Abstract
The fungal colony is a complex multicellular unit consisting of various cell types and functions. Asexual spore formation (conidiation) is integrated through sensory and regulatory elements into the general morphogenetic plan, in which the activation of the transcription factor BrlA is the first determining step. A number of early regulatory elements acting upstream of BrlA (fluG and flbA-E) have been identified, but their functional relations remain to be further investigated. In this report we describe FlbB as a putative basic-zipper-type transcription factor restricted to filamentous fungi. FlbB accumulates at the hyphal apex during early vegetative growth but is later found in apical nuclei, suggesting that an activating modification triggers nuclear import. Moreover, proper temporal and quantitative expression of FlbB is a prerequisite for brlA transcription, and misscheduled overexpression inhibits conidiation. We also present evidence that FlbB activation results in the production of a second diffusible signal, acting downstream from the FluG factor, to induce conidiation.
The mycelium is a very successful colony form capable of displaying different cell types and functions, and this attribute has led to the realization that it is governed by a complex chemosensitive system (43). The production of asexual spores is an integral part of the mycelial colony plan and the principal means of dispersal among the filamentous fungi (23, 55). Aspergillus nidulans is a homothallic ascomycete which has been used as a model system for molecular genetic studies due to its versatile life cycle, its amenability to genetic manipulation, and the open availability of its genome sequence (13, 24). It is also a reference for basic studies in fungal morphogenesis (12).
After the germination of a spore, an initial period of vegetative growth ensues, consisting of apical extension of the hyphae. Any of a number of suitable environmental stimuli (emergence to the atmosphere, osmotic and nutrient stress) provokes the halting of hyphal extension and subsequent formation of asexual spore-bearing structures called conidiophores (2, 6). Molecular analysis of this process has revealed that stimuli are sensed and transduced as intracellular signals that converge in the transcriptional activation of brlA, which encodes a C2H2 zinc finger transcription factor. This first common step also marks the start of an irreversible central developmental cascade of downstream regulators leading to sporogenesis (1, 10).
Among the various possible upstream regulatory branches responding to the above-mentioned stimuli, the principal one, involving air exposure, starts with the biosynthesis of an unidentified low-molecular-weight diffusible FluG factor (21). The FluG signal is proposed to inactivate a repressor of conidiation (SfgA) that negatively regulates a number of development-specific functions required to activate brlA (35). It has been proposed that such FluG-mediated derepression leads to the activation of other Flb (fluffy low brlA expression) components: FlbA, a protein which inhibits vegetative growth signaling mediated by Gα (FadA) (22, 51, 54), and potential transcription factors (TFs) FlbD, FlbB and FlbC (48, 2, 49). Simultaneous inhibition of vegetative signaling and activation of Flb TFs are required for the conidiation program to progress. However, detailed functional characterization of FlbB and FlbC remains to be done.
In this report, we present the identification and characterization of flbB (first described as vegA [4]) and demonstrate that FlbB is a potential transcriptional activator with a basic leucine zipper (bZIP) (44) and other conserved domains. The deletion of flbB results in the blockage of the synthesis of an extracellular signaling compound required for conidiation. The expression pattern of flbB shows elevated mRNA steady-state levels during the early phases of vegetative growth, which fall with the initiation of asexual development and remain low or undetectable during sexual development. FlbB initially localizes at the hyphal tip and then relocates to the nuclei proximal to the cell apex during vegetative growth. The complex regulatory roles played by FlbB in brlA transcriptional activation and developmental progression are further discussed.
MATERIALS AND METHODS
Strains, oligonucleotides, media, and culture conditions.
The strains of A. nidulans employed in this study are listed in Table 1. Plasmids were amplified in Escherichia coli strain DH5α or DH1 grown in Luria-Bertani medium with ampicillin (75 μg ml−1). Purification was carried out using GenElute plasmid miniprep or maxiprep kits (Sigma). Oligonucleotides used in this study are listed in Table 2.
TABLE 1.
Aspergillus nidulans strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| FGSCA4 | Wild type (veA+) | 31 |
| FGSC26 | biA1 veA1 | 17 |
| FGSCA68 | suA1adE20 yA2 adE20 acrA1 phenA2 pyroA4 lysB5 sB3 nicB8 riboB2 veA1 | 17 |
| FGSCA283 | suA1adE20 yA2 adE20 acrA1 galA1 pyroA4 (ssb+) facA303 sB3 nicB8 riboB2 veA1 | Fungal Genetics Stock Center |
| FGSC33 | biA1 pyroA4 | Fungal Genetics Stock Center |
| FGSC773 | pyrG89 wA3 pyroA4 | Fungal Genetics Stock Center |
| TTA127.4 | pabaA1 yA2 ΔfluG::trpC veA1 | 21 |
| RMS011 | pabaA1 yA2 ΔargB::trpCΔB trpC801 veA1 | 39 |
| GR5 | pyrG89 wA3 pyroA4 veA1 | 46 |
| MAD782 | pyrG89 pabaA1 biA1 yA2 veA1 | Eduardo Espeso (CIB-CSIC, Madrid, Spain) |
| TN02A3 | pyrG89 ΔnkuA::argB argB2 pyroA4 veA1 | 27 |
| TN02A21 | ΔnkuA::argB argB2 pyroA4 riboB2 veA1 | 27 |
| TN02A25 | pyrG89 pabaB22 ΔnkuA::argB argB2 riboB2 veA1 | 27 |
| BD14 (flbB100f) | biA1 flbB100 veA1 | This study |
| BD12 (flbB101f) | biA1 flbB101 veA1 | This study |
| BD11 (flbB102f) | biA1 flbB102 veA1 | This study |
| BD109 | pyrG89 pabaA1 yA2 argB2 flbB101 veA1 | This study |
| BD143 | pyrG89 ΔnkuA::argB argB2 ΔflbB::pyrG pyroA4 veA1 | This study |
| BD164 | pyrG89 pabaA1 argB2 ΔflbB::pyrG pyroA4 | This study |
| BD167 | pyrG89 ΔnkuA::argB argB2 flbB::gfp/pyrG pyroA4 veA1 | This study |
| BD183 | pyrG89 ΔnkuA::argB argB2 gfp::flbB pyroA4 veA1 | This study |
| TNI16.2 | biA1 alcA(p)::flbB::pyroA+ | This study |
| RNI24.5 | biA1 alcA(p)::flbB::pyroA+ | This study |
| TNI22.1 | biA1 pJW53::pyroA+ | This study |
| TJW113 | biA1 methG1 ΔflbB::argB+ | 18 |
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′) | Target or purpose |
|---|---|---|
| pRG3up | GAA TTC GAG CTC GGT ACC | Sequencing pRG3 insert flanking regions |
| pRG3 down | AAG CTT GCA TGC GCG GCC | Sequencing pRG3 insert flanking regions |
| AN7542-1 | GGT GAG GGT AGA AAG GG | flbB sequencing |
| AN7542-2 | GAC AGG GTG ATT CCA GC | flbB sequencing |
| AN7542-3 | GGG TTT TTC TGA CGT CG | flbB sequencing |
| flbB-GSP1 | CCT CTT GTG GAT GCA CTC TGG AAT GTG GGC | Obtaining flbB::gfp/pyrG |
