Abstract
The detection of ligand-binding sites is often the starting point for protein function identification and drug discovery. Because of inaccuracies in predicted protein structures, extant binding pocket-detection methods are limited to experimentally solved structures. Here, FINDSITE, a method for ligand-binding site prediction and functional annotation based on binding-site similarity across groups of weakly homologous template structures identified from threading, is described. For crystal structures, considering a cutoff distance of 4 Å as the hit criterion, the success rate is 70.9% for identifying the best of top five predicted ligand-binding sites with a ranking accuracy of 76.0%. Both high prediction accuracy and ability to correctly rank identified binding sites are sustained when approximate protein models (<35% sequence identity to the closest template structure) are used, showing a 67.3% success rate with 75.5% ranking accuracy. In practice, FINDSITE tolerates structural inaccuracies in protein models up to a rmsd from the crystal structure of 8–10 Å. This is because analysis of weakly homologous protein models reveals that about half have a rmsd from the native binding site <2 Å. Furthermore, the chemical properties of template-bound ligands can be used to select ligand templates associated with the binding site. In most cases, FINDSITE can accurately assign a molecular function to the protein model.
Keywords: pocket detection, protein structure prediction, ligand screening
To date, although the genomes of >500 organisms have been sequenced (1, 2), the biological function of many identified genes/gene products is unknown. This rapid accumulation of protein sequences of unknown structure and function has motivated the development of proteome-scale protocols for protein structure and function prediction (3–5). The detection of ligand-binding sites is often a starting point for structure-based function identification. Knowledge of the ligand-binding site is also essential for structure-based drug discovery (6). Existing approaches for ligand-binding site prediction can be roughly divided into sequence- and structure-based methods (see refs. 6 and 7). The main strength of sequence-based methods is their ability to identify a ligand-binding/interaction motif in proteins that may not have the same overall fold. However, motif-based searches frequently become ineffective if the binding region is nonlocal in sequence. Homology-based methods require related proteins with significant sequence identity to a protein in the Protein Data Bank (PDB) (8, 9) because the conservation of biochemical function drops rapidly for proteins sharing <35–40% sequence identity (10, 11). In that regard, a number of structure-based approaches have been developed to identify ligand-binding sites (6). Geometry-based methods locate binding residues by searching for cavities/pockets in a protein structure (12–15). Other methods consider blind docking of small molecules into the receptor structure (16, 17), calculate theoretical microscopic titration curves (18), or identify electrostatically destabilized residues (19). Finally, analysis of the spatial hydrophobicity distribution can identify sites on the protein surface involved in ligand binding (20). Among the best of these pocket-detection algorithms is the recently developed LIGSITECSC (14), an extension and implementation of LIGSITE (13). LIGSITECSC calculates surface accessibility for the protein's Connolly surface (21) and then reranks the identified pockets by the degree of conservation of identified surface residues.
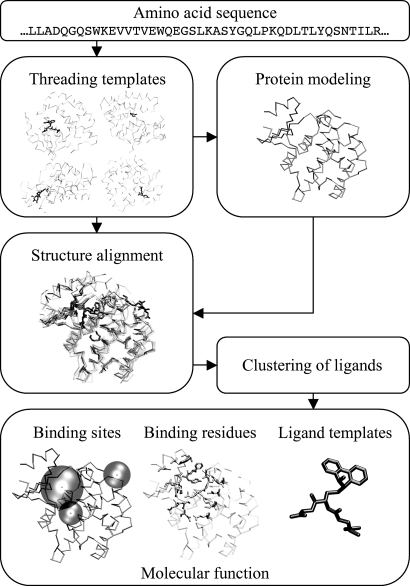
A systematic analysis of known protein structures grouped according to SCOP (22) reveals a general tendency of certain protein folds to bind substrates at a similar location, suggesting that analogous or very distantly homologous proteins can have common binding sites (11). If so, it should be possible to develop an approach for ligand-binding site identification that is less sensitive than pocket-detection methods to distortions in the modeled structures. In this spirit, we develop FINDSITE, a method for the prediction of ligand-binding sites and functional annotation based on binding-site similarity among superimposed groups of template structures identified from threading (23); a schematic overview of the methodology is shown in Fig. 1. For a given target protein, the PROSPECTOR_3 threading algorithm (24, 25) identifies ligand-bound structural templates. Then, holo-templates are superimposed onto the predicted (or experimental, if available) target protein structure by the TM-align (26) structure alignment program. Upon superimposition, the clustered centers of mass of the ligands bound to the threading templates identify putative binding sites, and the predicted sites are ranked according to the number of templates that share a common binding pocket. FINDSITE also specifies the chemical properties of the ligands that likely occupy detected binding sites. To assess its validity, we use a representative set of proteins that are weakly homologous to their templates and generate models using two state-of-the-art programs for protein structure modeling: TASSER (27–29), and MODELLER9v1 (30, 31). We demonstrate that FINDSITE operates satisfactorily in the “twilight zone” of sequence similarity (32), which covers roughly two-thirds of known protein sequences (30). Its main advantage is that no experimental structure of the target protein is required; the high accuracy of the prediction and the ability to correctly rank the identified binding sites is sustained when protein models instead of target crystal structures are used for template superimposition. In most cases, FINDSITE can accurately assign a molecular function to the protein model. Use of consensus ligands extracted from the binding sites is shown to be quite useful in ligand screening. These features should enhance the utility of low-to-moderate quality protein models in ligand screening and structure-based drug design.
