Prions are the infectious agents causing transmissible spongiform encephalopathies (TSEs), which comprise human Creutzfeldt–Jakob disease (CJD), scrapie of sheep, bovine spongiform encephalopathy (BSE), and several other rare ailments of various species. According to the protein-only hypothesis (1), prions are composed solely of PrPSc, a misfolded form of the cellular protein PrPC. PrPSc typically forms highly ordered fibrillary aggregates, also termed “amyloid.” The term “prion strain” denotes individual prion isolates sharing the same PrP sequence but giving rise to distinct, stable disease traits with different incubation periods and lesion profiles upon serial transmission in congenic hosts. The propagation of different strains in mice congenic with respect to their Prnp allelotypes is difficult to explain by the protein-only hypothesis because the epigenetic strain characteristics of prions appear to dominate over the primary prion protein sequence of the infected host (2, 3).
Circumstantial evidence suggests that strain phenotypes are encoded by distinct conformations of PrPSc (Fig. 1). This was first implied by experiments showing that distinct strains of transmissible mink encephalopathy went along with different protease-exposed sites within PrPSc (4). Great strides have been made since then, yet the final proof that conformational variants of PrPSc represent the biological basis of mammalian prion strains is still elusive. Distinct prion strains may bear highly divergent risks of transmission to humans: Sheep scrapie-derived strains may be mostly innocuous, whereas BSE-derived strains appear to induce variant CJD (vCJD) in humans. Also, two subtypes of sporadic CJD have been recently demonstrated to coexist in humans (5). Therefore, strain discrimination is not only a curious academic riddle but is also crucial for prion diagnostics and public health.
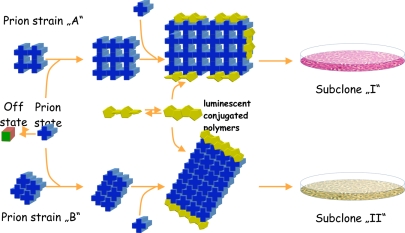
Fig. 1.
A model for prion strain propagation and detection. PrPc (green cube) exists in equilibrium with a misfolded monomeric isoform (blue cross). The latter can assemble into structurally heterogeneous, yet highly ordered aggregated forms (upper vs. lower assemblies) that replicate differentially in select cell lines. A panel of such lines, as provided by Weissmann and colleagues, may form the basis for classifying prions. When stained with luminescent-conjugated thiophene polymers, PrPSc aggregates stemming from distinct prion strains fluoresce in different colors.
Multiple TSE strains were historically distinguished by characteristic incubation periods in panels of differentially susceptible inbred mice (6). Different strains also differ in their capability to induce morphologically diverse aggregates ranging from tiny deposits to huge amyloid plaques (7), and they can target distinct brain regions (8, 9). Combinations of these methods were used to establish the uniqueness of the British BSE strain and its identity with the vCJD agent. However, strain determinations involving the inoculation of mice are unbearably slow and cumbersome and prohibitively expensive, and their reliability is based on merely correlative evidence (Table 1).
Table 1.
Synopsis of currently used prion strain differentiation assays
| Assay principle | Test substrate | Speed | Cost |
|---|---|---|---|
| Incubation period in indicator mice (6) | Mice | Years | +++ |
| Histological lesion profile (8, 9) | Mice | Years | +++ |
| Histoblots (24) | Immunohistology | Days | ++ |
| Conformation-dependent immunoassay (12) | ELISA | Days | + |
| Conformational stability assay (13) | Western blot | Days | ++ |
| PK cleavage site (4) | Western blot | Days | + |
| Detection with N-terminal antibodies (5) | Western blot | Days | + |
| Glycosylation profile on Western blot (11) | Western blot | Days | + |
| Amyloid detection by thioflavin and Congo red stains (7) | Histochemistry | Hours | + |
| Luminescent-conjugated polymers (21) | Histochemistry | Hours | + |
| Cell panel assay | Cell culture | Weeks | ++ |
Most assays sport high discriminatory power between few specific strains (e.g. glycosylation profiles for BSE and vCJD) but may perform poorly with other strains. +, low; ++, high; +++, extremely high.
