Abstract
In contrast to classical chemical phenomenology, theory suggests that proteins may undergo downhill folding without an activation barrier under certain thermodynamic conditions. Recently, the BBL protein was proposed to fold by such a downhill scenario, but discrepancies between experimental results found in different groups argue against this. After briefly reviewing the major experimental studies of the BBL folding mechanism, we show that simulations of both coarse-grained and atomistic models can reconcile the seemingly conflicting observations.
Keywords: energy landscape theory, protein folding, downhill folding, electrostatics, shielding
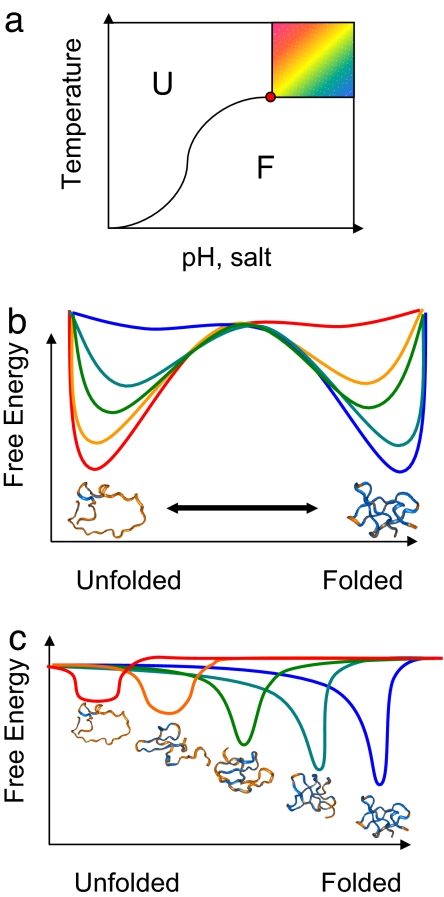
Evolution has selected sequences of natural proteins so that their energy landscape is globally funneled toward the native state (1–3). Nevertheless, to reach the native state, proteins generally must overcome at least one free energy barrier, giving rise to simple exponential kinetics for smaller proteins and encouraging the use of standard chemical kinetic phenomenology. Although the lack of symmetry between unfolded and folded ensembles generally allows such a barrier to exist, as in the nucleation of a liquid droplet from a vapor, the microscopic origin of the barrier is nontrivial (4–8). In analogy with the condensation of a liquid near a spinodal or critical point (Fig. 1a), under certain circumstances, it would seem possible that a protein may enter a regime in which the barrier disappears, giving no obvious separation of time scales (2). Such behavior is easily found in both on- and off-lattice model simulations of proteins. In the laboratory, near the midpoint of the thermodynamic transition, the folding free energy profile usually has two minima, separated by a barrier (Fig. 1b). To first order, changing the thermodynamic conditions simply biases the system toward the folded or unfolded well. Yet, for more extreme conditions, the protein may change the shapes of the wells, yielding a “critical point” or spinodal where the free energy barrier disappears, giving a single minimum (Fig. 1c). Now, the position of the minimum can be shifted from the folded state all of the way to the unfolded state in a continuous manner by tuning an external control variable such as temperature (2, 9). It is important to note that although theory suggests the possibility of downhill folding and although such behavior is seen in simulations of model systems, it may be the case that the environment of proteins cannot be perturbed enough to achieve it under physiologically relevant conditions (2). To thermally destabilize the denatured ensemble sufficiently may necessitate freezing of the surrounding water. It has been argued that the presence of barriers may not be a physicochemical necessity but may have evolved as an adaptation. A large folding barrier can guard against aggregation and proteolysis by making partially unfolded structures less populated (10–13). In keeping with this suggestion, recent surveys indicate that nearly downhill folding proteins, however, may not be as rare as has been thought (14). Nevertheless, further studies are necessary to determine whether these promising candidates are in fact truly downhill folders.
Fig. 1.
Schematic diagram and free energy profiles for protein folding. (a) Phase diagram consisting of a first-order transition between the folded and unfolded states, which corresponds to a two-state folding behavior. This regime continues until the critical point (red circle), after which the separation between the two states is not sharp, corresponding to a downhill folding mechanism. (b) Free energy profile for a two-state folding mechanism with two minima corresponding to the unfolded and folded states. (c) Free energy profiles for a downhill folding mechanism at different temperatures (red, high; blue, low), where each profile has a single minimum.
