C2, which had been implicated as guide RNAs that methylate rRNA by nonconventional methylation rules (46, 47). Later it was found that the Nms predicted to be guided by these RNAs are guided by other snoRNAs based on the conventional guiding rules (9, 10). Thus, the functions of these two RNAs remained puzzling. Primer extension suggests that these two RNAs are very abundant, much more abundant than any C/D involved in methylation, suggesting that they may function in rRNA processing. The clear effect of NOP58 silencing on the levels of these two RNAs suggests that they are indeed members of the C/D snoRNA class. Interestingly, NOP58 silencing had no effect on the level of SL RNA, suggesting that, in contrast to the H/ACA pathway (4), the C/D pathway is not essential for trans-splicing. NOP58-silenced cells served as a tool to identify novel C/D snoRNAs. Extensive bioinformatic searches were employed to identify the snoRNAs in the genome that could methylate the positions that were mapped (see Fig. S-9 in the supplemental material). Surprisingly, only three novel C/D snoRNAs, which had not been identified previously (35), were found (Fig. 8B). Two of the novel C/D snoRNAs (TB10Cs2′C1 and TB7Cs1C1) are encoded by a single-copy-number snoRNA gene. Their sequences, genomic locations, and potential base pairing with their target sites are presented in Fig. S-10 in the supplemental material and in Fig. 8Ba and b. To verify that these RNAs are indeed C/D snoRNAs, their levels in NOP58-silenced cells were examined; as expected, the levels of these RNAs were reduced upon silencing (Fig. 8Bc).
FIG. 8.
Effects of NOP58 knockdown. (A) Effects of NOP58 silencing on the levels of C/D snoRNAs and snRNAs. Total RNA was prepared from cells carrying the NOP58 RNAi construct without induction (−) or after 3 days of induction (+). Total RNA (10 μg) was subjected to primer extension with radiolabeled oligonucleotides complementary to the indicated snoRNA and to SL and U snRNAs. The oligonucleotides specific to these RNAs are listed in Fig. S-7 in the supplemental material. cDNA was separated on a 6% sequencing gel. To control for the level of RNA in each sample, primer extension was performed with primer TBsno-H-1, specific to TB9Cs2H1 H/ACA snoRNA. (B) Identification of novel C/D snoRNAs. (a) Schematic representations of the clusters encoding the snoRNAs. The positions of the snoRNAs in the genome are given on the left. The sizes of the snoRNAs and the intergenic regions (in nucleotides) are given below each diagram. The number of times the gene cluster (delineated by parentheses) is repeated is given on the right. (b) Potential for base pair interaction between the rRNA region and the snoRNA. The position of the modified Nm is indicated and underlined. (c) Level of the C/D snoRNA upon NOP58 silencing. Total RNA (20 μg) was prepared from cells carrying the NOP58 silencing construct without induction (−Tet) or after 3 days of induction (+Tet). The RNA was separated on a 10% polyacrylamide denaturing gel and electroblotted onto a nylon membrane. The blot was hybridized with oligonucleotides complementary to the designated snoRNAs. To control for the level of RNA in each sample, the blot was hybridized with an oligonucleotide complementary to the SL RNA.
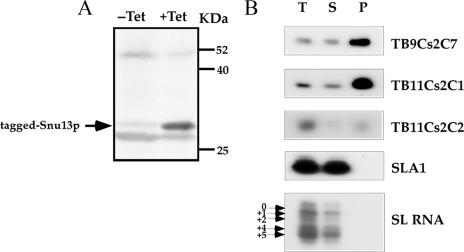
Another experimental approach to demonstrate that an RNA belongs to a certain group of RNPs is affinity selection of the RNA using antibodies to the cognate RNP protein. To be able to perform such experiments, we identified the homologue of SNU13 (another C/D snoRNP protein) in the T. brucei genome and TAP tagged it. A sequence alignment of the protein with its homologues is presented in Fig. S-5 in the supplemental material. The T. brucei SNU13 homologue shares 64% identity and 73% similarity with the S. cerevisiae protein. A cell line expressing the TAP-tagged version of the protein was prepared as described in Materials and Methods. The expression of the tagged protein was examined by Western blot analysis (Fig. 9A), and a fusion product of 34.5 kDa was observed. Extracts were prepared from the cell line and were subjected to affinity selection using IgG-coated beads. The RNA bound to the beads was identified by primer extension. The results, presented in Fig. 9B, indicate specific selection of 10 to 15% of the abundant C/D snoRNAs, TB11Cs2C1 and TB11Cs2C2, on the beads. The selection was specific, since the H/ACA SLA1 and SL RNAs were not selected in the experiment.
