Abstract
Multivalent binding allows high selectivity and affinity in a ligand–protein interaction. The N-end rule pathway is a ubiquitin (Ub)-dependent proteolytic system in which specific E3s, called N-recognins, mediate ubiquitylation through the recognition of types 1 and 2, destabilizing N-terminal residues of substrates. We recently identified a set of E3 Ub ligases (named UBR1–UBR7) containing the 70-residue UBR box, and we demonstrated that UBR1, UBR2, UBR4, and UBR5 can bind to destabilizing N-terminal residues. To explore a model of heterovalent interaction to the N-recognin family, we synthesized the small-molecule compound RF-C11, which bears two heterovalent ligands designed to target N-recognins, together with control molecules with two homovalent ligands. We demonstrate that heterovalent ligands of RF-C11 selectively and cooperatively bind cognate-binding sites of multiple N-recognins and thereby inhibit both types 1 and 2 N-end rule activities. Furthermore, the efficacy of heterovalent RF-C11 was substantially higher than homovalent inhibitors, which can target either a type 1 or type 2 site, providing the molecular basis of designing multivalent inhibitors for the control of specific intracellular pathways. In addition, RF-C11 exhibited higher efficacy and stability, compared with dipeptides bearing destabilizing N-terminal residues, which are known competitive inhibitors of the pathway. We also used the heterovalent compound to study the function of N-recognins in cardiac signaling. Using mouse and rat cardiomyocytes, we demonstrate that the N-end rule pathway has a cell-autonomous function in cardiac proliferation and hypertrophy, explaining our earlier results implicating the pathway in cardiac development and proteolysis of multiple cardiovascular regulators.
Keywords: N-recognin, protein degradation, ubiquitin, cardiovascular cardiomyocyte
The N-end rule pathway is a ubiquitin (Ub)-dependent proteolytic system, in which N-terminal residues of short-lived proteins function as an essential component of degradation signals (degrons) called N-degrons (Fig. 1A) (1–13). An N-degron can be created from a pre-N-degron through specific N-terminal modifications (13). In mammals, N-terminal Asn and Gln can act as destabilizing residues through deamidation into Asp and Glu, which is mediated by NTAN1 and a hypothetical enzyme called NTAQ1 (5). N-terminal Asp and Glu are conjugated with Arg by ATE1 R-transferase, creating N-terminal Arg, one of the primary destabilizing residues (4, 7). In mammals, N-terminal Cys can function as a tertiary destabilizing residue through its oxidation in a manner depending on nitric oxide and oxygen and subsequent arginylation by ATE1 (7, 9, 14). N-terminal Arg and other destabilizing N-terminal residues are recognized for protein ubiquitylation by specific E3 Ub ligases called N-recognins (3, 6, 8). Biochemical and genetic studies identified its physiological functions in cardiac development and signaling, angiogenesis, meiosis, and neurogenesis in mice; the pathogenesis in human genetic diseases; and peptide import and chromosome stability in Saccharomyces cerevisiae (6–8, 10, 12, 15–17).
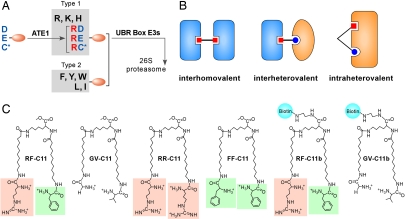
Fig. 1.
Heterobivalent inhibitor of the N-end rule pathway. (A) The mammalian N-end rule pathway. (B) Models of interhomovalent, interheterovalent, and intraheterovalent interactions. (C) Structures of RF-C11 and related compounds. Terminal Arg and Phe are indicated by orange and green backgrounds, respectively.
We previously identified the UBR box E3 family characterized by a 70-residue zinc finger-like domain, termed the UBR box (3, 6–8, 11–13). Different organisms have different sets of UBR proteins; the mammalian genome encodes at least seven UBR proteins named UBR1–UBR7, whereas S. cerevisiae encodes UBR1 and UBR2 (11). UBR proteins are generally heterogeneous in size and sequence, but contain, with the exception of UBR4, specific signatures unique to Ub ligases or a substrate-recognition subunit of the E3 complex: the RING domain in UBR1, UBR2, and UBR3; the HECT domain in UBR5; the F-box in UBR6; and the PHD domain in UBR7 (11–13). UBR1, UBR2, UBR4, and UBR5 were determined to bind to destabilizing N-terminal residues (8, 11–13), whereas the biochemical properties of UBR3, UBR6, and UBR7 as candidate N-recognins are largely unknown. N-terminal degradation determinants can be divided into type 1 (basic: Arg, Lys, and His) and type 2 (bulky hydrophobic: Phe, Leu, Trp, Tyr, and Ile) residues (13). The binding of N-end rule substrates to N-recognins can be competitively inhibited by specific dipeptides bearing destabilizing N-terminal residues (6, 8, 11, 18).