By illuminating the origin of folding barriers in its strictest form, searching for downhill folders is potentially relevant to understanding protein folding mechanisms in general. In addition, it has been emphasized that probing downhill folding proteins could, in principle, allow direct experimental access to the entire spectrum of states between the folded and unfolded states by continuously changing the position of the free energy minimum, whereas this direct view is hardly possible for strictly two-state systems (9, 15). Studying downhill folders is not easy, however, because they should fold near the diffusion-limited rate approximately on the order of 1 μs. Nevertheless, fast-folding experiments have given evidence for downhill folding in some proteins artificially designed to have small and weak hydrophobic cores such as the engineered mutants of the already fast-folding lambda repressor protein. This engineered system showed nonexponential kinetics and exhibited a broad, noncooperative folding transition as measured by several spectroscopic probes (16, 17).
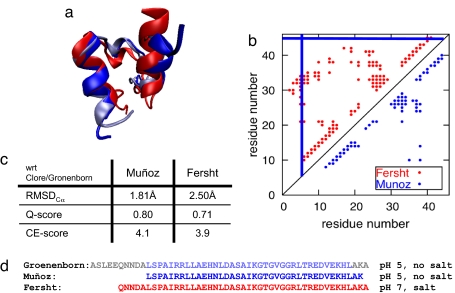
In contrast to such deliberately engineered systems, it has been hotly debated recently in the literature whether downhill folding occurs for natural proteins. Experimental studies of BBL by the Muñoz and Fersht groups have excited much work on this important question (18–21). BBL is an ≈40-residue, independently folding domain excised from the much larger 2-oxoglutarate dehydrogenase multienzyme complex (22). The domain is flanked by long sequences of unstructured residues. Clore, Gronenborn, and coworkers (23) isolated and resolved the structure of BBL at pH 5.3 using solution NMR. Although their protein construct contained 50 residues, only 37 were found to be structured (Fig. 2d). The final structure consists of two parallel α-helices separated by a long partially structured loop (Fig. 2a).
Fig. 2.
Comparisons of independently determined NMR structures for variant protein constructs of BBL. (a) Superimposed ribbon diagrams of the NMR structures of BBL as resolved by the groups of Clore and Gronenborn (light blue), Muñoz (blue), and Fersht (red). (b) Contact map of the Muñoz (blue) and Fersht (red) structures with a distance cutoff of 9.0 Å. The solid blue lines indicate where the Muñoz structure was truncated. (c) Table of the Cα-RMSD, Q-score, and CE-scores of the Muñoz and Fersht structures with respect to the Clore/Gronenborn structure. (d) Sequences of the structured residues in the NMR structures of the protein constructs obtained by the groups of Clore and Gronenborn (light blue, structured; gray, unstructured), Muñoz (blue), and Fersht (red).
Inspired by the experimental work, the groups of Chan (24) and Wang (25) performed purely additive Go-model simulations of BBL using the Clore/Gronenborn NMR structure (Protein Data Bank entry 1BBL). Their simulations gave no barrier. They also showed a broad specific heat vs. temperature profile with low cooperativity. However, it is known that pairwise additive native structure-based models underestimate free energy barrier heights (26, 27). Normally, one must incorporate nonadditivity into Go models to obtain realistic estimates of the free energy barrier heights. Even so, Chan et al. (24) argued that the lack of cooperativity observed in the folding of BBL is related to the low contact density per residue of the protein native topology (24), a feature we confirm. Wang et al. (25) also attributed the downhill folding behavior they found for the Clore/Gronenborn construct of BBL to an absence of long-range interactions in the native topology present in other two-state folding proteins.