Fig. 1.
Overview of the FINDSITE prediction methodology. Details are given in Materials and Methods.
Results
Ligand-Binding Site Prediction.
We evaluated the performance of the LIGSITECSC pocket-detection and FINDSITE threading-based approaches on a nonredundant benchmark set of 901 proteins in terms of the accuracy of ligand-binding site prediction and the ability to correctly rank identified pockets in both crystal structures and protein models. LIGSITECSC detects pockets using a geometric analysis of the target protein's surface, whereas FINDSITE uses the putative protein structure (either predicted or experimental, when available) as the reference for template structure superimposition. For both, the results are compared with randomly selected patches on the target's protein surface. The prediction success rate is assessed by the distance between the center of mass of the bound ligand and a single point representing the predicted pocket. In LIGSITECSC, this point represents the geometric center of the pocket's grid points. FINDSITE defines a pocket center as the center of mass of all ligands that occupy the consensus-binding site in the superimposed threading template structures. For a random surface patch, the center corresponds to the side-chain center of mass of the predicted binding residues. A cutoff distance of 4 Å is used as the hit criterion, because the average radius of gyration for ligand molecules in our dataset is 4.03 Å. To evaluate the results of ligand-binding site prediction for protein models, we transferred ligand molecules from crystal structures onto protein models by superposition of the binding residues. In no case is the sequence identity between the target and template sequences >35%. Thus, binding-site prediction is done by using the structures of weakly homologous template proteins.
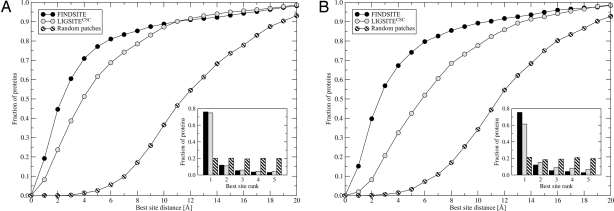
The results of ligand-binding site prediction carried out for the 901 benchmark proteins are shown in Fig. 2 and supporting information (SI) Fig. 5. In Fig. 2A, we use the target protein's crystal structure. For LIGSITECSC, the native structure is scanned to identify binding pockets, whereas for FINDSITE, the set of predicted template models (where the target has a sequence identity <35% to all of the selected template structures) is superimposed onto the crystal structure. FINDSITE performs better than the pocket-detection method in both overall accuracy and ranking ability of identified pockets. When the native crystal structure is used for the binding-site prediction procedure, the success rate using the best of top five identified binding pockets is 70.9% and 51.3% for FINDSITE and LIGSITECSC, respectively. For those proteins where a binding pocket is correctly identified, the ranking is comparable: 76.0% and 74.7% of the best pockets are ranked as the top solutions by FINDSITE and LIGSITECSC, respectively.
Fig. 2.
Performance of FINDSITE and LIGSITECSC compared with randomly selected patches on a target protein surface using target crystal structures (A) and TASSER models (B). The results are presented as the cumulative fraction of proteins with a distance between the center of mass of a ligand in the native complex and the center of the best of top five predicted binding sites, less than or equal to the distance displayed on the x axis and the rank of the best pocket selected from the top five predictions (Inset).