FIG. 9.
Affinity selection of C/D snoRNAs using TAP-tagged Snu13p protein. (A) Western blot analysis. Whole-cell extracts were prepared from uninduced (−Tet) or induced (+Tet) cells (50 ml; 107 cells/ml) carrying the Snu13-TAP construct. The extracts were fractioned on a 12% SDS-polyacrylamide gel and subjected to Western blot analysis with IgG antibodies. Arrow indicates tagged Snu13p. (B) Specific affinity selection of C/D snoRNAs with IgG beads. Whole-cell extracts were prepared from induced cells (500 ml; 107 cells/ml) carrying the Snu13-TAP construct as described in Materials and Methods. The extracts were subjected to affinity selection using IgG-Sepharose beads. RNAs from total-cell extracts (1/30), supernatants (1/30), and all of the selected RNA (lanes T, S, and P, respectively) were subjected to primer extension analysis with radiolabeled oligonucleotides complementary to the designated RNA. The RNA was separated on a 6% denaturing gel. The position of the Cap-4 nt on the SL RNA (the four hypermodified nt at the 5′ end of the SL RNA) is indicated.
DISCUSSION
This study demonstrates the role of the C/D pathway in rRNA processing and highlights the importance of Nm for the developmental regulation of the parasite life cycle. The factors affecting the differential levels of snoRNAs were explored experimentally and evaluated by a machine-learning algorithm, and prediction of snoRNA abundance was achieved. rRNA-processing defects were studied in cells depleted of C/D pathway proteins, and our results support the existence of C/D snoRNAs that function in trypanosome-specific processing to produce the srRNA fragments. MRP RNA, a ubiquitous processing snoRNA that was not previously detected in trypanosomes, was identified in this study as part of a cluster encoding snoRNAs. This is the first study to demonstrate that Nm modification and expression of snoRNA are developmentally regulated and may assist the parasite in coping with the temperature changes encountered during cycling between its insect and mammalian hosts.
rRNA processing is severely affected in the absence of functional C/D snoRNP.
One of the most intriguing outcomes of this study is the finding that specific rRNA-processing defects are elicited by depletion of the C/D pathway (Fig. 4). Our data suggest that C/D snoRNAs are expected to process srRNA-1, -6, and -4. The rest of the cleavages might be generated either by H/ACA RNAs or by RNase III endonucleases. Examination of the rRNA defects generated in CBF5-silenced cells indicates a specific defect in generating srRNA-2, suggesting that this cleavage is mediated by a trypanosome-specific H/ACA RNA (our unpublished data). This H/ACA RNA, as well the C/D snoRNAs predicted to exist based on this study, has yet to be discovered. Two abundant snoRNAs that were suggested to guide rRNA by nonconventional rules, TB11Cs2C1 (76 nt) and TB11Cs2C2 (92 nt), are potential candidates for the cleavage of at least some of the sites predicted above. Our preliminary results, based on bioinformatic predictions and snoRNAi inhibition of these RNAs, suggest that TB11Cs2C1 is most probably involved in 18S rRNA processing. On the other hand, TB11Cs2C2 is predicted to function in the processing of srRNA-6 (A. Hury, Y. Ziporen, and S. Michaeli, unpublished data).
Differential levels of snoRNAs are a consequence of multiple factors.
Our results demonstrate that different C/D snoRNA molecules have different abundances and that not all Nm sites are equally modified in the two life stages of the parasite.