| flbB-GSP1* | CGT TGA ACG ATT GTC GCA CAG TCG C | Obtaining flbB::gfp/pyrG |
| flbB-GSP2 | TGA ATA CAT CGT CTC ATC AGC ATG CCG GGT | Obtaining flbB::gfp/pyrG |
| flbB-GSP3 | TGA CCT GAC AGC TCG CTT TTT TTC TGA GCT TTC TAA TGC | Obtaining flbB::gfp/pyrG and ΔflbB |
| flbB-GSP4 | GAA AGG TGC GTG GGT TCG AAT CCC ACC | Obtaining flbB::gfp/pyrG, gfp::flbB and ΔflbB |
| flbB-PP1 | GTT TTC TGG TCC TCG GTC AAC CGG TGG | Obtaining gfp::flbB and ΔflbB |
| flbB-PP2 | CAT GGT GGT CGA GCT GTG AAT AGC GGA GAA | Obtaining gfp::flbB and ΔflbB |
| flbB-GFP1 | TCC CGG CAT GCT GAT GAG ACG ATG TAT TCA GGA GCT GGT GCA GGC GCT GGA GCC | Obtaining flbB::gfp/pyrG |
| flbB-GFP2 | GCA TTA GAA AGC TCA GAA AAA AAG CGA GCT GTC AGG TCA GTC TGA GAG GAG GCA CTG ATG CG | Obtaining flbB::gfp/pyrG and ΔflbB |
| flbB-SMP1 | TTC TCC GCT ATT CAC AGC TCG ACC ACC ATG ACC GGT CGC CTC AAA CAA TGC TCT | Obtaining ΔflbB |
| flbB-gfpSP | TTC TCC GCT ATT CAC AGC TCG ACC ACC ATG AGT AAA GGA GAA GAA CTT TTCACT GGA GTT | Obtaining gfp::flbB |
| flbB-gfpFP | GTT CAG ATC CAA GGG TAT TGG CCT ACT ACT GAT CGA AGT CAT TTT GTA TAG TTC ATC CAT GCC ATG TGT | Obtaining gfp::flbB |
| flbB-geneSP | ATG ACT TCG ATC AGT AGT AGG CCA ATA CCC TTG GAT CTG AAC | Obtaining gfp::flbB |
| OJA141 | CCG TTC TGC TTA GGG TA | 5′ end of alcA(p) |
| OJA106 | TTT GAG GCG AGG TGA TAG GAT TGG A | 3′ end of alcA(p) |
| OJA108 | CG GGA TCC AGT GGT TCG GTA ATC | alcA(p) 5′ nested with BamHI tail |
| OMN233 | TCC AAT CCT ATC ACC TCG CCT CAA A ATG ACT TCG ATC AGT AGG C | flbB overexpression 5′ with complementary alcA(p) tail |
| OMN212 | GTT AAC TAG TGG CAG TGC AAC C | flbB overexpression 3′ amplification |
| OMN234 | CG GGATCC GCT CGA TAA GGA GTG AG | flbB overexpression construct 3′ nested with BamHI |
| OJA142 | CTG GCA GGT GAA CAA GTC | Forward primer for brlA probe |
| OJA143 | AGA AGT TAA CAC CGT AGA | Reverse primer for brlA probe |
| OMN213 | TGA TCT GTC CAT AGA ACA TCG C | Forward primer for flbB probe |
| OMN227 | CGA GCT GTC AGG TCA TGA ATA C | Reverse primer for flbB probe |
| ONK20 | ATA TGA ATT CAT GAC TTC GAT CAG TAG TAGG | FlbB forward for TF activity test |
| ONK21 | ATA TGT CGA CTC ATG AAT ACA TCG TCT CAT C | FlbB forward for TF activity test |
| ONK68 | ATA TGA ATT CAT GGG ATC GGG ATT CCA CTC CAG | FlbB-C forward for TF activity test |
| ONK69 | ATA TGT CGA CTC CCA TCT GTC GCT CGG CTC TG | FlbB-N forward for TF activity test |
The strains were cultivated in minimal medium (MMA [17]) or complete medium (MMA plus 5 g liter−1 yeast extract) that was adequately supplemented in case of auxotrophy. Nutrient depletion experiments in solid medium involved dilution of glucose or sodium nitrate to one-fifth of the original concentration. Salt stress experiments involved the addition of KCl (0.6 M) and MES (2-[N-morpholino]ethanesulfonic acid, 0.05 M). Experiments in liquid medium with nutrient limitation were conducted as described by Skromne et al. (37). Briefly, strains were cultivated for 18 h, at 37°C and 250 rpm, and then transferred to minimal medium with KCl and MES, with or without glucose or sodium nitrate. The morphology of the mycelium was examined after 10 and 20 h of culture.
Extracellular complementation experiments were conducted by point inoculation of two strains onto the solid medium at a distance of 2 cm in the same petri dish. After 3, 4, and 5 days of cultivation, the contact zone was examined and photographed under a binocular microscope.
Time course experiments for the induction of development were conducted essentially as described previously (3, 20). After 18 h of culture in liquid MMA as described above, mycelium was filtered onto nitrocellulose membranes (0.45 μm; MicronSep; GE Water and Process Technologies), placed on solid medium, and cultured for 0, 6, 12, 24, and 48 h before being processed for Northern blot analysis.
The intracellular localization of FlbB was analyzed by inoculating 40 μl of a conidiospore suspension onto a coverslip submerged in a petri dish containing adequately supplemented liquid minimal medium. After incubation at room temperature for 16, 24, and 36 h, samples were fixed with 4% (wt/vol) p-formaldehyde in phosphate-buffered saline for 16 h at 4°C, washed three times with phosphate-buffered saline, and stained with DAPI (4′,6′-diamidino-2-phenylindole) (essentially as described in references 7, 30, and 38). The coverslip was inverted onto a slide, and FlbB and nucleus localizations were observed by fluorescence microscopy.
Fluorescein staining experiments for confirmation of autolysis in liquid starvation cultures were performed as described by Roncal et al. (33).
Cloning of flbB.
The gene library contained in the self-replicating plasmid pRG3-AMA-NotI (29) was amplified using competent E. coli DH5α library efficiency cells (Invitrogen). Preparation and transformation of protoplasts were carried out as previously reported (42) with the following adaptations: after 20 to 24 h of culture at 37°C, in order to avoid conidiation induced by osmotic stress, the protoplast regeneration cultures (MMA plus 1 M sucrose) were overlaid with 7 ml of complete medium. The cultures were then incubated for 24 h. Conidiating colonies which emerged through the overlay were selected as positive transformants, and plasmids were isolated and amplified for sequencing. Inserts were amplified using oligonucleotides pRG3up and pRG3down. In order to localize and define the three mutations, genomic DNA was extracted from the three aconidial mutants and FGSC26. Each flbB open reading frame (ORF) was amplified using oligonucleotides 7542-1, 7542-2, and 7542-3 and sequenced.
Genetic techniques and extraction and manipulation of DNA.
Meiotic crosses were performed according to Pontecorvo et al. (32). DNA extractions were carried out from 24-h liquid-culture-grown mycelium that had been filtered, washed, and lyophilized. The extraction process involved a GenElute plant genomic DNA extraction kit (Sigma), with the addition of 50 units of RNase (Sigma). Southern blot analyses were carried out using a digoxigenin High Prime II DNA labeling and detection starter kit (Roche). The cDNA sequence of flbB was PCR amplified using specific primers and a cloned avian myeloblastosis virus first-strand cDNA synthesis kit (Invitrogen). A 24-h wild-type total RNA sample was used as the template.
RNA isolation and analysis.
A mycelium sample (100 mg [dry weight]) was frozen in liquid nitrogen, and 1 ml of TRIreagent (Fluka) was added. RNA extraction from these samples was performed according to the Invitrogen protocol for RNA extraction using the TRIzol reagent. RNA concentrations were calculated with a QuBit assay system (Invitrogen). Northern blot analysis of RNA was carried out with a digoxigenin Northern blot starter kit (Roche).
Fusion PCR.