In their work indicating that BBL is a downhill folding protein, Muñoz and coworkers (18) studied its folding using differential scanning calorimetry (DSC), far-UV (UV) circular dichroism (CD), and fluorescence resonance energy transfer (FRET) experiments. Their protein construct had 39 residues (Fig. 2d), which included the entire structured region found by NMR in the Clore/Gronenborn study (Fig. 2d). Because the BBL construct lacks a natural fluorophore, they introduced a naphthyl-alanine by substituting an alanine at the N terminus (Naf-BBL). Because the labels are not stable at neutral pH, the labeled construct was studied at pH 5.3 without salt, as was done for the Clore/Gronenborn unlabeled protein. Solution NMR of this labeled form resulted in a structure in which all 39 residues were structured. The determined structure closely resembles the Clore/Gronenborn result both in secondary and tertiary structures. There is an RMSD of 1.8 Å between the labeled and unlabeled forms (Fig. 2c). The DSC experiments resulted in a broad unfolding temperature range, from ≈280 to 345 K, with a maximum at ≈322 K (18). The far-UV CD spectrum is consistent at low temperatures with that of an α-helical structure with random coils but at high temperatures is characteristic of a denatured protein. Between the temperature extremes, a gradual, continuous shift from the α-helical profile to the denatured profile is observed that is superficially consistent with downhill folding behavior. Nevertheless, an isodichroic point is seen at ≈203 nm, possibly indicating some cooperative behavior, but the degree of cooperativity is unclear. Recently, Muñoz and coworkers reported the denaturation curves for individual protons of 158 backbone and side chain protons (out of 204 possible) with about two-thirds resulting in a sigmoidal curve. The lack of synchrony of these curves seems to indicate that parts of the protein do not fold (or unfold) cooperatively with other parts of the protein (21).
Independent of the Muñoz work, Fersht and coworkers probed the folding of a construct of BBL without labels, and they came to different conclusions from the Muñoz group. Their construct included four additional residues at the N terminus plus one additional residue at the C terminus, as compared with the Muñoz construct (19, 20). Furthermore, the structure of this modestly longer unlabeled construct was determined at pH 7 under high-salt conditions. The resulting structure has many parallels to the structures of the Clore/Gronenborn and Muñoz constructs. Like those results, their protein construct consists of two α-helices separated by a long partially structured loop, but the Fersht structure is significantly more compact than the others (Fig. 2a). The thermal denaturation of this BBL construct was also studied by DSC, far-UV CD, 1H-NMR, and 13C-NMR. The heat capacity peak is reported to be narrower than that obtained for the Muñoz construct, and the maximum, corresponding to the melting temperature, is located at 328 K, which is somewhat greater than the 322 K measured for the Muñoz construct. Furthermore, the thermal denaturation curves of four backbone and two side chain carbons as measured by 13C-NMR were fitted to a two-state transition with a melting temperature of 324–329 K. We see that the cooperative behavior and melting temperature of the unlabeled and longer BBL as observed by Fersht and coworkers using multiple, independent experimental approaches contrasts with the findings of the labeled, shorter variant of BBL observed by Muñoz and coworkers. The increased stability of the larger construct suggests a higher free energy barrier for folding than does the Muñoz study and points to a two-state folding mechanism.
The differences between the constructs and the experimental protocols studied by the two experimental groups are so small that ordinarily they would be considered essentially the same protein. Yet, for the question of downhill folding, there are key differences, which we shall see can be sufficient to explain the discrepancies in their findings. First, the sequence lengths of the two protein constructs were different. The pH and ionic strength were also different in the two investigations. Furthermore, the shorter molecule contained relatively bulky fluorescent probes, whereas the construct studied by Fersht and coworkers did not. Although one may be tempted to think of these differences as negligible, because of the sparse three-dimensional connectivity of the protein, they may play a significant role in the folding mechanism. We will also show that the published structures differ precisely in the region that gives rise to the main bottleneck for folding. Indeed, although the secondary structures of the two are largely the same, the tertiary structures are different by multiple metrics (Fig. 2c). The Fersht structure is more compact than the Muñoz structure (Fig. 2a), resulting in far more long range interactions in the Fersht structure (Fig. 2b), and these seem to provide the extra cooperativity. Different refinement methods were used by the two groups, but it is not clear that this could account for these structural differences. Our simulations suggest that the differences may actually be due to the different solvent environments. Independent experimental studies of two different variants of BBL under different conditions do suggest different folding mechanisms and apparently result in modestly different structures of BBL.