As shown in Fig. 2 B and SI Fig. 5, using LIGSITECSC, the prediction accuracy falls off considerably if one uses modeled protein structures rather than the experimental structure in ligand-binding site prediction by pocket detection. LIGSITECSC's success rate decreases from 51.3% to 27.2%, 32.5%, and 25.7%, when PROSPECTOR_3 template structures and protein models generated by TASSER and MODELLER, respectively, are used. This decrease is accompanied by deterioration in the ability to correctly rank the binding site. For the top-ranked threading templates, TASSER and MODELLER models, only 59.1%, 61.4%, and 58.9% of the best pockets are assigned to rank 1, respectively. In contrast, with FINDSITE, both the high accuracy of ligand-binding site prediction and the ability to correctly rank the identified binding sites are sustained if models instead of native structures are used as reference structures for holo-template superimposition. The success rate is 67.0%, 67.3%, and 65.7% for the top-ranked PROSPECTOR_3 templates, TASSER and MODELLER models, respectively, with a corresponding ranking accuracy of 75.9%, 75.5%, and 75.7%. The high accuracy may be explained by TM-align's ability to find a similar structural alignment for crystal and modeled structures, especially for confidently predicted targets (25). Most proteins used here (811 of 901) are classified by PROSPECTOR_3 as targets for which good template structures and alignments can be identified (an overview of TM-align and PROSPECTOR_3 is provided in SI Text).
We note that for both native structures and predicted models, the results using random patches are much worse than for LIGSITECSC and FINDSITE.
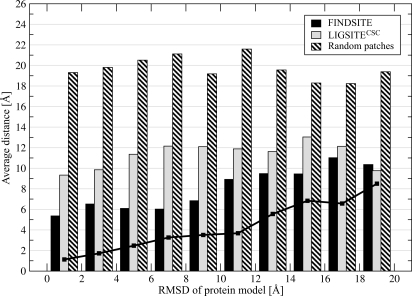
In Fig. 3, binding site prediction accuracy is evaluated in terms of the quality of the modeled protein structure used. FINDSITE tolerates structural inaccuracies in a protein model with a global rmsd from the crystal structure of 8–10 Å. In this interval, the modeled binding sites (solid line) usually do not have a rmsd >3 Å from the native binding sites, and the average distance between the center of mass of the native ligand pose and the center of the predicted binding site is ≈6 Å. However, the pocket-detection approach, LIGSITECSC, is far more sensitive to structural distortions. For protein models with a global rmsd from the crystal structure >4 Å, the average distance between the LIGSITECSC predicted and observed binding pockets is 10–13 Å. Again, if random patches are considered, the results are much worse than LIGSITECSC and FINDSITE.
Fig. 3.
Average distance between the native ligand center of mass upon superposition of a protein model onto the protein–ligand crystal complex with respect to the binding residues and the center of predicted binding pocket. The accuracy is presented for decreasing quality of TASSER models used in the prediction procedure, expressed by the global rmsd from the crystal structure. The solid line is the local rmsd calculated for the ligand-binding regions.
The relatively low accuracy of binding-site prediction by pocket detection applied to theoretical protein models compared with experimental structures motivated us to examine the possible structural distortions of ligand-binding regions in weakly homologous protein models. The rmsd from the crystal structure was calculated on the superposition of protein models onto the corresponding ligand-bound crystal structures with respect to the binding residues. In general, the overall structure of the ligand-binding regions was preserved, with a rmsd from native >2 Å found for approximately one-half of the protein models. The structural distortion of binding regions observed in the modeled structures may also account for the reduced ability to properly accommodate ligand molecules.
Confidence Index.
The overall accuracy of binding site prediction depends on the number of identified ligand-bound templates that share a common binding site (see SI Fig. 6). We used this observation to classify proteins as Easy (>125 threading templates, including homologous proteins for each template), Medium (25–125 templates), and Hard (<25 templates) targets for threading-based binding site prediction. The average distance between the centers of predicted and observed binding pockets calculated for top-ranked FINDSITE solutions is ≈2 Å, ≈5 Å, and ≈10 Å for Easy, Medium, and Hard targets. According to this classification, 9%, 47%, and 44% of proteins in the benchmark dataset are assigned by FINDSITE as Easy, Medium, and Hard targets, respectively. With a cutoff distance of 4 Å between predicted and observed binding sites, the hit rate for the top-ranked predictions is 90.0%, 71.7%, and 43.7% for Easy, Medium, and Hard targets, respectively.
Performance on a True Negative Dataset.