The machine-learning algorithm was used to examine parameters that might enable us to predict the levels of snoRNAs, which seem to vary considerably. We recently made a similar attempt to understand this phenomenon using 20 Leishmania major snoRNAs (31). Transcriptional regulation cannot explain the major differences that exist among levels of snoRNAs encoded by the same cluster. Two major factors may influence the levels of snoRNAs: the extent of processing and the strength with which these snoRNAs bind their cognate binding proteins. The gene copy number, structural features of the snoRNA such as the conservation of the boxes (especially C, since D is highly conserved in all snoRNA species), the internal K-turn in the molecule, and the ΔG of the snoRNA-target interaction can all affect snoRNA levels. Thus, snoRNA abundance may be governed by combinatorial effects of all these factors. To test this hypothesis in a quantitative manner, we employed the machine-learning algorithm to predict the C/D snoRNA levels. Four factors were found to be important: the conservation of the C box, the number of modifications each molecule can guide, the secondary structure of the domain at which the modification is situated, and the level of conservation of Nm modification among eukaryotes. Although the last two factors cannot be rationalized as directly affecting the level of snoRNA, they do differ among different snoRNAs. The conservation level may be an indicator of the importance of the modification and thus may invoke additional mechanisms, yet to be characterized, to enhance modifications at these sites. A model based on these parameters can successfully predict the level of cellular snoRNA in 85% of cases. As we mentioned above, additional elements such as transcriptional regulation could exist for snoRNA genes (34) and might explain the 15% of predictions that were inaccurate. We tried to explore whether additional parameters such as the K-turn and the strength of the extragenic elements flanking the snoRNA gene could also contribute to snoRNA levels. Interestingly, we found no significant correlation between the level of snoRNA and the ΔG of the extragenic elements, such as that recently described for L. major (31). Indeed, the extragenic stems of T. brucei snoRNAs are much shorter than those found for L. major.
snoRNA expression and Nm modification are developmentally regulated; existence of additional C/D snoRNAs in the trypanosome genome.
The degree of modification seems to be developmentally regulated, but why are different sites modified differently? The developmentally regulated Nms belong to different groups; a few are conserved among eukaryotes, but trypanosome-specific modifications also exist. As mentioned above, our survey for developmentally regulated Nm was limited to only 21% of the rRNA. The fact that we did not yet reveal any stage-specific modifications does not prove that no such modifications exist. A better way to identify such modifications may be to find snoRNAs whose expression is developmentally regulated. Very little is known about regulation of snoRNA expression. Most relevant to our experiments is a recent study that followed the expression of noncoding RNAs during C. elegans development. Both C/D and H/ACA RNAs were found to be poorly expressed at the egg-embryo stage, suggesting that snoRNA expression is developmentally regulated. In addition, the snoRNA level is elevated under stress, including heat shock and starvation (18). Transcriptional regulation can account for the differential expression of snoRNAs in nematodes, which are also intronic and are transcribed together with their host genes. Interestingly, however, the noncoding RNAs embedded within host genes do not always follow the pattern of the host gene expression, suggesting that RNA motifs of noncoding RNA may have a role in transcriptional activation.
We know very little about the regulation of snoRNA levels in trypanosomes. Although it is commonly believed that in trypanosomes, transcription is not the primary stage at which gene expression is regulated, it is possible that snoRNA genes are regulated by extragenic elements at the transcriptional level, as we reported for L. collosoma (34). It is also possible that transcription from these elements is activated under stress or is developmentally regulated, leading to high expression of snoRNA, as was observed for TB5Cs1C1 (Fig. 7b). It will be of great interest to perform microarray analysis on the different snoRNAs and to examine how many are developmentally regulated. Only when such snoRNAs are identified will it be possible to examine whether they guide trypanosome-specific or evolutionarily conserved modifications.
The observation that the level of Nm is increased in the bloodstream form supports our hypothesis that the high number of modifications and their level might assist the parasite in coping with the elevated temperature in the mammalian host.
The repertoire of C/D snoRNAs in trypanosomes.
Based on our previous publication (35) and the mapping data presented in this study, there should be at least 131 Nms on T. brucei rRNA. Earlier studies with Crithidia fasciculata suggest the presence of at least 100 Nms on rRNA (13), and recent results for Euglena gracilis, which is closely related to trypanosomes, suggest the presence of at least 200 Nms on its rRNA (48). However, our studies identified, to date, only 57 C/D snoRNAs (31, 35) that can potentially guide 84 Nms. Our searches to find additional snoRNAs that can guide most of these additional Nms failed. Such snoRNAs are not encoded by repeated genes, since all the repeated noncoding genes were examined by us and all are described in reference 35. Although we cannot exclude the possibility that some sequence information of the T. brucei genome is incomplete, other options may also explain the “missing” snoRNAs. For instance, there may be C/D-type RNAs whose boxes deviate strongly from the consensus but that still bind fibrillarin. Another option is the existence of guide RNAs that modify these Nms under different guiding rules. Only extensive genome comparative analysis to search for noncoding RNAs, as well as preparation of noncoding-RNA libraries from trypanosomes, which also include nonabundant RNAs, is expected to solve the mystery of these missing snoRNAs.