Nature employs multivalent interactions to increase selectivity and avidity of protein–protein or protein–ligand interactions, both thermodynamically (enhanced binding affinity) and kinetically (reduced dissociation rate). As such, synthetic molecules have been designed to employ cooperative interactions of multivalent ligands to target molecules. Most multivalent compounds synthesized to date are interhomovalent (Fig. 1B), in that two homovalent ligands target the same binding site of two identical proteins on the surface of viruses, bacteria, or cells (19). However, rapamycin, a natural compound produced from Streptomyces hygroscopicus, is interheterovalent (Fig. 1B), in that its two heterovalent ligands bind to cognate sites of two different proteins, FKBP12 (FK506-binding protein) and FRB (FKBP–rapamycine-binding domain), forming an FKBP–rapamycine–FRB ternary complex (20). In contrast to interhomovalent and interheterovalent interactions, multivalent interaction to a single intracellular protein remains largely unexplored perhaps because of the scarcity of appropriate model systems. As a test-of-concept study of intraheterovalent interaction, we previously tested whether overexpression of metabolically stable Arg-eΔK-β-gal (produced from the Ub-Arg-eΔK-β-gal fusion protein) and Leu-eΔK-β-gal (from Ub-Leu-eΔK-β-gal) in S. cerevisiae would inhibit the N-end rule activity (21). In the β-gal tetramer, two N termini of each dimer are oriented to the same direction so that ≈50% of β-gal dimers are heterodimers bearing N-terminal Arg and Leu. The coexpression of Arg-eΔK-β-gal and Leu-eΔK-β-gal in S. cerevisiae inhibited the degradation of a model N-end rule substrate more effectively than the expression of either Arg-eΔK-β-gal or Leu-eΔK-β-gal alone, which is indicative of a heterovalent interaction between β-gal tetramers and N-recognin.
In this study, we took advantage of two distinct substrate-binding sites of N-recognins to study a model of a small-molecule-based intraheterovalent interaction, compared with intrahomovalent interactions. We synthesized a model compound with two heterovalent ligands to N-recognins and demonstrate its selective and cooperative binding to multiple N-recognins, with higher efficacy, compared with homovalent control compounds. We also show that this heterovalent compound is more potent and has higher stability than dipeptide inhibitors of the pathway. By using the heterovalent compound, we demonstrate that the N-end rule pathway has a cell-autonomous function in cardiac proliferation and hypertrophy, explaining our earlier results implicating the pathway in cardiac development and proteolysis of multiple cardiovascular regulators.
Results
Design, Rationale, and Synthesis of the Heterovalent Inhibitor RF-C11 of the N-Recognin Family.
To explore a heterovalent interaction to the N-end rule pathway, we designed a model compound (RF-C11) whose heterovalent ligands, Arg and Phe, can target simultaneously and cooperatively cognate-binding sites of multiple N-recognins (Fig. 1C). In this heterovalent interaction with a flexible linker (≈15–30 Å) between two ligands (Fig. 1B), the bound Phe ligand to the type 2 site will prevent the unbound Arg ligand (of the same RF-C11 molecule) from moving away from the type 1 site, and thereby will increase the local concentration of Arg in the proximity of the type 1 site, resulting in enhanced Arg interaction to N-recognin. Reciprocally, the bound Arg, whose binding has been facilitated by the bound Phe, will in turn increase the local concentration of Phe in the proximity of the type 2 site, further facilitating the interaction of Phe to N-recognin. Through this mutual enhancement of local ligand concentrations, RF-C11 would exhibit higher overall affinity to N-recognins, compared with homovalent compounds. To experimentally test this model by using N-recognins, we conjugated Arg and Phe, as heterovalent ligands, to (nonproteinaceous) hydrocarbon linkers (C = 11), which in turn were linked to the core Lys moiety by its ε- and α-amine groups, respectively [Fig. 1C, supporting information (SI) Materials and Methods, and SI Fig. 7]. We also synthesized a structural control, GV-C11 (bearing Gly and Val at its termini), and two homovalent controls, RR-C11 (bearing Arg at its termini) and FF-C11 (bearing Phe at its termini). The distance between heterovalent ligands was designed to be ≈45 Å at the equilibrium conformation (Spartan version 4.0, Wavefunction) to reach both binding sites if they are on the same side of proteins. Lys, which has tripartite functional groups, is a core moiety whose reactive carboxylic acid end can be either esterified to curb further reaction or conjugated to biotin or other moieties.
Heterovalent Inhibitor RF-C11 Selectively Inhibits N-Recognins with Higher Efficacy than Homovalent Inhibitors.
