Abstract
The assembly mechanisms of amyloid fibrils, tissue deposits in a variety of degenerative diseases, is poorly understood. With a simply modified application of the atomic force microscope, we monitored the growth, on mica surface, of individual fibrils of the amyloid β25–35 peptide with near-subunit spatial and subsecond temporal resolution. Fibril assembly was polarized and discontinuous. Bursts of rapid (up to 300-nm−1) growth phases that extended the fibril by ≈7 nm or its integer multiples were interrupted with pauses. Stepwise dynamics were also observed for amyloid β1–42 fibrils growing on graphite, suggesting that the discontinuous assembly mechanisms may be a general feature of epitaxial amyloid growth. Amyloid assembly may thus involve fluctuation between a fast-growing and a blocked state in which the fibril is kinetically trapped because of intrinsic structural features. The used scanning-force kymography method may be adapted to analyze the assembly dynamics of a wide range of linear biopolymers.
Keywords: atomic force microscopy, beta-amyloid, growth dynamics, self-assembly
Amyloid fibrils are filamentous aggregates of various misfolded proteins (1–4). They are thought to assemble in a nucleation-dependent polymerization process, during which the slow formation of nuclei is followed by a more rapid extension phase (5). The kinetics of amyloid assembly are typically investigated in bulk assays that often hide details of the process. Recently, the growth of individual amyloid fibrils was observed by using atomic force microscopy (AFM) (6, 7) and fluorescence microscopic methods (8, 9). In some experiments, a stop-and-run phenomenon was observed, during which the fibril growth was temporarily halted (9, 10), whereas others pointed to a constant growth rate (8, 9). The lack of either high spatial or high temporal resolution, however, hid the greater detail of the assembly process.
In the present work, we explored the assembly dynamics of individual amyloid fibrils during template-assisted growth with exceptional spatial and temporal resolution. We focused on analyzing the dynamics of oriented growth of amyloid beta 25–35 (Aβ25–25) fibrils on mica (11). In addition, the aggregation of the full-length amyloid peptide, Aβ1–42 on graphite was monitored (12–14). Fibril assembly and disassembly were not continuous but proceeded in bursts of rapid steps interrupted with pauses that lasted for up to tens of seconds. The stepwise assembly dynamics suggest that the amyloid fibril fluctuates between structural states in which growth may be either permitted or inhibited.
Results and Discussion
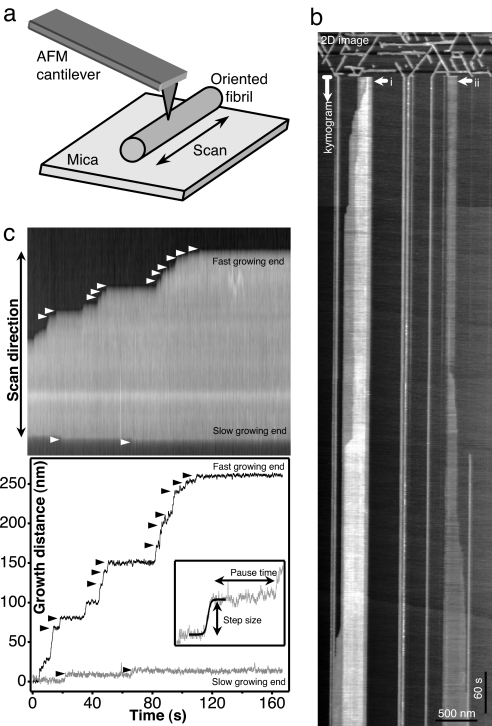
To explore the dynamics of amyloid self-assembly with high spatial and temporal resolution, we implemented a modified AFM method that we call scanning-force kymography (SFK). We took advantage of the oriented binding and growth of Aβ25–35 fibrils on mica [supporting information (SI) Fig. 4 and SI Movie 1] (11). Aβ25–35 is a fibril-forming peptide that represents the biologically active, toxic form of the full-length Aβ monomer (15, 16). In SFK, the cantilever tip is scanned repetitively along the fibril axis (Fig. 1a). The fibril-end position is detected as a sudden change in topographical height. Precision is limited by pixel resolution (typically 1 nm per pixel here) and is unaffected by tip convolution. Consecutive line scans, separated by a period dictated by the scanning frequency (up to 3.3 Hz here), are assembled into a scanning-force kymogram (Fig. 1b).