Results and Discussion
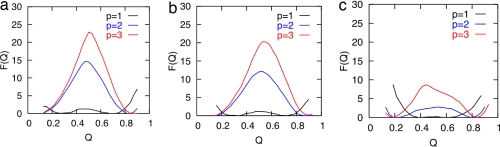
To explore whether the differences in the protein structures could lead to significantly different folding mechanisms, we began by performing purely additive native structure-based simulations for the full-length Fersht and Muñoz structures, as well as a truncated Fersht structure in which the sequence is shortened as in the Muñoz construct. In every case, the pairwise additive Go-model predicts a very low free energy barrier height (Fig. 3). Both the full-length Fersht and the truncated Fersht structures yield free energy barriers of ≈2 kBT, whereas the profile for the Muñoz structure has a truly negligible barrier.
Fig. 3.
Free energy profiles calculated from Go-model simulations using the different structures of BBL with varying degrees of nonadditivity. The free energy profiles for the full-length Fersht (a), truncated-Fersht (b), and Muñoz (c) constructs are shown for varying values of p.
Native topology-based simulations using the pairwise additive model underestimate the free energy barrier heights in many cases when compared with experiments (26, 27), sometimes giving no free energy barriers even when relatively slow folding rates are observed in the laboratory. Plotkin and coworkers (27) found that adding only ≈20–30% nonadditivity (i.e., cooperativity) to account for the side chains and solvent interactions that are absent in the purely additive models generally improves agreement of simulations with experimentally observed absolute folding rates and Φ-values. Following the protocol of Eastwood and Wolynes (26), we supplemented the purely additive Go model with many-body interactions. In this approach, there exists a continuous adjustable parameter, p, which controls the amount of nonadditivity added to the purely additive Hamiltonian. A p value of 1 is a purely additive Hamiltonian, and a p value of 2 gives a Hamiltonian having both two- and three-body interactions. The simulation free energy barriers for all three protein constructs increase with increasing nonadditivity. Interestingly, the barrier height increases much more rapidly for the Fersht construct as p is increased than it does for the Muñoz protein construct. The difference in the barriers becomes >14 kBT for p = 3 (Fig. 3).
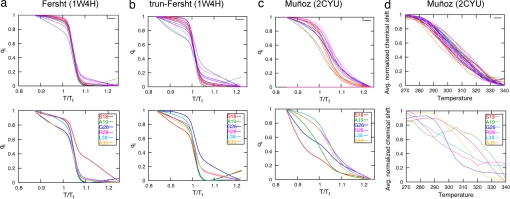
Whereas the range of nonadditivity that best reflects normal experimental thermodynamic conditions is known (≈20–30%), a more precise value can be estimated by benchmarking structural observables of the folding mechanism (e.g., Φ-values) from our simulations against those from experiments (27). Because no complete Φ-value analysis is yet available for any construct of BBL, we estimated the appropriate amount of nonadditivity in our model by comparing the temperature dependence of the folding of individual residues in the simulations with the melting curves from the 1H-NMR experiments of the Muñoz group (21). Although they observed that a large majority of the residues in their construct do fold synchronously, there were several notable exceptions to the synchrony of melting for individual residues (Fig. 4d). At a p value of 1.3, we observed in our simulations of the Muñoz construct that the outlier residues that the simulation suggests fold dissimilarly from the majority are the same as the outliers found in experiment (Fig. 4c). Furthermore, the predicted dispersion (i.e., slopes) of the melting profiles is qualitatively consistent with the experiments. In our view, this constitutes a remarkable agreement of the simulations with the site-specific experimental results given that one introduces only a single adjustable parameter into the Hamiltonian. In contrast, for the same p value, simulations of the full-length Fersht and truncated-Fersht constructs find that every single residue folds cooperatively. Clearly, truncation of the construct alone is not the origin of the change in the folding mechanism. In any case, the melting profiles from simulations with nonadditivity based on the Fersht structure are in good agreement with the cooperative 13C-NMR thermal denaturation curves of BBL observed by the Fersht group (see ref. 19). At this value of p, the simulations of full-length Fersht and truncated Fersht structures yielded free energy barriers of ≈4 kBT, whereas the free energy barrier predicted for the Muñoz structure is <0.5 kBT. Therefore, by incorporating nonadditivity in a plausible way, our models predict a higher cooperativity in the folding mechanism for the Fersht construct than for the Muñoz construct, but this difference relies on their observed structural differences at key locations.
Fig. 4.