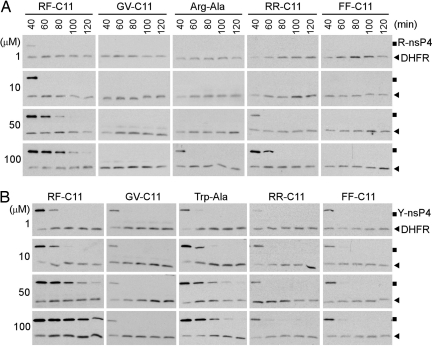
To determine the heterovalent interaction of synthesized compounds to N-recognins, we characterized the proteolysis of model N-end rule substrates in transcription translation-coupled rabbit reticulocyte lysates. The fDHFRh-UbR48-X-nsP4f fusion proteins [X = Met (stabilizing), Arg (type 1), Tyr (type 2), or Ala (type 3); h and f, HA and FLAG tags] (5–7) were expressed in the lysates, where its cotranslational cleavage by deubiquitylating enzymes (DUBs) at the UbR48-X junction yields the long-lived DHFRh-UbR48 reference and the X-nsP4f (X-nsP4) substrate (SI Fig. 8). Arg-nsP4, the full-length 69-kDa Sindbis virus RNA polymerase nsP4 bearing a type 1 N terminus, was rapidly degraded, which was inhibited by the dipeptide Arg-Ala, but not by Trp-Ala. The selectivity of dipeptides was not significantly affected at a concentration as high as 2 mM. Likewise, Tyr-nsP4 also was extremely short-lived and was selectively stabilized by Trp-Ala, but not by 2 mM Arg-Ala. In contrast, Met-nsP4 and Ala-nsP4, bearing stabilizing N termini, were virtually nondegradable in the lysates. Thus, the proteolysis of X-nsP4 in the lysates requires the presence of either type 1 or type 2 N-terminal residues, a degradation signal recognized by multiple N-recognins through two cognate, highly selective, and mutually exclusive binding sites.
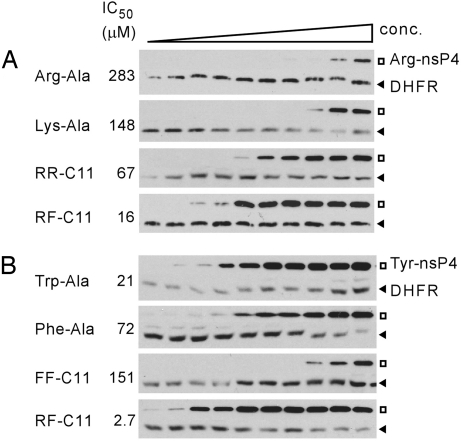
We then determined the effect of synthesized compounds on the proteolysis of X-nsP4. RR-C11 inhibited the degradation of Arg-nsP4, but not Tyr-nsP4; its efficacy (67 μM half-maximal IC50) was even higher than the dipeptide Arg-Ala (283 μM IC50), which has been widely used as a type 1 N-end rule inhibitor (Figs. 2 and 3A and SI Fig. 9). The activity of RR-C11 should be specific to the terminal moiety Arg because the structural control GV-C11 did not affect the degradation of Arg-nsP4 or Tyr-nsP4. These results identify a single amino acid linked to a nonproteinaceous hydrocarbon chain as an efficient and selective ligand to N-recognins. However, FF-C11 showed a (weak) inhibitory effect for Tyr-nsP4, but not for Arg-nsP4 (Fig. 2). Its efficacy (151 μM IC50) was substantially lower, compared with the type 2 dipeptide Trp-Ala (21 μM IC50) (Fig. 3B), suggesting that the terminal moiety Phe of FF-C11 is a selective, but poor, ligand to N-recognins. RF-C11 with heterovalent ligands effectively inhibited the degradation of both Arg-nsP4 and Tyr-nsP4 (Fig. 2 and SI Figs. 10 and 11). Moreover, the efficacy of RF-C11 (16 μM for Arg-nsP4, 2.7 μM for Tyr-nsP4) was significantly higher, compared with both RR-C11 (67 μM for Arg-nsP4) and FF-C11 (151 μM for Tyr-nsP4) (Fig. 3). The enhanced efficacy of RF-C11 should be due to a heterovalent interaction, rather than allosteric conformational change of binding sites, because mixtures of dipeptides or homovalent compounds did not give significantly additive effects (Fig. 2 and SI Figs. 10 and 11). RF-C11 strongly inhibited ubiquitylation of both Arg-nsP4 and Tyr-nsP4 (Fig. 4A), suggesting that it inhibits substrate ubiquitylation by N-recognins.
Fig. 2.
Heterovalent RF-C11 inhibits the proteolysis of both Arg-nsP4 (R-nsP4) and Tyr-nsP4 (Y-nsP4) with higher efficacy than homovalent compounds or dipeptides. fDHFRh-UbR48-X-nsP4f (X = Arg or Tyr) was expressed in reticulocyte lysates, where its cotranslational cleavage yields the DHFR-UbR48 reference (DHFR; lower rows in each gel) and the X-nsP4 substrate (upper rows in each gel). The effects of synthesized compounds and dipeptides on degradation of Arg-nsP4 (A) and Tyr-nsP4 (B) were monitored in the presence of 150 μM bestatin by using time course anti-biotin Western blotting.