Fig. 1.
Scanning-force kymography. (a) Schematics of the method. Rapid scanning is along the fibril axis, and the perpendicular slow scan is disabled. (b) Acquisition of a scanning-force kymogram. The 2D image is shifted to the kymogram upon disabling the slow scan (capped arrow). Rapid scan direction is horizontal. Grayscale intensity corresponds to topographical height. (c) Kymogram (Upper) converted to a time-dependent growth–distance plot (Lower). White and black arrowheads mark corresponding small steps. (Inset) Example of sigmoid fit onto a growth step.
As revealed by the scanning-force kymograms, Aβ25–35 fibrils grew on both ends, but the net rate was greater on one of them (fast-growing end). A similar polarized growth has been noted for amylin fibrils (7). The fast-growing end of different fibrils may point at opposite directions despite identical axial orientation (compare fibrils i and ii in Fig. 1b), indicating that polarization is independent of the surface and is probably due to the characteristics of the fibril. Fibrils grew on mica and the surface of an already existing fibril (fibril i in Fig. 1b and SI Fig. 5). The assembly dynamics of such fibrils were similar to those of the underlying one. The most prominent feature of the self-assembly dynamics was discontinuous growth. Bursts of rapid step-like growth were interrupted with pauses of variable time period (Fig. 1 b and c). In fact, net fibril growth was achieved via an assembly of consecutive steps and pauses.
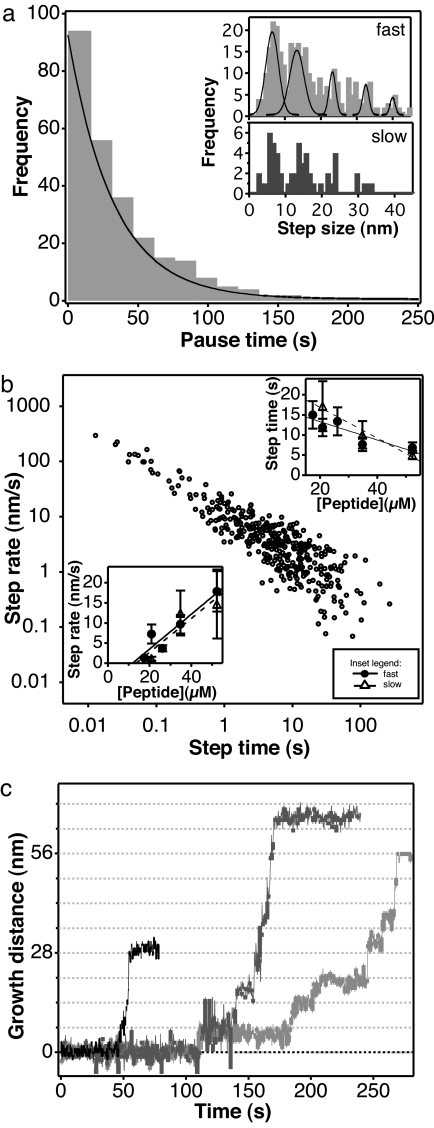
The quantitative features of discontinuous assembly were analyzed in images converted with user-developed routines (see SI Text) into growth distance (d) versus time (t) plots (Fig. 1c). Steps were fitted with the sigmoid function d = b + s [1 + e(thalf − t/τ)]−1, where b is start length, s is step size, thalf is the time at the half-step, and τ is time constant that correlates with fibril growth rate during the step. Considering that τ corresponds to the time during which the fibril has grown a distance of s/2e, step time (ts) was calculated as ts = 2eτ. The linear growth rate during the step (called step rate, rs) was then calculated as rs = s/ts. Pause time is the time between the end and start of consecutive steps, respectively. Pause times for the fast-growing end displayed an exponential distribution with a time constant of 32.3 s ± 0.8 s SEM (Fig. 2a). In the case of the slow-growing end, consecutive steps most often fell on successive kymograms, which precluded a precise measurement. Because acquiring a kymogram took ≈6 min, the pause time on the slow-growing fibril is at least an order of magnitude greater than on the fast-growing one. The step-size histogram (Fig. 2a Inset) displayed a multimodal distribution with peaks at integer multiples of ≈7 nm for both the fast- and slow-growing ends. Considering β-sheet spacing within the fibril [4.7 Å (17)], the 7-nm distance corresponds to a stretch of fibril containing ≈15 peptides along its length. Although it is difficult to rule out the presence of oligomeric species, it is unlikely that the growth steps were caused by the annealing of already existing assemblies to the fibril end. Rather, the 7-nm distance might correspond to a dimension related to the structural features of the fibril. The pairwise rs versus ts plot (Fig. 2b) showed an inverse relationship that correlates with the constant discrete values of the step size. Thus, fibril growth may be accelerated by reducing the time spent on making discrete constant-size steps as opposed to making greater steps at a constant step rate. The average step rate was 4.37 ± 0.09 nm/s SEM, and step rates in excess of 100 nm/s were also observed.