We next examine the true negative rate of LIGSITECSC and FINDSITE, namely how often each algorithm is likely to predict that a binding site is present, when in reality none occurs. The negative dataset consists of protein–protein interfaces that are assumed unlikely to have protein-ligand interactions. We considered a residue as binding a ligand if any of its heavy atoms lie within a distance of 4 Å from the predicted binding site center. The false positive rate is defined as the fraction of interface residues predicted to bind a ligand. The performance of LIGSITECSC and FINDSITE was evaluated for crystal structures of target proteins as well as for the top-ranked PROSPECTOR_3 templates and TASSER-refined models. The results obtained for crystal structures using LIGSITECSC and FINDSITE compared with randomly selected surface patches are presented in SI Fig. 7A. Both LIGSITECSC and FINDSITE perform equally well on the negative dataset, with the random patch calculation giving much worse results. For the top binding pockets, LIGSITECSC (FINDSITE) misclassified >5% interface residues as belonging to a binding pocket in 10.1% (8.4%) of the cases. Moreover, both approaches achieve satisfactory results on the negative dataset even if the worst of the top five predictions is considered as well as if predicted structures are used (see SI Fig. 7 B and C). In contrast, the fraction of proteins with >5% of interface residues assigned as ligand binding by randomly selecting surface patches is considerably higher, 41.4%.
Among the proteins present in the negative dataset, 128 bind a single ligand in a noninterfacial region. We used these proteins to assess the true positive rate of LIGSITECSC and FINDSITE on this dataset and compare the results with these obtained for the benchmark set described above. For crystal structures, the true positive rate considering the best of top five identified binding pockets was 49.2% and 36.7% for FINDSITE and LIGSITECSC, respectively, with the corresponding ranking accuracy of 57.8% and 57.0%. If protein models were used in the prediction procedure instead of crystal structures, the accuracy of FINDSITE and LIGSITECSC was 46.1% and 25.8%, with a ranking ability of 56.3% and 45.3%, respectively. We note that 4%, 38%, and 58% of proteins present in the negative dataset used to assess the true positive rate were assigned by FINDSITE as Easy, Medium, and Hard targets, respectively; this may explain the lower accuracy of ligand-binding site prediction for this dataset.
Comparison with PROSITE.
We compared consensus binding residues predicted for each target protein by FINDSITE with ligand-binding signatures detected by ScanProsite (33). We consider only the top-ranked threading consensus binding sites and the best PROSITE pattern (34). Performance is evaluated by several recognized metrics defined in SI Text. The overall accuracy of both methods is comparably high: 0.93 and 0.92 for FINDSITE and PROSITE, respectively. Similarly, both methods demonstrate comparable median specificity: 0.96 and 1.00. However, FINDSITE clearly covers more ligand-binding sites than PROSITE. The median sensitivity and Matthew's correlation coefficient between predicted and observed binding residues are 0.64 and 0.59, and 0.00 and 0.00, for FINDSITE and PROSITE, respectively (the zero values calculated for PROSITE result from small binding site coverage). Using FINDSITE, excellent agreement is found between the average numbers of predicted binding residues (20.0 ± 7.3 and 20.3 ± 6.8 for the best and the top identified pockets, respectively) and that observed in the crystal structures of protein–ligand complexes (19.6 ± 6.5).
Ligand Selection.
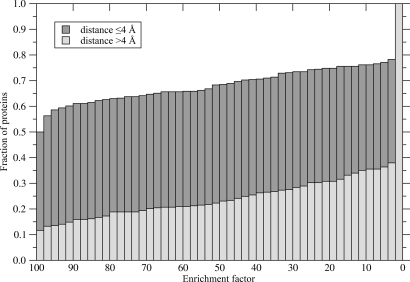
FINDSITE also provides information on the chemical properties of the binding ligands, termed here “template ligands.” We used this observation to select representative ligand molecules likely bind to the predicted site on the protein's surface. Subsequently, these molecules were used as ligand templates in a simple ligand-based virtual screening experiment against the KEGG (35) compound library. The results shown in Fig. 4 present the cumulative distribution of enrichment factors calculated for the target proteins (see Materials and Methods). The ligand templates selected by FINDSITE for accurately predicted binding sites (whose center of mass is within 4 Å of the experimental one) used in fingerprint-based similarity searching perform better than random in 78% of the cases. The ideal enrichment factor (all native-like compounds found in the top 1% of the ranked library) was observed for 50% of these target proteins. For less accurately predicted binding pockets, the quality of ligand templates is notably worse (the ideal enrichment factor was obtained for 12% of the cases and is better than random for 38% of the cases). We note that for a given target protein, template ligands can be selected even if the crystal structure is unavailable and its molecular function unknown. Finally, a case study presenting the performance of FINDSITE in ligand-based virtual screening for HIV-1 protease inhibitors is described in detail in SI Text (Case Study), with quite encouraging results. If only templates with <35% sequence identity are used, the enrichment factor of the top 1% of compounds is 40.