Residue melting profiles of BBL. The qi vs. temperature of residues in simulations for the BBL constructs of the Fersht (a), truncated-Fersht (b), and Muñoz (c) structures. Also shown for comparison are the 1H-NMR profiles obtained by Muñoz and coworkers for their construct. (Upper) Residues whose profiles are typical 1H-NMR profiles. (Lower) Residues whose profiles are dissimilar as compared with the typical profiles.
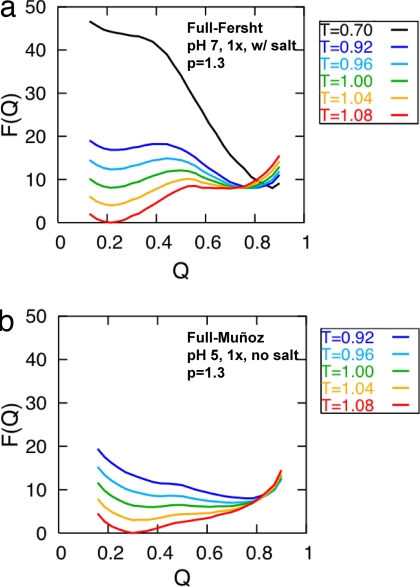
Because the Fersht and Muñoz constructs were studied under different conditions, we further supplemented our pure native contact-only models with models that also treat long-range electrostatics by incorporating crudely the effects of pH and salts. For each structure, the addition of electrostatics to the basic models does not significantly perturb the barrier heights at pH 7, even when we unrealistically increase the strength of the electrostatic contribution by 3-fold [see supporting information (SI) Fig. 7 d–f]. In addition, when shielding effects are included in our simulations, the changes in the free energy profile are modest (SI Fig. 7 a–c). Larger differences are seen when the simulations are performed at pH 5 but only when the electrostatic contributions are very large (SI Fig. 7 g–l). Clearly, this coarse-grained treatment of the electrostatics and shielding effects does not overcome the native tertiary interactions postulated in the native topology-based models.
We see that the rather modest tertiary structural differences between the Fersht and Muñoz constructs seem to be the origin of the discrepancy in whether downhill behavior applies. These differences are precisely located at the specific cluster of residues that through many-body interactions leads to the main folding barrier in the simulation models. As we discussed above, the main difference between the contact maps of the Fersht and Muñoz structures is the presence of long-range native contacts in the Fersht structure that are absent in the Muñoz structure (Fig. 2b). A careful analysis of the nuclear Overhauser effects (NOEs) in the Fersht structure shows that the NOEs associated with the sixth and eighth residues are relatively weakest (unpublished data). Those residues correspond to the first and third residues at the N-terminal of the Muñoz sequence. To test whether these contacts are the crucial ones for the barrier, we studied a model in which we eliminated the long-range contacts from the first three residues of the truncated Fersht structure (corresponding to the sixth to eighth residues in the full-length Fersht sequence), and we performed Go-model simulations with electrostatics. Now, we do see a significantly smaller free energy barrier in the truncated Fersht simulations (SI Fig. 8b) as compared with the free energy profile of the full-length Fersht simulations (SI Fig. 8a), although it is not as small as that of the free energy profile of the Muñoz simulations (SI Fig. 8c). The influence of electrostatics truncation and of deleting the contacts on the barrier is clearly far greater than the effect of simply truncating the sequence.
We also investigated the sensitivity of temperature to the free energy barriers for folding by extrapolating the free energy profile over a range of temperatures. In the case of the full-length Fersht construct simulations, the presence of a barrier persisted for a broad temperature range, and a downhill folding scenario is reached only at a relatively low temperature, below ≈0.7 Tf (Fig. 5a). Because the experimentally determined folding (or melting) temperature of the Fersht construct at pH 7 and high-salt conditions is 328 K (19, 20), if the hydrophobic forces did not weaken upon cooling, the Fersht construct would be predicted by our simulations to fold by a downhill scenario only at ≈230 K, which is an upper limit. Of course, the protein would be effectively immobilized at that temperature because of the freezing of the surrounding waters, and, accordingly, downhill folding would not be observed under physiologically relevant temperatures in the laboratory. In contrast, the miniscule free energy barrier of the Muñoz construct simulations disappears completely with only a minor change in temperature (Fig. 5b). Therefore, our simulations strongly suggest that the Muñoz construct is indeed a downhill folder under physiologically relevant temperatures, whereas the Fersht construct is not.