The DNA constructions for the generation of null mutants as well as green fluorescent protein (GFP) tagging were conducted as described by Yang et al. (50), using the Triplemaster PCR system (Eppendorf). Null mutants were generated using a modified version of the protocol of Yang et al. (50), by constructing a DNA fragment of 5.1 kb with 5′and 3′ regions of flbB flanking the auxotrophic marker pyrG of Aspergillus fumigatus. GFP tagging at the C-terminal region of FlbB was done as described by Yang et al. (50). In the case of N-terminal tagging, the GFP coding sequence was added behind the start codon using the overlapping PCR technique, and no marker was included. In this latter case, selection of transformants was conducted using 2 mg/ml fluoroorotic acid in an flbB-null strain, selecting for gene replacement by the A. fumigatus pyrG gene, which replaced the flbB coding sequence.
Construction of alcA(p)::flbB strain.
The alcA(p)::flbB construct was created as described previously (53) and cloned into the BamHI site of pJW53 (J. W. Bok and N. P. Keller, unpublished data) to generate pNI41. pNI41 was transformed into FGSC33 to yield strain TNI16.2. TNI16.2 was crossed with FGSC773 to produce strain RNI24.5. pJW53 was introduced into FGSC33 to give rise to the control strain TNI22.1.
Yeast strains and plasmids for the transactivation assay.
The Saccharomyces cerevisiae reporter strain L40 [MATα his3-D200 trp1-901 leu2-3,112 ade2 LYS::(LexAop)4-HIS3 URA3::(LexAop)8-LacZ] was obtained from Invitrogen. L40 was grown in synthetic dropout minimal medium (36) with various supplements. The medium composition (per liter) is as follows: 20 g glucose, 6.7 g of yeast nitrogen base (MP Biomedicals), 50 ml of 20× dropout solution (300 mg l-isoleucine, 1,500 mg l-valine, 500 mg l-phenylalanine, 200 mg l-arginine, 300 mg l-tyrosine, 300 mg l-threonine, 300 mg l-lysine, 200 mg l-methionine, and 500 mg l-adenine hemisulfate salt) with or without 10 ml of 100× nutrient solution (10 g leucine, 2 g tryptophan, and 2 g histidine per liter). The plasmid pTLexA (11) (kindly provided by Suhn-Kee Chae at Paichai University, Daejeon, Korea) carrying the LexA DNA binding domain (8) was generated by the modification of pHybLex/Zeo (Invitrogen) by inserting the TRP1 marker (obtained from pGBT9; Clontech).
Construction and transformation for testing FlbB transactivation activity.
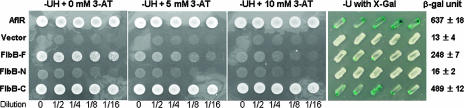
The cDNA-derived full-length FlbB (amino acids [aa] 1 to 426) and N-terminal (aa 1 to 172) and C-terminal (aa 166 to 426) regions of FlbB were PCR amplified using the primer pairs ONK20-ONK21, ONK20-ONK69, and ONK68-ONK21, respectively, from an A. nidulans cDNA library provided by Kwang-Yeop Jahng (Chonbuk University, Jeonju, Korea). Individual amplicons were digested with EcoRI and SalI and cloned into pTLexA, resulting in pNJ01, pNJ02, and pNJ03, respectively. Individual plasmids were then introduced into L40. Lithium acetate-polyethylene glycol-mediated yeast transformation (16) was carried out, and transformants were selected on synthetic dropout medium without tryptophan and uracil. To test FlbB transactivation activity, the transformants were inoculated on medium containing 3-amino-1,2,4-triazole (3-AT) or X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg/ml), and their growth and colony color were examined. To further confirm transactivation, more than five transformants of each plasmid were tested for β-galactosidase activity using a yeast β-galactosidase assay kit (Pierce). Results are expressed as means and standard deviations (see Fig. 7).
FIG. 7.
FlbB is a potential transcription factor. Yeast strains expressing the Gal4DBD::AflR (positive control) (28), pTLexA alone (negative control), LexADBD::FlbB-F (full length; 1 to 427 aa), LexADBD::FlbB-N (N-terminal region including the bZIP domain; aa 1 to 172) and LexADBD::FlbB-C (C-terminal region without the bZIP domain; aa 166 to 427) fusion proteins were spotted on the medium without uracil and histidine (−UH) in the presence of various concentrations (0, 5, and 10 mM) of 3-AT in serial dilutions. Plates were incubated at 30°C for 3 days. Yeast strains expressing LexADBD::FlbB-F and LexADBD::FlbB-C grew in the presence of 5 mM and 10 mM 3-AT, respectively, whereas those expressing LexADBD::FlbB-N or vector alone did not grow on the same medium. Five individual transformants for each construct were then tested for their β-galactosidase activities; mean values ± standard deviations are presented on the right. Rapid exhibition of blue color and high levels of β-galactosidase activity (489 ± 12) by the strains expressing LexADBD::FlbB-C confirms that the C-terminal region of FlbB contains a potential transactivation domain.
Light and fluorescence microscopy.
Liquid cultures were examined with a Nikon Optiphot microscope in bright-field or fluorescence mode. Images were recorded with a Nikon FX-35DX camera attached to the microscope. Direct examination of solid cultures was performed with a Nikon SMZ800 binocular microscope, attached to the above-mentioned camera.
GFP-labeled FlbB images were taken with an Eclipse E-600 Nikon microscope, equipped with a 60× Apo 1.4 numerical aperture oil immersion objective (Nikon), a 100-W mercury lamp, and B2-A (excitation, 495 nm; emission, 530 nm) and UV-2A (360 to 460 nm) filters (Nikon) for GFP and DAPI detection, respectively (7, 30, 38). Images were recorded with an ORCA-ER digital camera (Hamamatsu Photonics) coupled with Wasabi (Hamamatsu Photonics) and Metamorph (Universal Image) image analysis programs.
Sequence accession numbers.
Nucleotide and corresponding amino acid sequences for flbB were deposited in the EMBL database (http://www.ebi.ac.uk/embl/Submission/webin.html) under accession number AM494477.
RESULTS
Three aconidial mutants define distinct flbB mutant alleles.
Via chemical (nitrosoguanidine) or UV mutagenesis followed by visual screening of more than 64,000 survivors, we isolated three nonsporulating (fluffy) mutants. Conidiation of these mutant strains (BD11, BD12, and BD14) could be induced by growing them in contact with a conidiating strain. In turn, these mutant strains induced conidiation of fluffy nonconidial fluG mutant strains. The extracellular complementation of the fluG− strains, in which the corresponding diffusible FluG factor is absent, suggested that the mutations were not within the fluG gene.
Meiotic crosses and diploid analyses revealed that these three mutations were recessive and allelic and mapped to chromosome IV (results not shown). The recessive nature of the mutations allowed us to use an autoreplicative plasmid (pRG3/AMA/NotI)-based genomic DNA library (29). We constructed strain BD109 (Table 1), carrying the pyrG89 mutation from strain BD12, and used it for complementation-based gene cloning. All the plasmids isolated from conidiating (complemented) transformants contained inserts including the AN7542.3 gene (Broad Institute; http://www.broad.mit.edu/annotation/fungi/aspergillus).