Fig. 2.
Quantitative analysis of discontinuous amyloid fibril growth. (a) Distribution of pause time measured on the fast-growing end. Pauses are considered if they last for at least three consecutive line scans (≈1 s). The continuous line is the exponential fit. (Inset) Distribution of step size for the fast- and slow-growing fibril ends. Gaussian fits to the peaks in the fast-growing-end histogram are at 6.5 ± 0.2 nm, 13.3 ± 0.3 nm, 23.2 ± 0.3 nm, 32.5 ± 0.4 nm, and 40 ± 0.6 nm, respectively. (b) Step rate versus step time for the total number of observations (including fast and slow ends, n = 372). (Upper Inset) Step time versus total peptide concentration. (Lower Inset) Step rate versus total peptide concentration. (c) Kymograms displaying cooperative run-on events by the coalescence of ≈7-nm steps.
To probe the self-assembly mechanisms further, concentration-dependent experiments were carried out. We found that both pause time and step size are independent of total peptide concentration (SI Fig. 6). Step time decreased (Fig. 2b Upper Inset) and step rate increased (Fig. 2b Upper Inset) with increasing concentration for both the fast- and slow-growing fibril ends. The concentration dependence of rs indicates that the rapid assembly step follows second-order kinetics. From the x-intercept of the concentration-dependent rs plots, a critical concentration (Kc) of ≈10 μM is obtained for the rapid growth step at both ends. Previously, Kc values of 50 μM (18) and 100 μM (19) were measured for the assembly of Aβ1–40 fibrils in solution. The low Kc observed here is probably caused by a catalytic effect of the surface (13, 20). It is notable that both the critical concentrations and the rate constants are similar at the fast- and slow-growing ends despite their different assembly dynamics. Conceivably, the net growth rates are controlled by the significantly different lifetimes of the paused states at the different ends rather than by their growth-step dynamics. Step sizes as large as 100 nm were sometimes observed. In these cases, ≈7 nm substeps could often be resolved (Fig. 2c), suggesting that large growth steps evolve by fusion of smaller steps. Furthermore, the pause time separating the successive substeps sometimes becomes progressively smaller, pointing to the presence of cooperative mechanisms in the formation of the fibril-growth bursts.