Fig. 5.
Free energy profiles calculated from Go models at different temperatures. Shown are the free energy profiles for the full-length Fersht construct at pH 7 with salt (a) and Muñoz construct at pH 5 without salt (b) for p = 1.3.
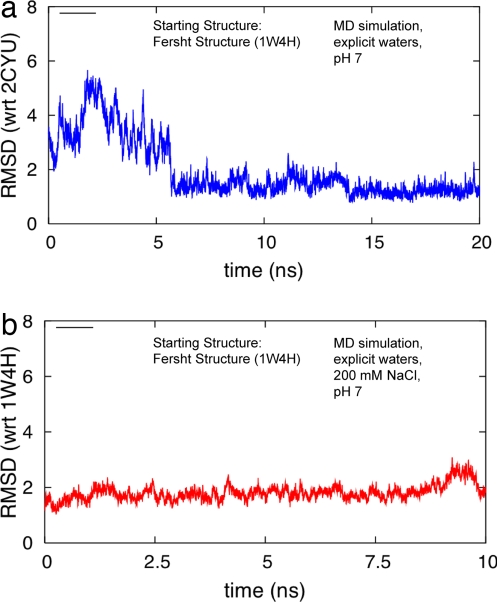
Is there a microscopic reason for the different structures found by the two groups? To try to answer this, we complemented our native structure-based simulations by performing all-atom empirical force field-based simulations. Normally, such simulations have difficulty probing a large range of conformations for folding studies owing to their computational cost, but we are most interested in exploring near the native basin. We thus performed two explicit water simulations: one with no salt (Fig. 6a) and one with ≈200 mM NaCl (Fig. 6b). In each simulation, the starting structure corresponded to the full-length structure found by the Fersht group, and the protonation state of the residues corresponds to pH 7. In the first simulation (Fig. 6a), corresponding to pH 7 and no salt, the structure undergoes large structural changes. Under these conditions, within ≈6 ns, the Fersht construct rearranges its configuration to closely correspond to the one found by the Muñoz group with Cα-RMSD < 2.0 Å with respect to the starting Fersht structure for the residues that are in common. This structure persisted throughout the rest of the simulation. We must note that because this is a single trajectory, resulting in a poor sampling of the conformational space, it must be taken with a grain of salt. We do not expect that all trajectories would result in the same outcome, and the observation of such a transition is clearly fortuitous. However, because no information regarding the Muñoz structure was provided to the simulation a priori, it is remarkable that this new native basin was indeed found and persisted for well over 10 ns.
Fig. 6.
All-atom simulations of BBL. (a) Cα-RMSD of BBL with respect to the native Muñoz structure during a 20-ns explicit water, pH 7 simulation of BBL starting with the native Fersht structure. (b) Cα-RMSD of BBL with respect to the native Fersht structure during a 20-ns 200 mM NaCl explicit water simulation of BBL starting with the native Fersht structure.
Because the Fersht construct was studied under higher salt, we repeated the simulation starting with the Fersht structure, except that now the simulation was performed at 200 mM NaCl (Fig. 6b). In this case, the Fersht construct structure was largely stable with the Cα-RMSD remaining largely less than 2.0 Å with respect to the starting Fersht structure for the 10-ns simulation. Apparently, far longer all-atom simulations would need to be performed to preclude the existence of a distant stable basin corresponding to the Muñoz structure under these conditions, but it is encouraging that, in contrast to the low-salt simulation, which converged to Muñoz's low-salt structure, the structure found by the Fersht laboratory is clearly stable for the duration of our simulation at the higher salt conditions used by them.