In summary, this study highlights the role of the C/D snoRNAs in conducting trypanosome-specific rRNA cleavage events and emphasizes the role of Nm modifications as a possible mechanism of adaptation to the temperature shift experienced by the parasite in cycling between the insect and mammalian hosts. The identification of the trypanosome MRP RNA increases our understanding of the repertoire of noncoding RNAs in this organism. The fact that this RNA was previously missed by bioinformatic tools that were suitable for identifying its homologues in other organisms further emphasizes the unique properties of small RNAs in this exotic parasite.
Supplementary Material
Acknowledgments
This research was supported by a grant from the Israel Science Foundation, a grant from the Israeli Ministry of Science and Technology to S.M and R.U., and an International Research Scholar's Grant from the Howard Hughes Foundation to S.M. S.M. holds the David and Inez Myers Chair in RNA silencing of diseases.
Footnotes
Published ahead of print on 2 November 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bachellerie, J. P., and J. Cavaille. 1997. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 22257-261. [DOI] [PubMed] [Google Scholar]
- 2.Bachellerie, J. P., J. Cavaille, and A. Hüttenhofer. 2002. The expanding snoRNA world. Biochimie 84775-790. [DOI] [PubMed] [Google Scholar]
- 3.Bakin, A. V., and J. Ofengand. 1998. Mapping of pseudouridine residues in RNA to nucleotide resolution. Methods Mol. Biol. 77297-309. [DOI] [PubMed] [Google Scholar]
- 4.Barth, S., A. Hury, X. H. Liang, and S. Michaeli. 2005. Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J. Biol. Chem. 28034558-34568. [DOI] [PubMed] [Google Scholar]
- 5.Cahill, N. M., K. Friend, W. Speckmann, Z. H. Li, R. M. Terns, M. P. Terns, and J. A. Steitz. 2002. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 213816-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, D. A., K. Kubo, C. G. Clark, and J. C. Boothroyd. 1987. Precise identification of cleavage sites involved in the unusual processing of trypanosome ribosomal RNA. J. Mol. Biol. 196113-124. [DOI] [PubMed] [Google Scholar]
- 7.Chu, S., R. H. Archer, J. M. Zengel, and L. Lindahl. 1994. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl. Acad. Sci. USA 91659-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decatur, W. A., and M. J. Fournier. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278695-698. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar, D. A., A. A. Chen, S. Wormsley, and S. J. Baserga. 2000. The genes for small nucleolar RNAs in Trypanosoma brucei are organized in clusters and are transcribed as a polycistronic RNA. Nucleic Acids Res. 282855-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunbar, D. A., S. Wormsley, T. M. Lowe, and S. J. Baserga. 2000. Fibrillarin-associated box C/D small nucleolar RNAs in Trypanosoma brucei. Sequence conservation and implications for 2′-O-ribose methylation of rRNA. J. Biol. Chem. 27514767-14776. [DOI] [PubMed] [Google Scholar]
- 11.Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14319-327. [DOI] [PubMed] [Google Scholar]
- 12.Ganot, P., M. L. Bortolin, and T. Kiss. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89799-809. [DOI] [PubMed] [Google Scholar]
- 13.Gray, M. W. 1979. The ribosomal RNA of the trypanosomatid protozoan Crithidia fasciculata: physical characteristics and methylated sequences. Can. J. Biochem. 57914-926. [DOI] [PubMed] [Google Scholar]
- 14.Hartshorne, T. 1998. Distinct regions of U3 snoRNA interact at two sites within the 5′ external transcribed spacer of pre-rRNAs in Trypanosoma brucei cells. Nucleic Acids Res. 262541-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartshorne, T., and N. Agabian. 1993. RNA B is the major nucleolar trimethylguanosine-capped small nuclear RNA associated with fibrillarin and pre-rRNAs in Trypanosoma brucei. Mol. Cell. Biol. 13144-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartshorne, T., and W. Toyofuku. 1999. Two 5′-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei. Nucleic Acids Res. 273300-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartshorne, T., W. Toyofuku, and J. Hollenbaugh. 2001. Trypanosoma brucei 5′ETS A′-cleavage is directed by 3′-adjacent sequences, but not two U3 snoRNA-binding elements, which are all required for subsequent pre-small subunit rRNA processing events. J. Mol. Biol. 313733-749. [DOI] [PubMed] [Google Scholar]
- 18.He, H., L. Cai, G. Skogerbo, W. Deng, T. Liu, X. Zhu, Y. Wang, D. Jia, Z. Zhang, Y. Tao, H. Zeng, M. N. Aftab, Y. Cui, G. Liu, and R. Chen. 2006. Profiling Caenorhabditis elegans non-coding RNA expression with a combined microarray. Nucleic Acids Res. 342976-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen, R. P., E. C. Hurt, H. Kern, H. Lehtonen, M. Carmo-Fonseca, B. Lapeyre, and D. Tollervey. 1991. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol. 113715-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen, B. C., D. L. Brekken, A. C. Randall, C. T. Kifer, and M. Parsons. 2005. Species specificity in ribosome biogenesis: a nonconserved phosphoprotein is required for formation of the large ribosomal subunit in Trypanosoma brucei. Eukaryot. Cell 430-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joachims, T. 1999. Making large-scale SVM learning practical, p. 169-184. In B. Scholkopf, C. Burges, and A. Smola (ed.), Advances in kernel methods: support vector learning. MIT Press, Cambridge, MA.