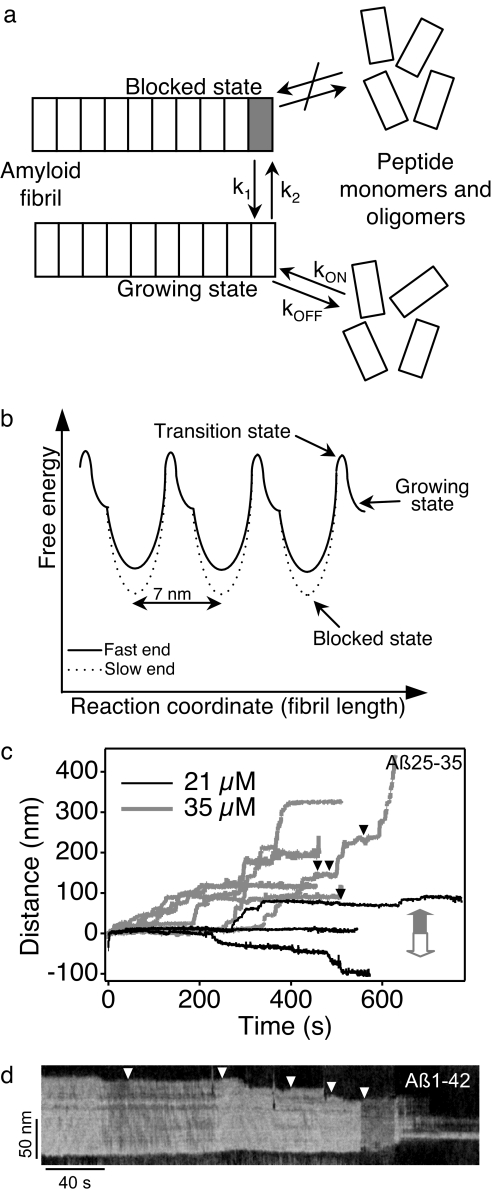
Our observations may be reconciled with a two-state model in which the fibril ends are either in growing or blocked state (Fig. 3). Transition from the blocked to the growing state, and hence the pause time, is independent of peptide concentration (SI Fig. 6a), indicating that the reaction does not involve the incorporation or removal of peptides. The blocked-to-growing transition is probably related to a structural change that proceeds with first-order kinetics. The fibril arrives at the subsequent blocked state with a concentration-dependent rate after growth of ≈7 nm or its integer multiples. This periodicity suggests that the blocked state is related to a structural feature of Aβ25–35 about which little is known at the present. It is unlikely that the periodicity is solely dependent on mica, considering mica's much smaller lattice spacing. One possibility is that the Aβ25-fibril adopts a helical structure with ≈7 nm periodicity, and, because growth would be associated with axial rotation under these conditions, an energetically unfavorable interaction with the surface might occur periodically. Although a helical protofilament structure with similar periodicity has been predicted for Aβ12–42 based on computer simulation (21), detailed fibril-structure data are needed to substantiate this hypothesis. Another intriguing possibility is that the ≈7-nm periodicity refers to a moiré pattern that arises because of a small mismatch between the spacing of β-strands within the Aβ25–35 β-sheets [pβ = 4.7 Å (17)] and that of the mica lattice (pm = 5.2 Å). However, the moiré pattern periodicity, calculated as pβ2/(pm − pβ), would be only 4.4 nm. Furthermore, discontinuous fibril growth dynamics occur not only on mica, but also on preexisting fibrils (Fig. 1b). Finally, the blocked state could be related to the “locked state” of the template-dependent dock-lock mechanism of amyloid propagation (22). The transition between docked and locked states might occur in cooperative units spanning several peptides. Whatever the structural basis of the blocked state, it probably represents a kinetically trapped configuration from which the fibril escapes at a slow rate. The kinetic trap is deeper for the slow-growing end than for the fast-growing one, resulting in longer paused-state lifetimes. In the growing state fibril assembly proceeds with a rate determined by peptide concentration. Because of the hypothesized shape of the fibril-assembly energy landscape (Fig. 3b), cooperative run-on events might be possible (see Fig. 2c) so that a fast-growing fibril may “jump” the kinetic trap. Below Kc, the fibrils are expected to disassemble. We often found that removing free peptides did not result in fibril disassembly (SI Movie 2). It is possible that the blocked state is accessible during depolymerization as well, supporting the idea that this state is related to the structural properties of the Aβ25–35 fibril. Interestingly, if the fibrils are exposed to high-speed scanning with enhanced pushing forces by the AFM cantilever, disassembly can be initiated, which proceeds in a stepwise fashion similarly to the assembly process (Fig. 3c and SI Movie 3).
Fig. 3.
Model of amyloid assembly dynamics. (a) Schematic diagram of fibril-end kinetics. k1 and k2 are first-order rate constants of the transition between the blocked and growing states and vice versa, respectively. kON and kOFF are second-order rate constants of rapid growth and depolymerization, respectively. (b) Hypothetic energy landscape of epitaxial amyloid fibril assembly. (c) Concentration-dependent kymograms of Aβ25–35 fibrils, demonstrating stepwise disassembly events. Gray and white block arrows indicate assembly and disassembly directions, respectively. Black arrowheads indicate backstep events. (d) Kymogram of Aβ1–42 on HOPG, revealing net stepwise disassembly. Grayscale intensity corresponds to the phase shift of the sinusoidally oscillated cantilever. White arrowheads indicate small assembly steps.