Meanwhile, a number of biochemical correlates for prion strains have been discovered. PrPSc from distinct prion strains differs in electrophoretic mobility (10), immunoreactivity to amino-proximal antibodies after proteolysis (5), and relative glycoform prevalence (11). Also, the PrPSc-capturing efficacy of conformational antibodies (12) and the stability of PrPSc to heat and chaotropes (13) are to some extent strain-dependent (Table 1). None of the above phenomena is conclusively discriminatory, yet they suggest that the structure of PrPSc aggregates might define prion strains (14). Alternative explanations have been put forward, including e.g., differential binding to non-PrPSc components (15).
Yeasts carry self-propagating elements consisting of ordered protein aggregates that share traits with mammalian prions, including strains. The physical basis for yeast prion strains has been convincingly traced to the supramolecular assembly of the respective protein aggregates (16). By extension, the diversity of mammalian prion strains may plausibly reside within the conformational heterogeneity of PrPSc. Yet in the absence of definitive knowledge about its physical substrate, all strain differentiation methods, be they based on animal inoculations or biochemical analyses, must be regarded as surrogate markers.
In this situation, any tool enabling strain discrimination from a new angle is welcome. In a recent issue of PNAS, members of the laboratory of Charles Weissmann (17) describe a panel of cell lines selectively permissive to distinct scrapie prion substrains. Weissmann and colleagues have availed themselves of their previously described scrapie cell assay (18) to determine the efficiency with which four murine prion strains were propagated on four selected cell lines. Infectivity titers of the prion strain preparations, as measured by endpoint dilutions in susceptible animals, were almost identical, yet their “response index,” the reciprocal of the dilution that results in a given proportion of infected cells under defined assay conditions, varied considerably between strains. As a consequence, at least four strains can be clearly distinguished on the cell panel.
Unexpectedly, sibling subclones from a single cell line show surprising variable relative susceptibilities to individual strains. This provides a powerful tool for identifying factors controlling strain permissivity. What could the identity of such factors be? Prion replication may be intrinsically controlled by the thermodynamics of aggregation but may also be modulated by cell-specific chaperones specifying the folding of PrP, or by putative “disaggregases” akin to Hsp104 of yeast (19) or the DnaK/ClpB system of bacteria (20). Detailed comparisons of the cell lines of the proteomes of Weissmann and colleagues (17) should help clarify these questions.
If the strain-specific properties are enciphered by PrPSc and the geometry of PrPSc amyloid retains affords sufficient degrees of freedom, cerebral PrPSc deposits of prion-infected individuals may exhibit subtle structural idiosyncrasies that are private to distinct strains. This is confirmed by the observation that luminescent conjugated polymers (LCPs) fluoresce in distinct colors upon binding to PrPSc aggregates associated with various prion strains (21). The modulation of fluorescence is caused by the rotational freedom bestowed by the single bonds between the thiophene building blocks of LCPs. Binding to PrPSc fixates the thiophenes in planar, orthogonal, or intermediate orientations, thereby altering their photophysical properties. Artificially assembled fibrils of pure, recombinant PrP also display conformation-dependent spectra, establishing that LCPs provide valid measurements of the supramolecular geometry of PrPSc.
Taken together, the two studies discussed above add to the evidence that the host PrPSc structure is determined by both the PrPSc conformation within the inoculum and constraints imposed by host factors. It is easy to imagine that important knowledge could be generated by combining Weissmann and colleagues's cell panel (17) and LCP-based techniques.
Amyloid strains may be of broader significance than prions. Strain-like amyloid conformational variants may occur in Alzheimer's disease (AD) (22), suggesting that the pathogenetic mechanisms operating in AD and prion to diseases have more in common than typically appreciated (23). Ordered aggregation of proteins was also found to occur in most instances of type II diabetes, chronic inflammatory conditions, and many disorders of skeletal muscle. Therefore, a full understanding of the prion strain phenomenon may help with devising a sensitive diagnostic procedure, and possibly also rational therapies, of many aggregation proteinopathies. Some of the latter diseases rank among the most prevalent chronic ailments of mankind.
Footnotes
The author declares no conflict of interest.