- 22.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109145-148. [DOI] [PubMed] [Google Scholar]
- 23.Kiss, T. 2004. Biogenesis of small nuclear RNPs. J. Cell Sci. 1175949-5951. [DOI] [PubMed] [Google Scholar]
- 24.Kiss-László, Z., Y. Henry, J. P. Bachellerie, M. Caizergues-Ferrer, and T. Kiss. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 851077-1088. [DOI] [PubMed] [Google Scholar]
- 25.Lafontaine, D. L., and D. Tollervey. 1999. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 5455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lafontaine, D. L., and D. Tollervey. 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 202650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitan, A., Y. Xu, C. Ben Dov, H. Ben Shlomo, Y. Zhang, and S. Michaeli. 1998. Characterization of a novel trypanosomatid small nucleolar RNA. Nucleic Acids Res. 261775-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, H. D., J. Zagorski, and M. J. Fournier. 1990. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 101145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, X., D. N. Frank, N. Pace, J. M. Zengel, and L. Lindahl. 2002. Phylogenetic analysis of the structure of RNase MRP RNA in yeasts. RNA 8740-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang, W. Q., and M. J. Fournier. 1995. U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev. 92433-2443. [DOI] [PubMed] [Google Scholar]
- 31.Liang, X. H., A. Hury, E. Hoze, S. Uliel, I. Myslyuk, A. Apatoff, R. Unger, and S. Michaeli. 2007. Genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Leishmania major indicates conservation among trypanosomatids in the repertoire and in their rRNA targets. Eukaryot. Cell 6361-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang, X. H., L. Liu, and S. Michaeli. 2001. Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J. Biol. Chem. 27640313-40318. [DOI] [PubMed] [Google Scholar]
- 33.Liang, X. H., Q. Liu, L. Liu, C. Tschudi, and S. Michaeli. 2006. Analysis of spliceosomal complexes in Trypanosoma brucei and silencing of two splicing factors Prp31 and Prp43. Mol. Biochem. Parasitol. 14529-39. [DOI] [PubMed] [Google Scholar]
- 34.Liang, X. H., A. Ochaion, Y. X. Xu, Q. Liu, and S. Michaeli. 2004. Small nucleolar RNA clusters in trypanosomatid Leptomonas collosoma. Genome organization, expression studies, and the potential role of sequences present upstream from the first repeated cluster. J. Biol. Chem. 2795100-5109. [DOI] [PubMed] [Google Scholar]
- 35.Liang, X. H., S. Uliel, A. Hury, S. Barth, T. Doniger, R. Unger, and S. Michaeli. 2005. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA 11619-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang, X. H., Y. X. Xu, and S. Michaeli. 2002. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA 8237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maden, B. E. 2001. Mapping 2′-O-methyl groups in ribosomal RNA. Methods 25374-382. [DOI] [PubMed] [Google Scholar]
- 38.Mandelboim, M., S. Barth, M. Biton, X. H. Liang, and S. Michaeli. 2003. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J. Biol. Chem. 27851469-51478. [DOI] [PubMed] [Google Scholar]
- 39.Morrissey, J. P., and D. Tollervey. 1993. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 132469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakanishi, K., and O. Nureki. 2005. Recent progress of structural biology of tRNA processing and modification. Mol. Cells 19157-166. [PubMed] [Google Scholar]
- 41.Newton, K., E. Petfalski, D. Tollervey, and J. F. Caceres. 2003. Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Mol. Cell. Biol. 238519-8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ni, J., A. L. Tien, and M. J. Fournier. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89565-573. [DOI] [PubMed] [Google Scholar]
- 43.Omer, A. D., S. Ziesche, H. Ebhardt, and P. P. Dennis. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA 995289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piccinelli, P., M. A. Rosenblad, and T. Samuelsson. 2005. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 334485-4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rashid, R., M. Aittaleb, Q. Chen, K. Spiegel, B. Demeler, and H. Li. 2003. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J. Mol. Biol. 333295-306. [DOI] [PubMed] [Google Scholar]