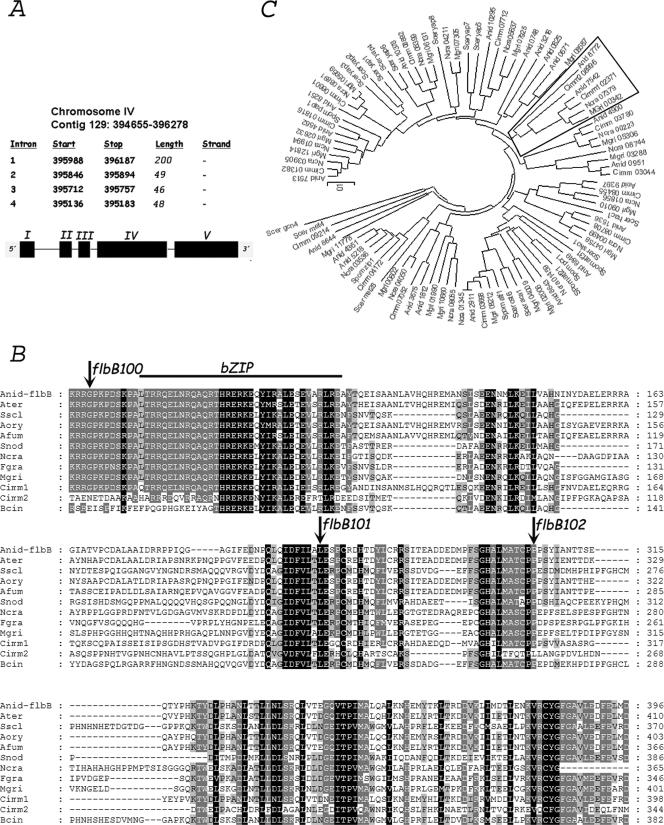
Subsequent genomic DNA sequence comparisons of the AN7542.3 region from the three mutants and a wild-type strain confirmed that the aconidial phenotype was caused by mutations in this gene (Table 3). The prediction for the coding region of AN7542.3 showed coincidence with preliminary cDNA data corresponding to flbB (= vegA), a gene implicated in the control of asexual conidiation, which had been partially characterized (4, 47). We amplified the cDNA of flbB, using specific primers, and confirmed previous data on gene structure (Fig. 1A). Accordingly, these mutant alleles are designated flbB100, flbB101, and flbB102. FlbB100 carries a single substitution, yielding G70R; flbB101 is predicted to have a mRNA splicing defect resulting in an FlbB101 protein truncated at amino acid 267 with a short (four-residue) out-of-frame tail. FlbB102 has the change P305L, prior to a frameshift adding 36 residues out of phase before a stop codon (Table 3 and Fig. 1B).
TABLE 3.
Mutations characterized in this study
| Allele | DNA change(s) | Mutant proteina | Change(s) in proteinb |
|---|---|---|---|
| flbB100 | G456A | aa 1-426 | G70R |
| flbB101 | G1143A | aa 1-267 + YVLR | L267 fs |
| flbB102 | C1255T, Δ1257 | aa 1-304 + L-35 | P305L, P306 fs |
YVLR and L-35 are residues added out of frame.
fs, frameshift followed by early termination.
FIG. 1.
General description of flbB. (A) Genome localization and general description of flbB. Black segments represent exons, and black lines designate introns. (B) Sequence alignment of FlbB homologues. Arrows indicate mutations which define changes in the amino acid sequence. Regions with low similarity are excluded. Genedoc software was used (version 2.6.003; www.psc.edu/biomed/genedoc). (C) Phylogenetic tree of bZIP domains identified in various fungal and yeast species. The bZIP branch including FlbB bZIP is framed. Abbreviations: Ater, A. terreus; Bcin, B. cinerea; Sscl, S. sclerotiorum; Aory, A. oryzae; Afum, A. fumigatus; Cimm, C. immitis; Snod, S. nodorum; Ncra, N. crassa; Fgra, F. graminearum; Mgri, M. grisea; Spom, S. pombe; Scer, S. cerevisiae. Phylogenetic and molecular evolutionary analyses were conducted using MEGA software, version 3.1 (neighbor-joining method, with a bootstrap of 50,000 replicates and amino p-distance substitution model).
flbB encodes a bZIP-type transcription factor.
The ORF of AN7542.3 consists of 1,624 bp (1,281 bp in the coding sequence), comprising five exons and four introns. Introns 2, 3, and 4 are of the usual sizes, 49 bp, 46 bp, and 48 bp, respectively, while intron 1 is much larger, 200 bp (Fig. 1A). The predicted FlbB protein is 426 amino acids in length with a predicted mass of 46.8 kDa and an isoelectric point of 5.98. Pfam database searches pointed to the presence of a bZIP-type DNA binding domain only between residues 79 and 113 (Pfam accession number PF00170) (Fig. 1B).
Protein database searches showed the existence of putative homologues in other Aspergillus species, such as A. terreus, A. fumigatus, and A. oryzae, as well as other filamentous fungi, such as Magnaporthe grisea, Fusarium graminearum, Neurospora crassa, Botrytis cinerea, Coccidioides immitis, Sclerotinia sclerotiorum, and Stagonospora nodorum (Fig. 1B). Bcin and Cimm2 homologues have less similarity with respect to FlbB in the region before the bZIP than in the conserved regions of the C-terminal half (Fig. 1B). Thus, it is possible to differentiate a group of FlbB homologues showing poor sequence conservation in this region. On the other hand, we could not detect similar genes in yeasts, such as Saccharomyces cerevisiae, Candida albicans, and Schizosaccharomyces pombe, or higher eukaryotes, such as Homo sapiens and Arabidopsis thaliana, indicating a restricted phylogenetic distribution of FlbB homologues. To further explore this idea, we searched for all putative bZIP-containing proteins in the genomes of different filamentous fungi (A. nidulans, C. immitis, M. grisea, and N. crassa) and yeasts (S. cerevisiae and S. pombe). We found 23 proteins in A. nidulans, and functions have been described for the following ones: AN1812.3/JlbA (40), AN2911.3/AtfA (5), AN3675.3/CpcA (15), AN4361.3/MetR (26), AN4900.3/MeaB (31), AN8251.3/HapX (41), and AN9397.3/Hac1 (34). The derived phylogenetic tree shows that the FlbB bZIP belongs to a branch where yeasts are excluded (Fig. 1C). Our in silico analyses strongly suggest a highly specialized filamentous fungal function for at least FlbB and probably for other bZIP-containing proteins, such as AN4900.3/MeaB and AN0951.3.
Phenotypic analysis of flbB mutants.
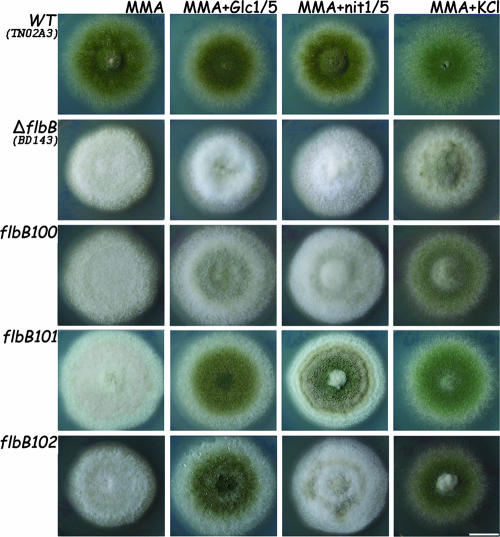
To further analyze the phenotypes of the isolated mutations, we constructed an flbB-null strain by gene replacement using the A. fumigatus pyrG gene (see Fig. S1 in the supplemental material). Strains carrying mutant and null alleles of flbB were further analyzed in parallel. Surface cultures (Fig. 2) of the strains carrying the different flbB mutant alleles yielded the aconidial phenotype in solid MMA. We then investigated whether carbon or nitrogen starvation or salt stress might have a suppressing effect on the aconidial phenotype displayed by the flbB mutant alleles (Fig. 2), as it was previously shown to promote conidiation in the wild type (2, 37). A reduction in glucose content provoked low levels of conidiation accompanied by some autolysis at the oldest part (the center) of the ΔflbB colony. The flbB100 mutant showed increased conidiation compared to the null mutant, and the response was much more pronounced in flbB101 and flbB102 mutants. Nitrogen starvation did not induce conidiation in the ΔflbB or flbB100 mutant. In contrast, conidiation was sparse in the flbB102 mutant and considerably stronger in the flbB101 mutant, with visible conidiation rings in both mutants. Finally, under salt stress conditions (0.6 M KCl), the ΔflbB mutant showed sparse conidiation over a mainly fluffy colony pattern. However, the flbB102 and flbB101 mutants conidiated profusely, similar to the wild type, and the flbB100 mutant responded to this stimulus but to a noticeably lesser degree.