Conclusions
By combining both experimental and theoretical perspectives, much deeper explorations of protein folding mechanisms are now possible than years ago. Although the basic nature of protein folding has now come into focus, there is still much we have yet to understand, as the BBL story indicates. The experimental groups of Fersht and Muñoz studied the BBL protein from independent viewpoints, and they arrived at different pictures of its folding mechanism. Their meticulous and nearly exhaustive efforts yielding seemingly contradictory interpretations presented a great opportunity for theoretical studies. The present theoretical study cannot decisively classify any BBL construct as a bona fide downhill folding protein with absolutely no barrier. Nevertheless, consistent with the observation in the Muñoz laboratory, the Muñoz construct of BBL exhibits in our simulations the expected behavior of a downhill folding protein. In particular, the free energy barrier height from simulations of the Muñoz construct of BBL is exceedingly small (≈0.5 kBT) and well under the 3 kBT estimate by Gruebele and coworkers (28) of the beginnings of the downhill folding regime. At the same time, the simulations based on the structure of the Fersht construct exhibit a notably more cooperative behavior, in good agreement with the observation made in the Fersht laboratory, with barrier heights of ≈4 kBT. These simulations strongly suggest that the source of the discrepancy seen in seemingly conflicting observations of the folding behavior of the BBL variants lies in the fine differences in the native basin of BBL (and therefore the funneled landscape) under different solvent conditions that result in different levels of cooperativity in the folding mechanism. This sensitivity is the result of the subtle origins of the folding barrier in nonadditive forces, in this case, coming from a mere handful of tertiary contacts.
Materials and Methods
Go-Model Simulations.
To perform folding simulations of the BBL constructs, we supplemented the basic Go-model Hamiltonian with electrostatic effects. The full Hamiltonian used in our study is
where the first two terms comprise the basic Go-model Hamiltonian (26, 29). The next two terms describe the electrostatic contributions from the charged residues at different pH. The final collapse term accounts for general hydrophobic collapse. A detailed description of the full Hamiltonian is presented in SI Methods. The free energy profiles were generated with the weighted histogram analysis method (WHAM), using the fraction of structural overlap, Q, as an order parameter (30).
Empirical Force Field-Based Simulations.
To explore the dynamics of the Fersht BBL protein construct at a microscopic level, we performed explicit solvent molecular dynamics simulations with a standard protocol, which we describe in detail in SI Methods. The starting coordinates were obtained from the lowest-energy structure of the published NMR ensemble (19). Two simulations were performed, one in which the solvent was pure water and another in which the solvent was composed of ≈200 mM NaCl.
Supplementary Material
ACKNOWLEDGMENTS.
We are grateful for fruitful discussions with Alan Fersht and Victor Muñoz that inspired us and led us to carefully examine the structural differences between their BBL constructs. We thank Bill Eaton, Martin Gruebele, and José Onuchic for a careful reading of the manuscript, and Charlie Brooks and Vijay Pande for preliminary advice on setting up and running the empirical force field-based MD simulations. This work was supported by National Institutes of Health Grant 5R01 GM44557 and the National Science Foundation-sponsored (Grants PHY-0216576 and PHY-0225630) Center for Theoretical Biological Physics. S.S.C. is supported by a Ruth L. Kirschstein National Research Service Award from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709376104/DC1.
References
- 1.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Bryngelson JD, et al. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 3.Oliveberg M, Wolynes PG. The experimental survey of protein-folding energy landscapes. Q Rev Biophys. 2005;38:245–288. doi: 10.1017/S0033583506004185. [DOI] [PubMed] [Google Scholar]
- 4.Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 5.Wolynes PG. In: Three Paradoxes of Protein Folding, in Protein folds: A Distances Based Approach. Bohr H, Brunak S, editors. Boca Raton, FL: CRC; 1996. pp. 3–17. [Google Scholar]
- 6.Plotkin SS, Wang J, Wolynes PG. Statistical mechanics of a correlated energy landscape model for protein folding funnels. J Chem Phys. 1997;106:2932–2948. [Google Scholar]
- 7.Takada S, Wolynes PG. Microscopic theory of critical folding nuclei and reconfiguration activation barriers in folding proteins. J Chem Phys. 1997;107:9585–9598. [Google Scholar]