- 46.Roberts, T. G., J. M. Dungan, K. P. Watkins, and N. Agabian. 1996. The SLA RNA gene of Trypanosoma brucei is organized in a tandem array which encodes several small RNAs. Mol. Biochem. Parasitol. 83163-174. [DOI] [PubMed] [Google Scholar]
- 47.Roberts, T. G., N. R. Sturm, B. K. Yee, M. C. Yu, T. Hartshorne, N. Agabian, and D. A. Campbell. 1998. Three small nucleolar RNAs identified from the spliced leader-associated RNA locus in kinetoplastid protozoans. Mol. Cell. Biol. 184409-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell, A. G., M. N. Schnare, and M. W. Gray. 2006. A large collection of compact box C/D snoRNAs and their isoforms in Euglena gracilis: structural, functional and evolutionary insights. J. Mol. Biol. 3571548-1565. [DOI] [PubMed] [Google Scholar]
- 49.Schmitt, M. E., and D. A. Clayton. 1993. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 137935-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnaufer, A., N. L. Ernst, S. S. Palazzo, J. O'Rear, R. Salavati, and K. Stuart. 2003. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell 12307-319. [DOI] [PubMed] [Google Scholar]
- 51.Schultz, A., S. Nottrott, N. J. Watkins, and R. Lührmann. 2006. Protein-protein and protein-RNA contacts both contribute to the 15.5K-mediated assembly of the U4/U6 snRNP and the box C/D snoRNPs. Mol. Cell. Biol. 265146-5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szewczak, L. B., S. J. Degregorio, S. A. Strobel, and J. A. Steitz. 2002. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 91095-1107. [DOI] [PubMed] [Google Scholar]
- 53.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9337-342. [DOI] [PubMed] [Google Scholar]
- 54.Tollervey, D., H. Lehtonen, M. Carmo-Fonseca, and E. C. Hurt. 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 10573-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tollervey, D., H. Lehtonen, R. Jansen, H. Kern, and E. C. Hurt. 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72443-457. [DOI] [PubMed] [Google Scholar]
- 56.Tran, E. J., X. Zhang, and E. S. Maxwell. 2003. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ ’ RNPs. EMBO J. 223930-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uliel, S., X. H. Liang, R. Unger, and S. Michaeli. 2004. Small nucleolar RNAs that guide modification in trypanosomatids: repertoire, targets, genome organisation, and unique functions. Int. J. Parasitol. 34445-454. [DOI] [PubMed] [Google Scholar]
- 58.Vapnik, V. N. 1995. The nature of statistical learning theory. Springer, New York, NY.
- 59.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33261-311. [DOI] [PubMed] [Google Scholar]
- 60.Walker, S. C., and J. M. Avis. 2005. Secondary structure probing of the human RNase MRP RNA reveals the potential for MRP RNA subsets. Biochem. Biophys. Res. Commun. 335314-321. [DOI] [PubMed] [Google Scholar]
- 61.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 27540174-40179. [DOI] [PubMed] [Google Scholar]
- 62.Watkins, N. J., A. Dickmanns, and R. Lührmann. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 228342-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinstein, L. B., and J. A. Steitz. 1999. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 11378-384. [DOI] [PubMed] [Google Scholar]
- 64.White, T. C., G. Rudenko, and P. Borst. 1986. Three small RNAs within the 10 kb trypanosome rRNA transcription unit are analogous to domain VII of other eukaryotic 28S rRNAs. Nucleic Acids Res. 149471-9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, P., J. S. Brockenbrough, A. C. Metcalfe, S. Chen, and J. P. Aris. 1998. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J. Biol. Chem. 27316453-16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao, S., F. Scott, C. A. Fierke, and D. R. Engelke. 2002. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 71165-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu, Y., L. Liu, C. Lopez-Estraño, and S. Michaeli. 2001. Expression studies on clustered trypanosomatid box C/D small nucleolar RNAs. J. Biol. Chem. 27614289-14298. [DOI] [PubMed] [Google Scholar]
- 68.Yang, Z. R. 2004. Biological applications of support vector machines. Brief. Bioinform. 5328-338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.