FIG. 2.
Mutant characterization in solid medium. Phenotypes of wild-type (WT; TN02A3), ΔflbB (BD143), flbB100, flbB101, and flbB102 strains grown on solid MMA, MMA with reduced glucose, MMA with reduced nitrate, and MMA with KCl (see Materials and Methods) for 72 h are shown. Bar = 1 cm.
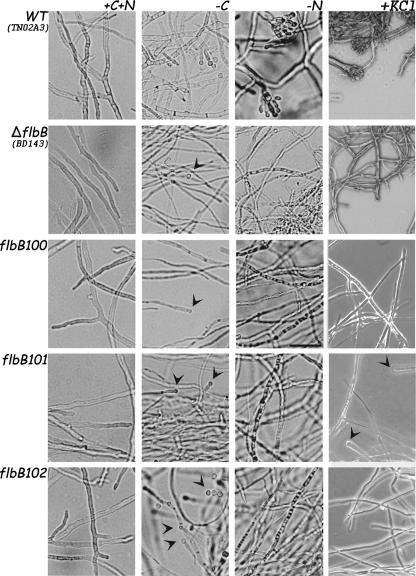
The growth and conidiation patterns of the null mutant and the three original mutants were also assessed in liquid cultures at 10 and 20 h after transfer to glucose- or nitrogen-free medium (Fig. 3). Cultures transferred to liquid MMA maintained a vegetative growth pattern. Under glucose limitation, however, the wild type produced reduced conidiophores, as previously reported (37) (Fig. 3). In contrast, the ΔflbB and flbB100 mutants produced narrow hyphae, approximately 1.5 μm in diameter, from which single conidium-like structures rarely emerged (Fig. 3). The flbB101 and flbB102 mutants showed an intermediate response between those of the wild type and ΔflbB (or flbB100) mutants. Under nitrogen starvation conditions, the ΔflbB mutant and the three flbB mutants showed no conidiation response but rather exhibited an autolytic phenotype confirmed by the absence of staining with the vital stain fluorescein, while a wild-type strain generated conidiophores (37 and this work). In liquid medium with a high salt concentration, neither the ΔflbB mutant nor the three flbB mutants showed any conidiation response, while the wild-type strain generated conidiophores. Only in the case of the flbB101 mutant were some vesicles seen after 20 h of incubation.
FIG. 3.
Mutant characterization in liquid media. Phenotypes of wild-type (WT; TN02A3), ΔflbB (BD143), flbB100, flbB101, and flbB102 strains after 18 h of culture in MMA, followed by transfer to MMA-glucose, MMA-nitrate, or MMA plus KCl and MES for a further 20 h (see Materials and Methods), are shown. Arrowheads indicate conidium-like structures. Bar = 30 μm.
Taken together, the molecular and genetic results with the obtained mutants and their comparisons with the null mutant lead us to conclude that the former bear partial loss-of-function mutations.
FlbB is associated with the production of a diffusible compound needed for conidiation.
Earlier studies had shown that wild-type strains extracellularly restored conidiation in all flb mutants (flbA-E) upon direct physical contact or through a dialysis membrane (21). flb mutants, in turn, unidirectionally restored conidiation in fluG loss-of-function mutants (47). We found that conidiation of flbB mutants was rescued by growing them in contact with a wild-type strain (Fig. 4) or through a dialysis membrane (data not shown). Moreover, all flbB mutants could extracellularly rescue conidiation in a ΔfluG strain. These results indicate that FlbB may function in the synthesis of a diffusible signal compound that is necessary for normal conidiation and distinct from the FluG factor.
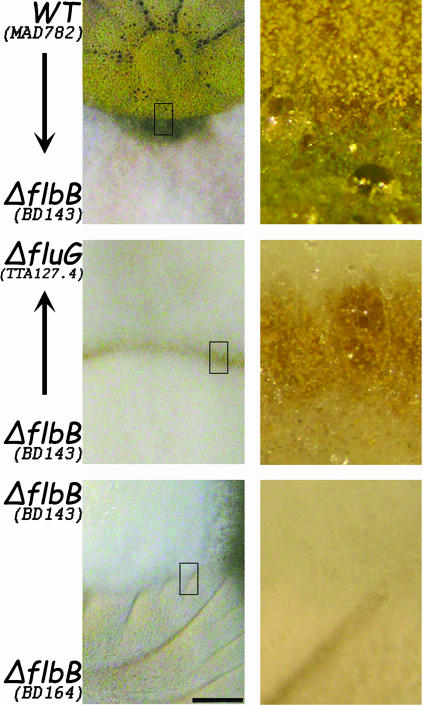
FIG. 4.
Extracellular complementation. The ΔflbB (BD143) mutant acts as a donor of a signaling molecule to a ΔfluG (TTA127.4) strain at zones of contact between colonies. In turn, ΔflbB (BD143) conidiates when in contact with a sporulating strain (MAD782). The arrows indicate the complemented strains. There is no complementation between two flbB deletion strains (BD143 and BD164). All other flbB mutants described in this study display similar behavior. Right panels show close-up views (∼×10 magnification of left panel) of the framed zones. Bar = 0.6 cm.
flbB expression precedes and determines brlA activation.
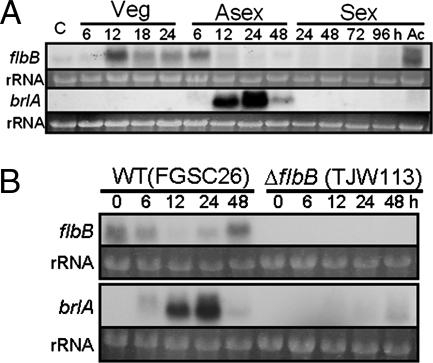
The expression of flbB during the A. nidulans life cycle started with vegetative growth and continued into early phases of asexual development (Fig. 5A). The decline of flbB mRNA steady-state levels coincided with the start of brlA transcription. In addition, flbB expression resumed 12 h after asexual induction and was also observed in conidia and ascospores (sexual spores).
FIG. 5.
flbB and brlA mRNA expression during the A. nidulans life cycle. (A) Steady-state mRNA levels of flbB and brlA during the life cycle of strain FGSC4. C and Ac represent conidia (asexual spores) and ascospores (sexual spores), respectively. Numbers indicate the time (hours) of incubation in liquid MMA with glucose (Veg) or solid MMA with glucose under conditions inducing asexual development (Asexual) or sexual development (Sexual). Note that flbB is expressed during vegetative growth and early phase of asexual development as well as in ascospores, while brlA is expressed as expected, during the early and medium stages of asexual development. (B) flbB and brlA mRNA expression levels, during asexual development, in the null mutant compared to the wild type (WT).
Since the above result did not clarify whether FlbB activated or repressed brlA expression, the ΔflbB mutant was examined by Northern blotting (Fig. 5B). The absence of detectable brlA mRNA accumulation demonstrated that FlbB is necessary for brlA expression and formally confirmed the designation of the gene as an flb (fluffy with low bristle expression) gene.
Finally, no significant differences could be detected in the flbB expression pattern between a veA wild-type (FGSC4) strain (Fig. 5A) and a veA mutant (veA1; FGSC26) strain (Fig. 5B), suggesting that flbB expression is not conditioned by the light-dependent conidiation regulator VeA.
Localization of FlbB at the hyphal tip and in the nucleus.