Fig. 4.
Cumulative distribution of enrichment factors resulting from the ligand-based virtual screening experiment against the KEGG compound library using ligand templates selected by FINDSITE. Target proteins are divided into the two subsets with respect to the accuracy of binding pocket prediction (the distance between the top-ranked pocket and the center of mass of the native ligand ≤4 and >4 Å).
Molecular Function Prediction.
The relatively high accuracy of the ligand selection procedure encouraged us to investigate the transferability of specific functions of the threading templates to the target. We use Gene Ontology (GO; ref. 36) to describe protein function. GO is based on three organizing principles: cellular components, biological processes, and molecular functions. The latter consider molecular events, such as catalytic or binding activities; therefore, we use GO molecular function terms. From the benchmark set, we selected 753 proteins for which a GO annotation is provided by GO (36) or UniProt (37). The procedure for molecular function prediction uses the superimposed group of holo-templates selected by threading as previously used for binding-site detection and ligand selection. Only predicted threading templates with <35% sequence identity to a target protein are used for the purpose of functional inference.
Function transferability is assessed by well known metrics (defined in SI Text). For each target protein, all GO annotations are identified for the threading templates that share the top-ranked predicted binding site using the GO and UniProt databases. Then, the target protein is assigned a function with a probability that corresponds to the fraction of threading templates annotated with the particular molecular function. For a probability threshold of 0.5 (at least one-half of the threading holo-templates must be annotated with the same GO term to transfer it to the target protein), the maximal Matthew's correlation coefficient of 0.64 is found. This corresponds to a precision of 0.76 with a sensitivity (recall) of 0.54. In addition, we calculated predictive metrics with respect to individual GO identifiers. FINDSITE distinguishes between the enzymatic and nonenzymatic character of an action that occurs at the predicted binding site, with a precision and sensitivity of 0.93 and 0.89, respectively. Moreover, many molecular functions are accurately transferable from the templates selected by threading to the target proteins. See SI Table 1 for an assessment of the best predictions. These cover a broad spectrum of molecular events including both enzymatic and binding activities. The full set of predicted functions can be found at http://cssb.biology.gatech.edu/skolnick/files/FINDSITE.
Discussion
Ligand-binding site identification is usually the first step in inferring the biological role of proteins of unknown structure and function. The development of accurate algorithms for ligand-binding site prediction in modeled protein structures is of importance, because protein models are increasingly used in the identification of protein function and in screens for new ligands (38, 39). Their main limitation is that high-quality structures are usually required for good prediction accuracy. Performance falls off considerably if one uses modeled protein structures. To improve the overall prediction accuracy on experimental structures as well as to develop an approach suitable for lower-quality predicted models, we developed FINDSITE, a threading-based method for the prediction of ligand-binding sites and functional annotation based on binding-site similarity across superimposed groups of threading templates. The ability to detect and correctly rank ligand-binding regions in weakly homologous protein models (<35% identity to any selected template) is the most pronounced practical advantage of FINDSITE, whose performance on the negative control dataset confirms its high specificity. Comparison with PROSITE reveals that FINDSITE covers more binding sites with similar accuracy and specificity. When no information concerning potential ligands is available for a given target protein, the chemical properties of template-bound ligands that occupy consensus binding pockets can be used to select ligand templates for virtual screening with encouraging results.
Function prediction based on homology to previously characterized proteins is frequently used (40, 41). However, current methods using global sequence alignment and local sequence motif identification frequently fail, as the sequence identity lies within and below the “twilight zone” of sequence identity (9). To overcome this limitation, a method for the prediction of enzymatic function based on 3D descriptors of specific protein functions, termed fuzzy functional forms, was developed (42) and shown to provide high-confidence novel annotations (43). However, approaches using geometrical active-site descriptors typically require high-quality target protein structures as well as high structural conservation of functional sites and thus far have been successfully applied only to enzymes. Our effective ligand selection procedure motivated us to test the possibility of direct functional annotation by inferring functional similarity from threading templates. In most of cases, molecular function according to the GO (36) classification can be inferred with a satisfactory precision from holo-templates selected by threading, even if the sequence similarity to the target protein is <35%. We found that weakly homologous templates identified by PROSPECTOR_3 can be used by FINDSITE to precisely distinguish between the enzymatic and nonenzymatic character of a predicted binding site. The superimposition of threading templates and the clustering of binding pockets serves as an additional filter that facilitates the extraction of molecular functions associated with common sites in the threading templates; this method may also capture more general functions for which binding pocket activity is one manifestation. Finally, these results suggest that a threading procedure that uses a strong sequence profile component (25) works by detecting very remote, yet evolutionary related homologues.