Fig. 3.
Evaluation of efficacy of synthesized compounds and dipeptides. Degradation of Arg-nsP4 (A) and Tyr-nsP4 (B) was monitored in the lysates in the presence of RF-C11 and other compounds (0.5, 1, 5, 10, 25, 50, 75, 100, 150, and 250 μM). IC50 values are indicated.
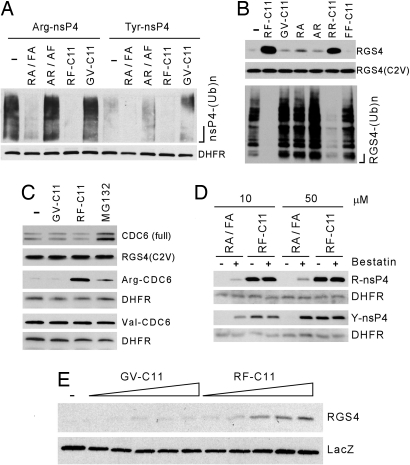
Fig. 4.
Characterization of synthesized compounds for their stability and effect on proteolysis of N-end rule substrates. (A) X-nsP4 proteins were expressed in reticulocyte lysates for 20 min in the presence of 5 μM MG132 and 50 μM other compounds, followed by anti-Ub immunoprecipitation and anti-biotin Western blotting. DHFR, the DHFR-Ub control. RA, Arg-Ala; FA, Phe-Ala; AR, Ala-Arg; AF, Ala-Phe. (B) The effect of 15 μM compounds on proteolysis (Upper) and ubiquitylation (Lower) of RGS4 expressed in reticulocyte lysates for 30 min, with RGS4(C2V) (9) as a control. (C) The effects of 50 μM synthesized compounds and 5 μM MG132 on the proteolysis of CDC6, Arg-CDC6128−589 (Arg-CDC6) and Val-CDC6128−589 (Val-CDC6) expressed for 45 min, with long-lived RGS4(C2V) (9) and DHFR-UbR48 as controls. (D) The effect of 150 μM bestatin on the efficacy of RF-C11 and a mixture of Arg-Ala (RA) and Phe-Ala (FA) on the degradation of X-nsP4 expressed for 45 min (for Arg-nsP4) or 60 min (for Tyr-nsP4). (E) RGS4 and lacZ were expressed in embryonic fibroblasts, followed by the treatment of RF-C11 and GV-C11 (0.1, 10, 50, 75, and 100 μM) for 3 h and anti-RGS4 immunoblotting.
We determined the effect of synthesized compounds on the proteolysis of other N-end rule substrates. We recently demonstrated that RGS4, a regulator of cardiovascular G protein signaling (9), is ubiquitylated through sequential N-terminal modifications, including oxidation and arginylation of N-terminal Cys-2, a process that yields N-terminal Arg (9, 14). RF-C11 efficiently inhibited ubiquitylation and degradation of this type 1 physiological substrate, whereas no effect was observed with GV-C11 or FF-C11. Moreover, the efficacy of RF-C11 was significantly higher than those of RR-C11 and Arg-Ala (Fig. 4B). We next determined whether RF-C11 affects non-N-end rule E3 pathways by using the replication licensing factor CDC6, a substrate of the APC–E3 complex (22). The full-length CDC6 was short-lived in reticulocyte lysates, and its degradation was inhibited by the proteasome inhibitor MG132, but not by RF-C11 (Fig. 4C). It has been shown that CDC6 can be cleaved by caspase 3 in mammalian cells to produce a C-terminal Asp-CDC6128−589 bearing N-terminal Asp (22). We found that Asp-CDC6128−589, produced by DUB-dependent cleavage of DHFRh-UbR48-Asp-CDC6, is rapidly degraded by ATE1-dependent N-end rule pathway in reticulocyte lysates (J.Y.A. and Y.T.K., unpublished data). Arg-CDC6128−589 (bearing N-terminal Arg, instead of Asp), but not Val-CDC6128−589, was rapidly degraded, and its degradation was virtually abolished by RF-C11 (Fig. 4C). Taken together, heterovalent interaction of RF-C11 to multiple N-recognins leads to strong inhibition of both types 1 and 2 N-end rule activities, with higher efficacy compared with homovalent compounds.
Heterovalent Compound RF-C11 Is a Potent Inhibitor of the N-End Rule Pathway.