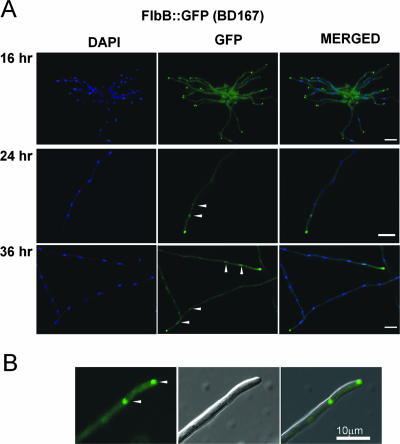
In order to determine the subcellular distribution of FlbB, strains expressing either FlbB::GFP or GFP::FlbB proteins were constructed (see Materials and Methods and Fig. S2 in the supplemental material). Microscopic examination of the strains in liquid static cultures revealed that FlbB is in the cytoplasm and accumulates at the hyphal tip at 16 h (Fig. 6A) and then (at 24 to 36 h) into the nuclei, as predicted by the putative transcriptional regulatory function of FlbB, despite the absence of an identifiable nuclear localization signal (Fig. 6A). Interestingly, in most of the screened cells, the nucleus closest to the cell apex exhibited the highest level of fluorescence, indicating the high degree of regulation in localization or compartmentalization of FlbB (see Discussion). The vast majority of the hyphae showed this localization for either C- or N-terminally tagged FlbB proteins, up to 36 h of culture (Fig. 6A; data not shown for GFP::FlbB). Blast searches using different programs available at www.expasy.org (HMMTOP, PredictProtein, SOSUI, TMAP, TMHMM, Tppred, TopPred, and PSORT) showed an absence of predictable transmembrane domains or cysteine residues susceptible to prenylation (PrePS), suggesting that FlbB is probably not anchored to the membrane and that its apical localization responds to other, as-yet-unknown factors (Fig. 6B).
FIG. 6.
Intracellular localization of FlbB. (A) The FlbB-GFP protein accumulates at the hyphal apex after 16 h of static culture in liquid MMA. Fluorescence at the cytoplasm is also visible, although in these images the contrast has been intentionally modified to show that no nuclei are fluorescent at 16 h. After 24 h and 36 h, fluorescence is also present in the proximal nuclei, as determined by DAPI staining. Arrowheads indicate fluorescent nuclei. (B) Proximity of FlbB to the tip of the cell. Arrowheads indicate the localization of FlbB at the apex and in the proximal nucleus. Bar = 10 μm.
FlbB is a potential transcriptional activator.
To test whether FlbB can function as a TF likely activating downstream genes necessary for conidiation, the ability of the FlbB protein to activate two reporters was examined. The full-length protein (FlbB-F), the N-terminal region including the bZIP domain (FlbB-N), and the C-terminal region without the bZIP domain (FlbB-C) were fused with LexADBD (DNA binding domain), and individual fusion proteins were expressed in S. cerevisiae. As shown in Fig. 7, when the transformants were tested on the medium lacking two nutrients (uracil and histidine) in the presence of various concentrations of 3-AT for the HIS3 reporter, those expressing LexADBD::FlbB-F and LexADBD::FlbB-C grew on the medium with 5 mM and 10 mM 3-AT, respectively. However, the strains with pTLexA alone or LexADBD::FlbB-N were unable to form colonies. In the X-Gal test, those expressing LexADBD::FlbB-C showed blue color within 4 h after inoculation, similar to the PJ69-4A strains expressing Gal4DBD::AflR (28, 52). The strains expressing LexADBD::FlbB-F began to exhibit blue color after 1 day. Moreover, when multiple transformants for each construct were tested for their β-galactosidase activity (Fig. 7, right), the strains expressing LexADBD::FlbB-C and LexADBD::FlbB-F showed high levels of β-galactosidase activity (489 ± 12 and 248 ± 7 units, respectively). These results indicate that the C-terminal region of FlbB has a transactivation activity and that the N-terminal region may be involved in a modulating capacity, not excluding inhibitory effects on transactivation.
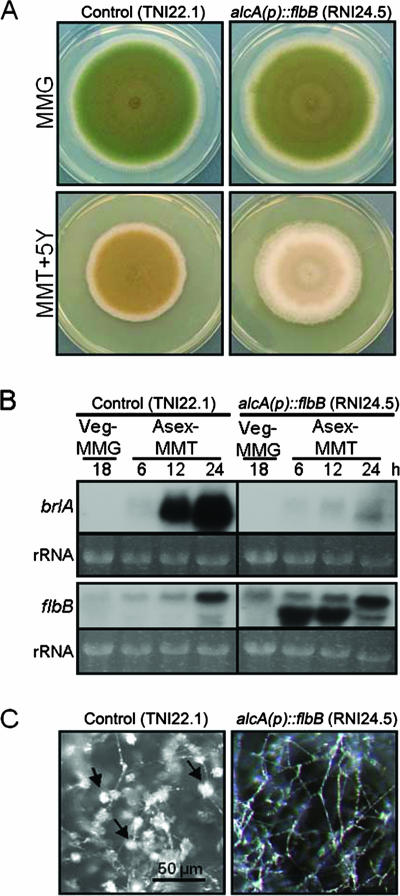
Overexpression of flbB inhibits conidiation.
To further examine the regulatory role of flbB in conidiation, we generated an flbB overexpression mutant by fusing the flbB ORF with the inducible alcA promoter (14). Since the above-mentioned data support the positive regulatory roles for FlbB in conidiation, a hyperconidiating phenotype was expected. However, overexpression of flbB resulted in severely reduced conidiation (Fig. 8A). Examination of brlA and flbB mRNA levels revealed that overexpression of flbB abolished the proper accumulation of brlA mRNA (Fig. 8B). This is consistent with the observation that the alcA(p)::flbB mutant showed hyphal growth only after synchronized asexual developmental induction on threonine medium, whereas a control strain produced plenty of conidiophores (Fig. 8C). These findings suggest that FlbB may be coupled with another factor(s) in a finely balanced stoichiometry, requiring a correct dosage in order to form an activating (heteromeric) complex (see Discussion).
FIG. 8.
Overexpression of flbB inhibits conidiation. (A) Photographs of the colonies of control (TNI22.1) and alcA(p)::flbB (RNI24.5) strains grown on MMA plus glucose (MMG; noninducing) and MMA plus yeast extract (MMT+5Y; threonine-inducing alcA promoter, with 5 g/liter yeast extract enhancing growth) for 5 days are shown. Overexpression of flbB resulted in a blockage in conidiation. (B) Control (TNI22.1) and alcA(p)::flbB (RNI24.5) strains were grown in liquid glucose medium at 37°C and 250 rpm for 18 h and then transferred onto solid medium with threonine (MMT; inducing) as the sole carbon source. Samples were collected at 18 h of vegetative growth (Veg) and 6, 12, and 24 h after asexual developmental induction (Asex) on MMT. Total RNA was isolated and subjected to Northern blot analyses for the levels of brlA and flbB mRNAs. Equal loading of total RNA was evaluated by ethidium bromide staining of rRNA. (C) Close-up views of control (TNI22.1) and alcA(p)::flbB (RNI24.5) strains grown on MMT for 48 h after transfer onto solid MMT. The control strain produced conidiophores abundantly (arrows), whereas alcA(p)::flbB strain (RNI24.5) formed elongated aerial hyphae without conidiophores.