Materials and Methods
Benchmark Set of Protein–Ligand Complexes.
The structures of protein–ligand complexes used here were selected from the Protein Data Bank (PDB) (44). First, ligand-bound forms are identified, where noncovalently bound organic molecules, cofactors, nucleotides, and short peptides composed of standard or modified amino acids are considered as ligands if the number of atoms was ≥6 and ≤100. We remove proteins having more than one ligand in the binding pocket. Because proteins >400 residues cannot be modeled using TASSER (27–29) in a reasonable amount of computer time, these are excluded. No two proteins in the dataset share >35% sequence identity. More details concerning dataset creation are given in SI Text. The benchmark set consists of 901 protein–ligand complexes and may be found in SI Text as well as at http://cssb.biology.gatech.edu/skolnick/files/FINDSITE.
Negative Dataset of Protein–Protein Interfaces.
A set of protein–protein interfaces is used as a negative control to supplement the positive dataset of protein–ligand complexes described above. The underlying assumption is that interfacial residues are not likely to bind small ligands. The negative dataset consists of 281 protein–protein dimeric interfaces formed by 562 nonredundant protein chains (45). The following criteria are applied to select multichain crystal structure entries from the PDB: The minimum number of interfacial contacts between chains is 30, where interfacial contacting residues are defined as a pair of residues from different chains with at least one pair of heavy atoms within 4.5 Å. Proteins with >40 residues are accepted; the set of dimers have <35% sequence identity with each other.
Template–Consensus-Binding Pockets.
The flowchart of the FINDSITE approach is presented in Fig. 1. For a given target sequence, template structures are identified from a nonredundant PDB library by the PROSPECTOR_3 threading algorithm (24, 25). From threading templates with a Z-score ≥4, we use only those with <35% sequence identity to the target protein. In addition, we expand the set of identified threading templates by including homologous proteins for each template. Again, proteins homologous to threading templates with a sequence identity to the target sequence >35% are rejected. The results are frequently improved by including template homologues (see SI Fig. 6). Among all templates, structures containing a bound ligand molecule are identified and superimposed onto the target structure using TM-align (26). The performance of FINDSITE was assessed for either the target crystal structure or the protein model (TASSER- or MODELLER-generated or the top-ranked PROSPECTOR_3 template) used as a reference structure for template superimposition. Subsequently, the centers of mass of ligands bound to threading templates are clustered according to their spatial proximity, using an 8-Å cutoff distance. This cutoff maximizes the ranking accuracy and accommodates some structural distortions. The geometrical center of each cluster corresponds to the center of a putative binding site. Finally, the predicted binding sites are ranked according to the number of threading templates that share a common binding pocket (cluster multiplicity). If two or more pockets have the same number of templates, the average Z-score of threading templates is used as an additional ranking criterion. For each target, we select the top five identified ligand-binding sites for further analysis.
Template–Consensus-Binding Residues.
For each predicted binding site, binding residues are identified in those threading templates that share a common pocket based on interatomic contacts reported by LPC (46). Using the sequence alignment provided by PROSPECTOR_3, for each residue in the aligned target sequence, the fraction of templates that have a residue in the corresponding position in contact with the ligand is calculated. A consensus-binding residue is defined as a residue in contact with a ligand in at least 25% of the threading templates. This criterion maximizes Matthew's correlation coefficient between predicted and observed binding residues and reduces overpredictions. Here, the chemical properties of binding residues are ignored. Subsequently, consensus binding residues identified for each target sequence are compared with the results of ligand-binding motif prediction using the PROSITE database (34) and ScanProsite tool (33). The accuracy of predictions made with FINDSITE and ScanProsite is assessed by standard evaluation metrics. Details are given in SI Text.
Ligand Selection.