- 8.Qi X, Portman JJ. Excluded volume, local structural cooperativity, and the polymer physics of protein folding rates. Proc Natl Acad Sci USA. 2007;104:10841–10846. doi: 10.1073/pnas.0609321104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muñoz V. Thermodynamics and kinetics of downhill protein folding investigated with a simple statistical mechanical model. Int J Quant Chem. 2002;90:11311–11316. [Google Scholar]
- 10.Schindler T, et al. Extremely rapid protein folding in the absence of intermediates. Nat Struct Biol. 1995;2:663–673. doi: 10.1038/nsb0895-663. [DOI] [PubMed] [Google Scholar]
- 11.Otzen DE, Kristensen O, Oliveberg M. Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: a structural clue to amyloid assembly. Proc Natl Acad Sci USA. 2000;97:9907–9912. doi: 10.1073/pnas.160086297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobson CM. Experimental investigation of protein folding and misfolding. Methods. 2004;34:4–14. doi: 10.1016/j.ymeth.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Gruebele M. Downhill protein folding: evolution meets physics. C R Biol. 2005;328:701–712. doi: 10.1016/j.crvi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Naganathan AN, Doshi U, Muñoz V. Protein folding kinetics: Barrier effects in chemical and thermal denaturation experiments. J Am Chem Soc. 2007;129:5673–5682. doi: 10.1021/ja0689740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubelka J, Hofrichter J, Eaton WA. The protein folding “speed limit.”. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Yang WY, Gruebele M. Folding lambda-repressor at its speed limit. Biophys J. 2004;87:596–608. doi: 10.1529/biophysj.103.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, Gruebele M. Tuning lambda6–85 towards downhill folding at its melting temperature. J Mol Biol. 2007;370:574–584. doi: 10.1016/j.jmb.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Mira MM, et al. Experimental identification of downhill protein folding. Science. 2002;298:2191–2195. doi: 10.1126/science.1077809. [DOI] [PubMed] [Google Scholar]
- 19.Ferguson N, et al. One-state downhill versus conventional protein folding. J Mol Biol. 2004;344:295–301. doi: 10.1016/j.jmb.2004.09.069. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson N, et al. Ultra-fast barrier-limited folding in the peripheral subunit-binding domain family. J Mol Biol. 2005;353:427–446. doi: 10.1016/j.jmb.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 21.Sadqi M, Fushman D, Muñoz V. Atom-by-atom analysis of global downhill protein folding. Nature. 2006;442:317–321. doi: 10.1038/nature04859. [DOI] [PubMed] [Google Scholar]
- 22.Packman LC, Perham RN. Chain folding in the dihydrolipoyl acyltransferase components of the 2-oxo-acid dehydrogenase complexes from Escherichia coli—identification of a segment involved in binding the E3 subunit. FEBS Lett. 1986;206:193–198. doi: 10.1016/0014-5793(86)80979-6. [DOI] [PubMed] [Google Scholar]
- 23.Robien MA, et al. 3-Dimensional solution structure of the E3-binding domain of the dihydrolipoamide succinyl transferase core from the 2-oxoglutarate dehydrogenase multienzyme complex of Escherichia coli. Biochemistry. 1992;31:3463–3471. doi: 10.1021/bi00128a021. [DOI] [PubMed] [Google Scholar]
- 24.Knott M, Chan HS. Criteria for downhill protein folding: calorimetry, chevron plot, kinetic relaxation, and single-molecule radius of gyration in chain models with subdued degrees of cooperativity. Proteins Struct Funct Bioinformatics. 2006;65:373–391. doi: 10.1002/prot.21066. [DOI] [PubMed] [Google Scholar]
- 25.Zuo GH, Wang J, Wang W. Folding with downhill behavior and low cooperativity of proteins. Proteins Struct Funct Bioinformatics. 2006;63:165–173. doi: 10.1002/prot.20857. [DOI] [PubMed] [Google Scholar]
- 26.Eastwood MP, Wolynes PG. Role of explicitly cooperative interactions in protein folding funnels: a simulation study. J Chem Phys. 2001;114:4702–4716. [Google Scholar]
- 27.Ejtehadi MR, Avall SP, Plotkin SS. Three-body interactions improve the prediction of rate and mechanism in protein folding models. Proc Natl Acad Sci USA. 2004;101:15088–15093. doi: 10.1073/pnas.0403486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 29.Weinkam P, Zong CH, Wolynes PG. A funneled energy landscape for cytochrome c directly predicts the sequential folding route inferred from hydrogen exchange experiments. Proc Natl Acad Sci USA. 2005;102:12401–12406. doi: 10.1073/pnas.0505274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho SS, Levy Y, Wolynes PG. P versus Q: Structural reaction coordinates capture protein folding on smooth landscapes. Proc Natl Acad Sci USA. 2006;103:586–591. doi: 10.1073/pnas.0509768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.