Dipeptides with destabilizing N-terminal residues have been widely used as competitive inhibitors of N-recognins (13), but are highly unstable perhaps because of the cleavage by endopeptidases, hampering their application in mammalian cells. Having shown heterovalent inhibition of N-recognins, we compared the efficacy and stability of RF-C11 with those of known dipeptide inhibitors. The overall efficacy of RF-C11 for inhibiting the proteolysis of X-nsP4 and RGS4 was substantially higher, compared with any of types 1 and 2 dipeptides tested (Fig. 2 and SI Figs. 9–11). Notably, type 2 dipeptides Trp-Ala and Phe-Ala exhibited higher efficacy than type 1 dipeptides Arg-Ala and Lys-Ala, suggesting that N-recognins may generally have higher affinity to bulky hydrophobic amino acids than basic ones. We next characterized the stability of RF-C11 and dipeptides by monitoring X-nsP4 degradation in the presence or absence of the endopeptidase inhibitor bestatin (Fig. 4D). The inhibitory activity of Arg-Ala and Phe-Ala was almost completely abolished when bestatin was omitted in the reactions. In contrast, the efficacy of RF-C11 was not significantly affected by the presence or absence of bestatin. Given the higher stability of RF-C11 in the lysates, we next asked whether RF-C11 could inhibit the pathway in mammalian cells (Fig. 4E). RGS4 was rapidly degraded in mouse embryonic fibroblasts, and its degradation was significantly inhibited by RF-C11. In contrast, GV-C11 and dipeptides exhibited no significant effect on the stability of RGS4 (Fig. 4E). These results suggest that heterovalent RF-C11 is not only more potent than known homovalent inhibitors, but also the first inhibitor effective in mammalian cells.
Direct Interaction of RF-C11 to N-Recognins.
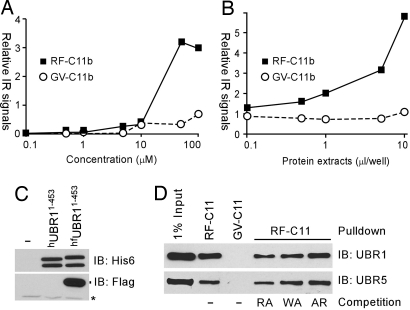
To determine whether RF-C11 directly targets N-recognins, we synthesized derivative compounds, RF-C11b and GV-C11b, where biotin was conjugated to the core Lys moiety (Fig. 1C and SI Fig. 12). Biotinylation did not significantly affect the inhibitory activities of synthesized compounds (data not shown). We then developed binding assays where hUBR11−453, an N-terminal His-6-tagged 52-kDa-UBR1 fragment containing the UBR box (Fig. 5C), was expressed in wheat germ lysates, immobilized onto microplates, and incubated with synthesized compounds. The ligand–target interaction was monitored by measuring streptavidin-infrared (IR) signals bound to the RF–C11b–UBR complex. We observed dose-dependent binding of RF-C11b, but not GV-C11b, to the immobilized UBR1 fragment (Fig. 5A); its binding was not significantly affected by 500 μM dipeptides (data not shown). RF–C11–UBR1 interaction was further confirmed by using reciprocal assays (Fig. 5B), where synthesized compounds were immobilized, incubated with hfUBR11−453 containing N-terminal His-6 and C-terminal FLAG tags (Fig. 5C), followed by incubation with anti-FLAG antibody and monitoring IR signals conjugated to anti-mouse IgG antibody. We next determined whether RF-C11 could pull down endogenous N-recognins from mammalian tissue extracts. Rat testes extracts were mixed with RF-C11b or GV-C11b conjugated to NeutrAvidin-coated beads, followed by Western blotting of precipitated proteins. Anti-UBR1 and UBR5 immunoblotting detected the presence of these N-recognins in precipitates by RF-C11b, but not by GV-C11b (Fig. 5D). The binding of RF-C11 to UBR1, which recognizes types 1 and 2 N termini, was partially inhibited by Arg-Ala and Trp-Ala, but not by Ala-Arg. In contrast, the binding of RF-C11 to UBR5, which preferentially binds to type 1 N termini, was significantly inhibited by Arg-Ala, but not by Trp-Ala or Ala-Arg (Fig. 5D). Our results demonstrate that RF-C11 directly binds to multiple N-recognins possibly through the UBR box.
Fig. 5.
RF-C11 interacts with N-recognins. (A) hUBR11−453 was immobilized onto Ni++-coated microplates and incubated with RF-C11b or GV-C11b, followed by monitoring streptavidin-IR signals. (B) RF-C11b or GV-C11b were immobilized onto streptavidin-coated microplates and incubated with hfUBR11−453, and anti-FLAG antibody-conjugated IR signals were measured. (C) hUBR11−453 and hfUBR11−453 were expressed in wheat germ extracts and subjected to anti-His-6 and FLAG immunoblotting. (D) RF-C11b can pull down endogenous N-recognins. RF-C11b or GV-C11b, conjugated to NeutrAvidin-coated microbeads, was incubated with rat testes extracts in the presence or absence of 2 mM dipeptides, followed by precipitation, washing, separation, and immunoblotting for UBR1 and UBR5.
RF-C11, but Not GV-C11, Inhibits Cardiac Proliferation and Hypertrophy.