DISCUSSION
In a mycelium, vegetative hyphae extend radially at the periphery, and some rise into the atmosphere. Recent evidence indicates that the recognition of this status is determined by an increase in extracellular levels of the FluG factor (33, 48). At this stage, some hyphae undertake conidiophore morphogenesis (1), but at least one additional signal seems to be required to confirm this particular fate, as previously shown (47) and confirmed in this study. The flbB-D genes appear to intervene at this morphogenetic stage, as a mutation in any one of them blocks the synthesis of the second diffusible signal (47). Epistasis studies show that the sequence FlbE → FlbD → FlbB → BrlA (FlbC acting in parallel) is operational (55), FlbB being the last reported element in the sequence before the activation of brlA. In addition, FlbB is a transcription factor located at the hyphal apex near the growing point of the cell. Such a location suggests that distinct but as-yet-undetermined changes at the growing tip may be involved in its activation. Finally, FlbB is required for the synthesis of the second signal downstream of FluG, which is necessary for conidiation induction.
Consistent with these observations, flbB displays unique characteristics as a transcription factor, i.e., a highly conserved N-terminal bZIP domain and various conserved regions, which participate in the function of the protein and could participate in transcriptional activation, as found in this study.
The Gly residue, where the missense mutation FlbB100 (G70R) lies, is situated nine residues upstream from the starting Leu of the bZIP domain. Previous reports have shown the presence a similar sequence in HapX, where this Gly is universally conserved (41). This region is necessary for the recruitment of HapX to the Hap complex and consequent recognition of CCAAT sequence-containing promoter regions (25). The apparent necessity of this flanking region for the functionality of FlbB suggests that the protein may form a (heterogenic) complex to activate conidiation, although an altered function of the bZIP DNA binding domain cannot be excluded.
The conserved FlbBK67-A78 and C-terminal regions were all required for normal conidiation in solid air-exposed cultures. However, in liquid culture, where stimuli can be studied separately, a differential response was revealed. Under carbon source-limiting conditions, which are known to induce conidiation in liquid cultures (37), a functional FlbBK67-A78 region is essential. In contrast, the C-terminal region does not appear to play a fundamental role. Under nitrogen starvation, however, all domains are required for conidiation in liquid culture, whereas in solid medium, truncating the protein from residue 267 onwards can result in notable conservation of activity. A more restricted truncation, affecting only residues 305 onwards, has a considerably greater effect, however, and this paradox raises the possibility of complex interactions between domains and indeed other interacting proteins. Under salt stress, mutants affected in the C-terminal region still showed a substantial conidiation response in solid medium, in sharp contrast to the mutant affected in Gly70, which resembled the practically fluffy phenotype of the null mutant. This indicates that the conserved region adjacent to the DNA binding domain, and possibly affecting bZIP function, appears to be essential for the response to salt stress. In contrast, in liquid medium all the regions characterized are necessary for conidiation to occur.
brlA expression starts just when flbB expression ends, and this could lead to the conclusion that FlbB could act as a brlA transcriptional repressor. However, no brlA expression was detected in the flbB-null mutant, showing that FlbB is required, directly or indirectly, for proper brlA transcription regulation. This suggests that the regulation of flbB expression must be precisely controlled for a normal asexual life cycle. The observation that flbB expression starts again at 48 h after asexual development induction suggests that FlbB could act in a cyclic way, having a sensory role, in order to confirm or remind that conidiation should continue. To confirm this, flbB and brlA expression should be analyzed for a longer period after asexual induction. Finally, flbB expression is not conditioned by VeA activity, and the reason for flbB mRNA presence in ascospores should be analyzed in more detail.
The abovementioned evidence, combined with the apical localization of the transcription factor before its purported activation, points to a potential sensory function of FlbB, effected through an as-yet-unidentified complex located at the hyphal apex. This evidence allows an interpretation of the aconidial phenotype observed in the overexpression strain. Like many bZIP proteins (45), FlbB may form a heteromeric complex. When flbB is overexpressed, this results in the misscheduled and unbalanced presence of the FlbB protein. This aberrant expression could block proper complex formation and function, resulting in an aconidial phenotype. Since hypofunctional flbB mutants are not affected in growth pattern or kinetics, the role of this regulatory mechanism appears to be strictly limited to the control of conidiation.
The nature of the stimulus triggering FlbB relocalization to the nucleus was not clarified in this study. Among the various possible cues is reactive oxygen species, which is associated with the cessation of growth and has been reported to have a role in fungal morphogenesis (19). Moreover, FlbB shares a number of conserved cysteines in its C-terminal region with another bZIP transcription factor, Pap1 of S. pombe, which is capable of sensing moderate levels of oxidative stress through these residues (9). This and other likely possibilities are under examination.
Supplementary Material
Acknowledgments
We express our admiration and gratitude to T. H. Adams, J. Aguirre, and J. K. Wieser for their pioneering work in the field of conidiation induction, and especially with flb mutants, which served as a basis for the findings of this investigation. We are also grateful to J. Clutterbuck for provision of strains and advice and T. Roncal and S. Cordobés for their assistance with mutant screening.
The work carried out at UW-Madison was supported by Hatch (WIS04667) and National Science Foundation (MCB-0421863 and IOS-0640067) grants to J.H.Y. E.A.E thanks Ministerio de Educación y Ciencia for support through grant BFU2006-04185. U.U. thanks Ministerio de Educación y Ciencia for grant BFU2004-03499/BMC and UPV/EHU for grant GIU05/36. O.E received a doctoral grant from UPV/EHU, and A.G. was a contract researcher under the research program of Gipuzkoako Foru Aldundia/Diputación Foral de Gipuzkoa. The work was also supported by the Max-Planck-Institute for Terrestrial Microbiology, Marburg, Germany.
Footnotes
Published ahead of print on 9 November 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adams, T. H., M. T. Boylan, and W. E. Timberlake. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54353-362. [DOI] [PubMed] [Google Scholar]
- 2.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 6235-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguirre, J. 1993. Spatial and temporal controls of the Aspergillus brlA developmental regulatory gene. Mol. Microbiol. 8211-218. [DOI] [PubMed] [Google Scholar]
- 4.Aguirre, J., R. Ortiz, J. Clutterbuck, R. Tapia, and M. Cardenas. 1993. vegA and cfwA define two new developmental genes in Aspergillus nidulans. Fungal Genet. Newsl. 40A:68. [Google Scholar]
- 5.Aguirre, J., M. Rios-Momberg, D. Hewitt, and W. Hansberg. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13111-118. [DOI] [PubMed] [Google Scholar]
- 6.Axelrod, D. E., M. Gealt, and M. Pastushok. 1973. Gene control of developmental competence in Aspergillus nidulans. Dev. Biol. 349-15. [DOI] [PubMed] [Google Scholar]
- 7.Bernreiter, A., A. Ramon, J. Fernandez-Martinez, H. Berger, L. Araujo-Bazan, E. A. Espeso, R. Pachlinger, A. Gallmetzer, I. Anderl, C. Scazzocchio, and J. Strauss. 2007. Nuclear export of the transcription factor NirA is a regulatory checkpoint for nitrate induction in Aspergillus nidulans. Mol. Cell. Biol. 27791-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brent, R., and M. Ptashne. 1985. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell 43729-736. [DOI] [PubMed] [Google Scholar]
- 9.Castillo, E. A., J. Ayte, C. Chiva, A. Moldon, M. Carrascal, J. Abian, N. Jones, and E. Hidalgo. 2002. Diethylmaleate activates the transcription factor Pap1 by covalent modification of critical cysteine residues. Mol. Microbiol. 45243-254. [DOI] [PubMed] [Google Scholar]
- 10.Chang, Y. C., and W. E. Timberlake. 1993. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics 133:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho, J. H., S. S. Yun, Y. K. Jang, M. J. Cha, N. J. Kwon, and S. K. Chae. 2003. Identification and cloning of jipA encoding a polypeptide that interacts with a homolog of yeast Rad6, UVSJ in Aspergillus nidulans. J. Microbiol. 4146-51. [Google Scholar]
- 12.Fischer, R., and U. Kües. 2006. Asexual sporulation in mycelial fungi, p. 263-292. In U. Kües and R. Fischer (ed.), The Mycota, vol. I: growth, differentiation and sexuality. Springer-Verlag, Berlin, Germany.