FINDSITE also selects representative molecules that bind to a particular binding site by exploiting the chemical properties of template-bound ligands that occupy consensus binding pockets. For a given target protein, these can be used to construct ligand templates for use in ligand-based virtual screening when no other information concerning potential binders is available. We use the 1,024-bit version of Daylight fingerprints (47) to calculate the Tanimoto coefficient (TC)† that expresses the chemical similarity between two compounds. First, all molecules that occupy the top-ranked binding pocket in identified ligand-bound threading templates are clustered using the TC cutoff of 0.7. Then, representative molecules selected from the clusters are used as ligand templates to rank a compound library using the sum of weighted TCs:
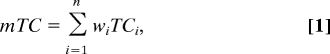 |
where n is the number of chemically dissimilar ligand clusters obtained for a given FINDSITE binding pocket, wi is the fraction of ligands belonging to cluster i, and TCi is the Tanimoto coefficient calculated for the ligand template selected from this cluster and a library compound. In this manner, larger clusters have potentially higher impact on the fingerprint similarity score, and thus, on the ranking of library compounds. The results were assessed by calculating the enrichment factor for each target protein: EF=(Fs/Ns)/(Ft/Nt), where Fs is the number of native-like compounds in the top-ranked sample of Ns compounds (here we consider top 1% of the ranked library), Ft and Nt is the total number of native-like and all compounds in the library, respectively. A native-like compound is defined as a molecule with TC ≥ 0.7 to the native ligand. In the ligand-based virtual screening experiment, we used the KEGG compound library (35) (release 44.0+/10–15, Oct07, the total of 12,478 compounds).
Protein Structure Modeling.
For each target protein, weakly homologous protein models were generated using the PROSPECTOR_3 templates, TASSER (27–29) and MODELLER9v1 (30, 31) models. The protocols for protein structure modeling are given in SI Text.
Pocket Detection.
We used LIGSITECSC for ligand-binding site prediction by pocket detection (14). LIGSITECSC is provided with the target protein's crystal structure, the top-ranked PROSPECTOR_3 templates and theoretical protein models generated by TASSER and MODELLER. In each case, we consider the top five identified binding pockets for further analysis.
Molecular Function Prediction.
We used the GO classification (36) to investigate the specific function transferability from threading templates to target proteins. We selected 753 proteins from the benchmark dataset for which a GO annotation is provided by GO (version 1.12) (36) or UniProt (37) (release 12.0). The procedure for transferring molecular function uses the previously described superimposed group of holo-templates selected by PROSPECTOR_3. Each target protein is annotated with the set of GO terms identified in GO and UniProt databases from the threading templates that share the top-ranked predicted binding site. All parent terms for a specific GO identifier are traced to explore the more general ontology classes. Annotation accuracy is assessed by the precision (positive predictive value), sensitivity (recall), and Matthew's correlation coefficient (MCC) as defined in SI Text. To assess the overall performance of FINDSITE functional annotation, the predictive metrics were calculated for all 7,825 molecular function terms available from the GO website (www.geneontology.org). We have also calculated predictive metrics with respect to individual GO numerical identifiers that appear in the benchmark dataset.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Dr. Adrian K. Arakaki for help with the PROSITE analysis and Dr. Huiling Chen (Center for the Study of Systems Biology, Georgia Institute of Technology) for the protein dimer dataset. This work was supported by National Institutes of Health Grant No. GM-48835.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707684105/DC1.
Tanimoto TT, IBM Internal Report, November 17, 1957.