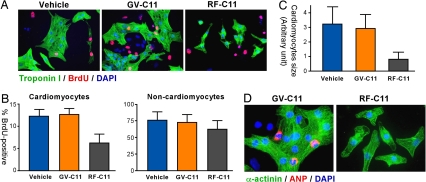
We previously showed that ATE1−/− and UBR1−/−UBR2−/− embryos are impaired in cardiac development and angiogenesis (7, 10) and the ATE1-UBR1/UBR2 circuit ubiquitylates RGS4, RGS5, and RGS16, regulators of cardiovascular G protein signaling (9), suggesting that the N-end rule pathway plays a role in cardiovascular development and signaling. However, it is unknown whether the pathway has a cell-autonomous function in cardiac proliferation. To address a possible role for N-recognins in cardiomyocyte proliferation, we cultured primary cardiac cells isolated from mouse embryonic hearts at embryonic day 13.5 (E13.5) and monitored the effect of RF-C11 on BrdU incorporation into newly synthesized DNA. Cardiac proliferation was measured by immunostaining cardiac troponin I specific for cardiomyocytes (green) and BrdU for cells in S-phase (red). The S-phase index of untreated cardiomyocytes was determined to be 12.1%, which was reduced to 6.3% by the treatment of 25 μM RF-C11. GV-C11 showed no significantly effects (Fig. 6 A and B). However, RF-C11 exhibited less significant effects on the proliferation of noncardiomyocytes. The effect of RF-C11 on the proliferation of cartdiomyocytes should not be due to cytotoxicity because their viability was not significantly affected by 25 μM RF-C11 and GV-C11 in 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) and TUNEL assays (data not shown).
Fig. 6.
RF-C11 inhibits the proliferation and hypertrophy of cardiomyocytes. (A) Cells from mouse embryonic hearts were treated with BrdU for 12 h in the presence of 25 μM compound (RF-C11 or GV-C11) or solvent alone (vehicle; 0.25% EtOH in PBS), followed by immunostaining troponin I (for cardiomyocytes, green) and BrdU (for S-phase cells, red). (B) BrdU indices were determined by counting BrdU-positive cells for cardiomyocytes (troponin I-positive) and noncardiomyocytes (troponin I-negative). At least 400 cells per group were counted for the BrdU incorporation assay. (B and C) Data represent mean ± SEM obtained from three independent experiments. (C) The surface areas of troponin I-positive cells were determined by using >1,000 cells per group by using ImageJ software version 1.34s (National Institutes of Health). (D) Cardiac cells treated with RF-C11 or GV-C11 were immunostained for sarcomeric α-actinin (for cardiomyocytes, green) and ANP (for hypertrophism, red) with DAPI counterstaining.
Notably, 16 h after the treatment of RF-C11, the surface area of troponin I-positive cardiomyocytes was found to be smaller, compared with untreated cells. GV-C11 did not induce a significant change (Fig. 6 A and C). Because embryonic cardiomyocytes in culture can undergo not only proliferation (cell division), but also hypertrophy (cell enlargement without cell division) (23), RF-C11-treated cardiomyocytes could have been impaired in hypertrophic response. To test this possibility, we determined the expression of atrial natriuretic peptide (ANP), a hallmark of genetic reprogramming during hypertrophy in cardiomyocytes and a prognostic indicator of clinical severity of hypertrophy (23). The treatment of 25 μM RF-C11 noticeably reduced the perinucleic expression of ANP, which was not observed in GV-C11-treated cells (Fig. 6D). In analogous assays using rat neonatal hearts, the treatment of RF-C11, but not GV-C11, significantly reduced the proliferation and cell size of cardiomyocytes (SI Fig. 13). These results indicate a role for N-recognins in cardiac proliferation and hypertrophy, unveiling a previously unknown function of the N-end rule pathway.
Discussion
In the current study, we synthesized a set of small-molecule compounds to study a heterovalent interaction to recognize E3 components of the N-end rule pathway. Our results (Figs. 2–5) collectively suggest that the model heterovalent compound inhibits types 1 and 2 N-end rule activities through selective, simultaneous, and cooperative binding to mutually exclusive, distinctive sites on multiple N-recognins. With appropriate control compounds, we demonstrate that a heterovalent interaction permits higher ligand-target affinity, compared with a homovalent interaction, providing a molecular basis for a heterovalent interaction to control a specific intracellular signaling pathway. The structural feature of RF-C11 (Fig. 1C) with interchangeable components (ligand, linker, core, and modifier) might be applied for heterovalent inhibitors to target other intracellular proteins containing two ligand-binding sites in the proximity. We also report that this prototype compound is more potent than dipeptide inhibitors and is the first inhibitor effective in mammalian cells (Figs. 2–4). Because dipeptides have been used in numerous studies for biochemical and physiological dissection of the pathway, our heterovalency-based compound could be used in some of these studies.