- 13.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 4381105-1115. [DOI] [PubMed] [Google Scholar]
- 14.Gwynne, D. I., F. P. Buxton, S. Sibley, R. W. Davies, R. A. Lockington, C. Scazzocchio, and H. M. Sealy-Lewis. 1987. Comparison of the cis-acting control regions of two coordinately controlled genes involved in ethanol utilization in Aspergillus nidulans. Gene 51205-216. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann, B., O. Valerius, M. Andermann, and G. H. Braus. 2001. Transcriptional autoregulation and inhibition of mRNA translation of amino acid regulator gene cpcA of filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 122846-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kafer, E. 1965. Origins of translocations in Aspergillus nidulans. Genetics 52217-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kellner, E. M., and T. H. Adams. 2002. Mutations in sfdA and sfdB suppress multiple developmental mutations in Aspergillus nidulans. Genetics 160159-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lalucque, H., and P. Silar. 2003. NADPH oxidase: an enzyme for multicellularity? Trends Microbiol. 119-12. [DOI] [PubMed] [Google Scholar]
- 20.Law, D. J., and W. E. Timberlake. 1980. Developmental regulation of laccase levels in Aspergillus nidulans. J. Bacteriol. 144509-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, B. N., and T. H. Adams. 1994. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 8641-651. [DOI] [PubMed] [Google Scholar]
- 22.Lee, B. N., and T. H. Adams. 1994. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol. Microbiol. 14323-334. [DOI] [PubMed] [Google Scholar]
- 23.Mah, J.-H., and J.-H. Yu. 2006. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 51585-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinelli, S. D. 1994. Aspergillus nidulans as an experimental organism. Prog. Ind. Microbiol. 2933-58. [PubMed] [Google Scholar]
- 25.McNabb, D. S., and I. Pinto. 2005. Assembly of the Hap2p/Hap3p/Hap4p/Hap5p-DNA complex in Saccharomyces cerevisiae. Eukaryot. Cell 41829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Natorff, R., M. Sienko, J. Brzywczy, and A. Paszewski. 2003. The Aspergillus nidulans metR gene encodes a bZIP protein which activates transcription of sulphur metabolism genes. Mol. Microbiol. 491081-1094. [DOI] [PubMed] [Google Scholar]
- 27.Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil, S. L. Murray, M. J. Hynes, S. A. Osmani, and B. R. Oakley. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 1721557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni, M., and J.-H. Yu. 2007. A novel regulator couples sporogenesis and trehalose biogenesis in Aspergillus nidulans. PLoS ONE 2e970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osherov, N., J. Mathew, and G. S. May. 2000. Polarity-defective mutants of Aspergillus nidulans. Fungal Genet. Biol. 31181-188. [DOI] [PubMed] [Google Scholar]
- 30.Peñalva, M. A. 2005. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet. Biol. 42963-975. [DOI] [PubMed] [Google Scholar]
- 31.Polley, S. D., and M. X. Caddick. 1996. Molecular characterisation of meaB, a novel gene affecting nitrogen metabolite repression in Aspergillus nidulans. FEBS Lett. 388200-205. [DOI] [PubMed] [Google Scholar]
- 32.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. McDonald, and A. W. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5141-238. [DOI] [PubMed] [Google Scholar]
- 33.Roncal, T., U. O. Ugalde, and A. Irastorza. 1993. Calcium-induced conidiation in Penicillium cyclopium: calcium triggers cytosolic alkalinization at the hyphal tip. J. Bacteriol. 175879-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saloheimo, M., M. Valkonen, and M. Penttila. 2003. Activation mechanisms of the HACI-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 471149-1161. [DOI] [PubMed] [Google Scholar]
- 35.Seo, J.-A., Y. Guan, and J.-H. Yu. 2006. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics 1721535-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 1943-21. [DOI] [PubMed] [Google Scholar]
- 37.Skromne, I., O. Sanchez, and J. Aguirre. 1995. Starvation stress modulates the expression of the Aspergillus nidulans brlA regulatory gene. Microbiology 14121-28. [DOI] [PubMed] [Google Scholar]
- 38.Stinnett, S. M., E. A. Espeso, L. Cobeno, L. Araujo-Bazan, and A. M. Calvo. 2007. Aspergillus nidulans VeA subcellular localization is dependent on the importin alpha carrier and on light. Mol. Microbiol. 63242-255. [DOI] [PubMed] [Google Scholar]
- 39.Stringer, M. A., R. A. Dean, T. C. Sewall, and W. E. Timberlake. 1991. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 51161-1171. [DOI] [PubMed] [Google Scholar]
- 40.Strittmatter, A. W., S. Irniger, and G. H. Braus. 2001. Induction of jlbA mRNA synthesis for a putative bZIP protein of Aspergillus nidulans by amino acid starvation. Curr. Genet. 39327-334. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka, A., M. Kato, T. Nagase, T. Kobayashi, and N. Tsukagoshi. 2002. Isolation of genes encoding novel transcription factors which interact with the Hap complex from Aspergillus species. Biochim. Biophys. Acta 1576176-182. [DOI] [PubMed] [Google Scholar]
- 42.Tilburn, J., C. Scazzocchio, G. G. Taylor, J. H. Zabicky-Zissman, R. A. Lockington, and R. W. Davies. 1983. Transformation by integration in Aspergillus nidulans. Gene 26205-221. [DOI] [PubMed] [Google Scholar]
- 43.Ugalde, U. O. 2006. Autoregulatory signals in mycelial fungi, p. 203-213. In U. Kües and R. Fischer (ed.), The Mycota, vol. I: growth, differentiation and sexuality. Springer-Verlag, Berlin, Germany.
- 44.Vinson, C. R., P. B. Sigler, and S. L. McKnight. 1989. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 246911-916. [DOI] [PubMed] [Google Scholar]
- 45.Vinson, C., A. Acharya, and E. J. Taparowsky. 2006. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim. Biophys. Acta 17594-12. [DOI] [PubMed] [Google Scholar]
- 46.Waring, R. B., G. S. May, and N. R. Morris. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin coding genes. Gene 79119-130. [DOI] [PubMed] [Google Scholar]
- 47.Wieser, J., B. N. Lee, J. Fondon III, and T. H. Adams. 1994. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 2762-69. [DOI] [PubMed] [Google Scholar]
- 48.Wieser, J., and T. H. Adams. 1995. flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes Dev. 9491-502. [DOI] [PubMed] [Google Scholar]
- 49.Wieser, J. 1997. Genetic requirements for initiation of Aspergillus nidulans conidiophore development. Ph.D. thesis. Texas A&M University, College Station, TX.
- 50.Yang, L., L. Ukil, A. Osmani, F. Nahm, J. Davies, C. P. C. De Souza, X. Dou, A. Perez-Balaguer, and S. A. Osmani. 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 31359-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, J.-H., J. Wieser, and T. H. Adams. 1996a. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 155184-5190. [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, J.-H., R. A. Butchko, M. Fernandes, N. P. Keller, T. J. Leonard, and T. H. Adams. 1996b. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 29549-555. [DOI] [PubMed] [Google Scholar]
- 53.Yu, J. H., Z. Hamari, K.-H. Han, J.-A. Seo, Y. Reyes-Dominguez, and C. Scazzocchio. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41973-981. [DOI] [PubMed] [Google Scholar]
- 54.Yu, J.-H. 2006. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J. Microbiol. 44145-154. [PubMed] [Google Scholar]
- 55.Yu, J.-H., J.-H. Mah, and J.-A. Seo. 2006. Growth and developmental control in the model and pathogenic aspergilli. Eukaryot. Cell 51577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.