See companion article on page 20908 in issue 52 of volume 104.
References
- 1.Prusiner SB. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Aguzzi A, Polymenidou M. Cell. 2004;116:313–327. doi: 10.1016/s0092-8674(03)01031-6. [DOI] [PubMed] [Google Scholar]
- 3.Aguzzi A, Heikenwalder M. Nature. 2003;423:127–129. doi: 10.1038/423127a. [DOI] [PubMed] [Google Scholar]
- 4.Bessen RA, Marsh RF. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polymenidou M, Stoeck K, Glatzel M, Vey M, Bellon A, Aguzzi A. Lancet Neurol. 2005;4:805–814. doi: 10.1016/S1474-4422(05)70225-8. [DOI] [PubMed] [Google Scholar]
- 6.Fraser H, Dickinson AG. J Comp Pathol. 1968;78:301–311. doi: 10.1016/0021-9975(68)90006-6. [DOI] [PubMed] [Google Scholar]
- 7.Sigurdson CJ, Manco G, Schwarz P, Liberski P, Hoover EA, Hornemann S, Polymenidou M, Miller MW, Glatzel M, Aguzzi A. J Virol. 2006;80:12303–12311. doi: 10.1128/JVI.01120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser H, Dickinson AG. J Comp Pathol. 1973;83:29–40. doi: 10.1016/0021-9975(73)90024-8. [DOI] [PubMed] [Google Scholar]
- 9.Bruce ME, McBride PA, Farquhar CF. Neurosci Lett. 1989;102:1–6. doi: 10.1016/0304-3940(89)90298-x. [DOI] [PubMed] [Google Scholar]
- 10.Bessen RA, Marsh RF. J Virol. 1992;66:2096–2101. doi: 10.1128/jvi.66.4.2096-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill AF, Joiner S, Beck JA, Campbell TA, Dickinson A, Poulter M, Wadsworth JD, Collinge J. Brain. 2006;129:676–685. doi: 10.1093/brain/awl013. [DOI] [PubMed] [Google Scholar]
- 12.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 13.Peretz D, Williamson RA, Legname G, Matsunaga Y, Vergara J, Burton DR, DeArmond SJ, Prusiner SB, Scott MR. Neuron. 2002;34:921–932. doi: 10.1016/s0896-6273(02)00726-2. [DOI] [PubMed] [Google Scholar]
- 14.Aguzzi A. Nat Cell Biol. 2004;6:290–292. doi: 10.1038/ncb0404-290. [DOI] [PubMed] [Google Scholar]
- 15.Weissmann C. Nature. 1991;352:679–683. doi: 10.1038/352679a0. [DOI] [PubMed] [Google Scholar]
- 16.Toyama BH, Kelly MJ, Gross JD, Weissman JS. Nature. 2007;449:233–239. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 17.Mahal SP, Baker CA, Demczyk CA, Smith EW, Julius C, Weissmann C. Proc Natl Acad Sci USA. 2007;104:20908–20913. doi: 10.1073/pnas.0710054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klohn PC, Stoltze L, Flechsig E, Enari M, Weissmann C. Proc Natl Acad Sci USA. 2003;100:11666–11671. doi: 10.1073/pnas.1834432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 20.Bukau B, Weissman J, Horwich A. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Sigurdson CJ, Nilsson KPR, Hornemann S, Manco G, Polymenidou M, Schwarz P, Hammarström P, Wüthrich K, Aguzzi A. Nat Methods. 2007;4:1023–1030. doi: 10.1038/nmeth1131. [DOI] [PubMed] [Google Scholar]
- 22.Meyer-Luehmann M, Coomaraswamy J, Bolmont T, Kaeser S, Schaefer C, Kilger E, Neuenschwander A, Abramowski D, Frey P, Jaton AL, et al. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 23.Aguzzi A, Haass C. Science. 2003;302:814–818. doi: 10.1126/science.1087348. [DOI] [PubMed] [Google Scholar]
- 24.Taraboulos A, Jendroska K, Serban D, Yang SL, DeArmond SJ, Prusiner SB. Proc Natl Acad Sci USA. 1992;89:7620–7624. doi: 10.1073/pnas.89.16.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]