Our results suggest that the amino acid, Arg, connected to the hydrocarbon chain, a previously unknown type of ligand to N-recognins, has higher affinity than N-terminal Arg of the dipeptide Arg-Ala (Figs. 2A and 3A). We previously showed that UBR3 does not bind to destabilizing N-terminal residues, although it is similar to UBR1 and UBR2 in sequence, domains (including the UBR box), and specificity to E2 Ub-conjugating enzymes (12). Taken together, one function of the N-end rule pathway might be the ability of N-recognins to recognize small-molecule ligands (e.g., neurotransmitters, hormones, metabolic products, or drugs) whose structures are similar to destabilizing N-terminal residues. In this model, the binding of these ligands to N-recognins regulates the E3 activity toward a short-lived protein bearing an internal degron (I-degron). If the I-degron-dependent substrate is a regulator of the homeostasis of the bound ligand, the N-recognin–substrate–ligand pathway might form a molecular circuit controlling the homeostasis of N-ligand function, such as its biosynthesis, transport, and metabolism. We predict the presence of a variety of N-recognin-specific ligands that function through their structural similarity to destabilizing N-terminal residues.
We demonstrate that RF-C11 has the ability to inhibit cardiac proliferation and hypertrophy in cultured cardiomyocytes (Fig. 6). Cardiac hypertrophy is an adaptive response to various stresses mediated by specific proliferation pathways. Myocardial hypertrophy, associated with hypertension, cardiac valvular disease, or ischemia, is typically followed by serious myocardial diseases, accounting for the leading causes of death in Western society. One prominent pathway underlying cardiac proliferation and hypertrophy is Gq-dependent signaling, whose function is tightly controlled by RGS proteins (24). RGS proteins are GTPase-activating proteins for Gα subunits and negatively regulate the G protein-coupled receptor signaling (24). RGS4, RGS5, and RGS16 are known regulators of Gq-activated signaling critical for the control of myocardial growth and vascular maturation/integrity. Various studies implicated RGS4 in cardiac hypertrophy (24, 25). We previously showed that ATE1−/− and UBR1−/−UBR2−/− mouse embryos are impaired in cardiac development, and that the ATE1–UBR1/UBR2 pathway mediates the proteolysis of RGS4, RGS5, and RGS16 (7, 10). Taken together, our data (Fig. 6) suggest that the N-end rule pathway has a cell-autonomous function in the control of cardiac signaling through ubiquitylation of multiple cardiovascular regulators, including a set of RGS proteins. In contrast to other E3 systems that recognize substrates based on protein–protein interface, N-recognins recognize single amino acids as a degradation determinant, making them an attractive target for small-molecule inhibitors. As such, heterovalent inhibitors targeting recognition E3 components of the N-end rule pathway might be a useful tool to control certain pathophysiological conditions.
Materials and Methods
Synthesis and Characterization of RF-C11 and Related Compounds.
The synthesis and characterization of RF-C11 and related compounds are described in SI Materials and Methods and SI Figs. 7 and 12.
Protein Degradation Assays.
Test proteins were expressed and biotin-labeled, in the absence or presence of dipeptides or synthesized compounds, by using the TNT Quick Coupled Transcription/Translation System (Promega), followed by anti-biotin Western blotting (9, 26). As the metabolically stable Met-nsP4 accumulates rapidly during the first ≈30 min of the transcription-translation reaction and reaches a plateau at ≈30 min, the reaction (of a short-lived protein) beyond ≈30 min can be a version of chase in a pulse–chase experiment (26). Ubiquitylation of test proteins was similarly characterized in the presence of 5 μM MG132, followed by anti-Ub immunoprecipitation and anti-biotin Western blotting (9). IC50 was determined by using a sigmoidal dose-response model based on the amounts of X-nsP4, relative to the amounts of DHFR-Ub at the same time points and normalized to the amount of the same substrate treated with RF-C11 for 60 min.
Interaction Assays Between RF-C11 and N-Recognins.
The hUBR11−453 fragment with an N-terminal His-6 tag was expressed in wheat germ lysates by using Rapid Translation System (Roche) and immobilized onto Ni++-coated 96-well plates (Pierce), followed by incubation with RF-C11b or GV-C11b and monitoring biotin-bound streptavidin-IR signals by using the Odyssey Infrared Imaging System (LI-COR). Alternatively, RF-C11b or GV-C11b was immobilized on streptavidin-coated microwells and incubated with extracts expressing hfUBR11−453, with N-terminal His-6 and C-terminal FLAG tags, followed by incubation with anti-FLAG antibody and monitoring IR signals conjugated to anti-mouse IgG antibody.
Pulldown of Endogenous N-Recognins with RF-C11b.
The biotin moiety of RF-C11b and GV-C11b was cross-linked to UltraLink NeutrAvidin beads (Pierce). Rat testes extracts were subjected to pulldown assays in the presence or absence of 2 mM dipeptides as described (11). Precipitated proteins were subjected to immunoblotting for UBR1 (6) or UBR5 (11).