References
- 1.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Baker D, Sali A. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez R, Pieper U, Melo F, Eswar N, Marti-Renom MA, Madhusudhan MS, Mirkovic N, Sali A. Nat Struct Biol. 2000;7(Suppl):986–990. doi: 10.1038/80776. [DOI] [PubMed] [Google Scholar]
- 5.Shah M, Passovets S, Kim D, Ellrott K, Wang L, Vokler I, LoCascio P, Xu D, Xu Y. Bioinformatics. 2003;19:1985–1996. doi: 10.1093/bioinformatics/btg262. [DOI] [PubMed] [Google Scholar]
- 6.Laurie AT, Jackson RM. Curr Protein Pept Sci. 2006;7:395–406. doi: 10.2174/138920306778559386. [DOI] [PubMed] [Google Scholar]
- 7.Campbell SJ, Gold ND, Jackson RM, Westhead DR. Curr Opin Struct Biol. 2003;13:389–395. doi: 10.1016/s0959-440x(03)00075-7. [DOI] [PubMed] [Google Scholar]
- 8.Devos D, Valencia A. Proteins. 2000;41:98–107. [PubMed] [Google Scholar]
- 9.Wilson CA, Kreychman J, Gerstein M. J Mol Biol. 2000;297:233–249. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]
- 10.Todd AE, Orengo CA, Thornton JM. J Mol Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 11.Russell RB, Sasieni PD, Sternberg MJ. J Mol Biol. 1998;282:903–918. doi: 10.1006/jmbi.1998.2043. [DOI] [PubMed] [Google Scholar]
- 12.Glaser F, Morris RJ, Najmanovich RJ, Laskowski RA, Thornton JM. Proteins. 2006;62:479–488. doi: 10.1002/prot.20769. [DOI] [PubMed] [Google Scholar]
- 13.Hendlich M, Rippmann F, Barnickel G. J Mol Graphics Model. 1997;15:359–363. 389. doi: 10.1016/s1093-3263(98)00002-3. [DOI] [PubMed] [Google Scholar]
- 14.Huang B, Schroeder M. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang J, Edelsbrunner H, Woodward C. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hetenyi C, van der Spoel D. FEBS Lett. 2006;580:1447–1450. doi: 10.1016/j.febslet.2006.01.074. [DOI] [PubMed] [Google Scholar]
- 17.Oshiro CM, Kuntz ID, Dixon JS. J Comput Aided Mol Des. 1995;9:113–130. doi: 10.1007/BF00124402. [DOI] [PubMed] [Google Scholar]
- 18.Ondrechen MJ, Clifton JG, Ringe D. Proc Natl Acad Sci USA. 2001;98:12473–12478. doi: 10.1073/pnas.211436698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elcock AH. J Mol Biol. 2001;312:885–896. doi: 10.1006/jmbi.2001.5009. [DOI] [PubMed] [Google Scholar]
- 20.Brylinski M, Konieczny L, Roterman I. Bioinformation. 2006;1:127–129. doi: 10.6026/97320630001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connolly M. J Appl Crystallogr. 1983;16:548–558. [Google Scholar]
- 22.Murzin AG, Brenner SE, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 23.Jones DT, Hadley C. In: Bioinformatics: Sequence, Structure and Databanks. Higgins D, Taylor WR, editors. Heidelberg, Germany: Springer; 2000. pp. 1–13. [Google Scholar]
- 24.Skolnick J, Kihara D. Proteins. 2001;42:319–331. [PubMed] [Google Scholar]
- 25.Skolnick J, Kihara D, Zhang Y. Proteins. 2004;56:502–518. doi: 10.1002/prot.20106. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Skolnick J. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Arakaki AK, Skolnick J. Proteins. 2005;61(Suppl 7):91–98. doi: 10.1002/prot.20724. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Skolnick J. Biophys J. 2004;87:2647–2655. doi: 10.1529/biophysj.104.045385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Skolnick J. Proc Natl Acad Sci USA. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 31.Sali A, Blundell TL. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 32.Rost B. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 33.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. Nucleic Acids Res. 2006;34:W362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M, Goto S. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. Nucleic Acids Res. 2006;34:D187–191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bissantz C, Bernard P, Hibert M, Rognan D. Proteins. 2003;50:5–25. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 39.Evers A, Hessler G, Matter H, Klabunde T. J Med Chem. 2005;48:5448–5465. doi: 10.1021/jm050090o. [DOI] [PubMed] [Google Scholar]
- 40.Groth D, Lehrach H, Hennig S. Nucleic Acids Res. 2004;32:W313–317. doi: 10.1093/nar/gkh406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zehetner G. Nucleic Acids Res. 2003;31:3799–3803. doi: 10.1093/nar/gkg555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fetrow JS, Skolnick J. J Mol Biol. 1998;281:949–968. doi: 10.1006/jmbi.1998.1993. [DOI] [PubMed] [Google Scholar]
- 43.Baxter SM, Rosenblum JS, Knutson S, Nelson MR, Montimurro JS, Di Gennaro JA, Speir JA, Burbaum JJ, Fetrow JS. Mol Cell Proteom. 2004;3:209–225. doi: 10.1074/mcp.M300082-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, Skolnick J. Biophys J. 2007 doi: 10.1529/biophysj.107.114280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 47.Anonymous . Daylight Theory Manual. Aliso Viejo, CA: Daylight Chemical Information Systems, Inc; 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.