Proliferation and Hypertrophy of Cardiomyocytes.
Primary cardiac cells isolated from E13.5 mouse embryonic hearts were treated with 30 μM BrdU for 12 h in the presence or absence of 25 μM RF-C11 or GV-C11 as described (25), followed by immunostaining for BrdU and troponin I with DAPI counterstaining. Cardiomyocyte size was determined by the surface area of troponin I-positive cells by using ImageJ software 1.34s (National Institutes of Health).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Jai Wha Seo for genotyping and maintaining mouse strains; Barry Gold, Xiang Gao, Ramalinga Kuruba, and Sam Poloyac for critical discussions; members of the graduation committee of M.J.L. for helpful advice; and Susanne Mumby (University of Texas Southwestern, Dallas, TX) for the RGS4 antibody. This work was supported by a Council of Scientific and Industrial Research (India) doctoral fellowship (to K.P. and S.R); National Institutes of Health Grants GM69482, GM074000, and HL083365 (to Y.T.K.); the American Heart Association (Y.T.K.); the Departments of Science and Technology, India (R.B.); and the Department of Biotechnology, India (R.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708465105/DC1.
References
- 1.Bachmair A, Finley D, Varshavsky A. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon YT, Reiss Y, Fried VA, Hershko A, Yoon JK, Gonda DK, Sangan P, Copeland NG, Jenkins NA, Varshavsky A. Proc Natl Acad Sci USA. 1998;95:7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon YT, Kashina AS, Varshavsky A. Mol Cell Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon YT, Balogh SA, Davydov IV, Kashina AS, Yoon JK, Xie Y, Gaur A, Hyde L, Denenberg VH, Varshavsky A. Mol Cell Biol. 2000;20:4135–4148. doi: 10.1128/mcb.20.11.4135-4148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon YT, Xia Z, Davydov IV, Lecker SH, Varshavsky A. Mol Cell Biol. 2001;21:8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon YT, Kashina AS, Davydov IV, Hu R-G, An JY, Seo JW, Du F, Varshavsky A. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 8.Kwon YT, Xia ZX, An JY, Davydov IV, Seo JW, Xie Y, Varshavsky A. Mol Cell Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee MJ, Tasaki T, Moroi K, An JY, Kimura S, Davydov IV, Kwon YT. Proc Natl Acad Sci USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.An JY, Seo JW, Tasaki T, Lee MJ, Varshavsky A, Kwon YT. Proc Natl Acad Sci USA. 2006;103:6212–6217. doi: 10.1073/pnas.0601700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tasaki T, Mulder LCF, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tasaki T, Sohr R, Hellweg R, Hörtnagl H, Varshavsky A, Kwon YT. J Biol Chem. 2007;282:18510–18520. doi: 10.1074/jbc.M701894200. [DOI] [PubMed] [Google Scholar]
- 13.Tasaki T, Kwon YT. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Hu RG, Sheng J, Xin Q, Xu Z, Takahashi TT, Varshavsky A. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 15.Zenker M, Mayerle J, Lerch MM, Tagariello A, Zerres K, Durie PR, Beier M, Hülskamp G, Guzman C, Rehder H, et al. Nat Genet. 2005;37:1345–1350. doi: 10.1038/ng1681. [DOI] [PubMed] [Google Scholar]
- 16.Turner GC, Du F, Varshavsky A. Nature. 2000;405:579–583. doi: 10.1038/35014629. [DOI] [PubMed] [Google Scholar]
- 17.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Nature. 2001;410:955–960. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 18.Baker RT, Varshavsky A. Proc Natl Acad Sci USA. 1991;88:1090–1094. doi: 10.1073/pnas.88.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huskens J. Curr Opin Chem Biol. 2006;10:537–543. doi: 10.1016/j.cbpa.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 21.Kwon YT, Levy F, Varshavsky A. J Biol Chem. 1999;274:18135–18139. doi: 10.1074/jbc.274.25.18135. [DOI] [PubMed] [Google Scholar]
- 22.Pelizon C, d'Adda di Fagagna F, Farrace L, Laskey RA. EMBO Rep. 2002;3:780–784. doi: 10.1093/embo-reports/kvf161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu ZP, Olson EN. Proc Natl Acad Sci USA. 2002;99:2043–2048. doi: 10.1073/pnas.261708699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers JH, Tamirisa P, Kovacs A, Weinheimer C, Courtois M, Blumer KJ, Kelly DP, Muslin AJ. J Clin Invest. 1999;104:567–576. doi: 10.1172/JCI6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamirisa P, Blumer KJ, Muslin AJ. Circulation. 1999;99:441–447. doi: 10.1161/01.cir.99.3.441. [DOI] [PubMed] [Google Scholar]
- 26.Davydov IV, Varshavsky A. J Biol Chem. 2000;275:22931–22941. doi: 10.1074/jbc.M001605200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.