To investigate whether discontinuous dynamics is a general feature of epitaxial amyloid assembly, we examined the formation of fibrils from the full-length Aβ1–42 peptide on a highly oriented pyrolytic graphite (HOPG) surface. We took advantage of the trigonally oriented assembly of Aβ1–42 on HOPG (SI Fig. 7) (12–14). After a lag phase that lasted up to 60 min, oriented Aβ1–42 fibrils with lengths up to several hundred nanometers were formed, which maintained a dynamic equilibrium characterized by stepwise length fluctuations. Kymograms of such fibrils (Fig. 3d) contained stepwise assembly and disassembly events and were often dominated by a net disassembly due most likely to the mechanical effect of the scanning AFM tip and to the weak interaction between Aβ1–42 and graphite. Notably, the net length changes are partitioned according to fast and slow fibril ends, similarly to the Aβ25–35 peptide. The observations suggest that amyloid assembly is a highly dynamic process, and the rapid stepwise dynamics may be a general feature of epitaxial amyloid growth.
The method we used, scanning-force kymography, provided a glimpse at the dynamics of individual amyloid fibril self-assembly with unprecedented detail. Considering its simplicity and richness of high-resolution information, the technique may be used to analyze the assembly dynamics of a wide variety of linear biopolymers.
Methods
Sample Preparation.
Synthetic Aβ25–35 peptide was dissolved in DMSO and transferred to Na-phosphate buffer [10 mM Na-phosphate (pH 7.4), 140 mM NaCl, 0.02% NaN3] at a final concentration of ≈2.5 mg/ml. Insoluble aggregates were removed by centrifugation at 300,000 × g for 2 h. Supernatant was diluted for further use. Peptide concentration was measured with the quantitative bicinchoninic acid assay (23). Seeds were established on freshly-cleaved mica surface by incubating peptides at ≈12 μM concentration in Na-phosphate for 10 min, then removing unbound peptides by washing with buffer. Fibril growth was initiated by the addition of peptides at a preadjusted total concentration in Na-phosphate buffer supplemented with 10 mM KCl to inhibit the generation of new seeds. Synthetic Aβ1–42 peptide was dissolved in HFIP, then transferred to 10 mM Na-citrate (pH 5) at a final concentration of ≈0.5 mg/ml and sonicated to disperse undissolved aggregates. Peptides were added to freshly cleaved HOPG surface at a final concentration of 10–50 μM in Na-citrate (pH 5).
Scanning-force Kymography.
Non-contact mode images of amyloid fibrils were acquired in buffer with an MFP3D (Asylum Research) AFM instrument, using silicon nitride cantilevers (Olympus BioLever; typical resonance frequency, ≈30 kHz). The scan angle was adjusted so that the fast scan direction was parallel with the fibril orientation. Slow scan along the direction perpendicular to the fibril axis was disabled, and consecutive line scans were assembled into a distance versus time image. Two-micrometer images were typically acquired at a 3-Hz scanning rate into 2048 × 1024 images, which corresponds to 1 nm per pixel and 333 ms per pixel spatial and temporal resolution, respectively. Images were converted to data points, using custom-written algorithms, and further analyzed with commercial software. For further detail, see SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
We thank László Grama, Mihály Kovács, András Málnási-Csizmadia, Attila Nagy, and Miklós Nyitrai for discussions and critical comments on the manuscript. This work was supported by Hungarian Science Foundation Grant OTKA T049591; Hungarian National Office for Research and Technology Grants OMFB-01600/2006, OMFB-01627/2006, OMFB-00198/2007, KFKT-1–2006-0021, and RET 08/2004; the South TransDanubian Cooperative Research Center; and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704305105/DC1.
References
- 1.Dobson CM. Nature. 2003;426:884–889. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Pepys MB. Annu Rev Med. 2006;57:1–19. doi: 10.1146/annurev.med.57.121304.131243. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe DJ. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 4.Stefani M, Dobson CM. J Mol Med. 2003;81:678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 5.Lomakin A, Teplow DB, Kirschner DA, Benedek GB. Proc Natl Acad Sci USA. 1997;94:7942–7947. doi: 10.1073/pnas.94.15.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackley HK, Sanders GH, Davies MC, Roberts CJ, Tendler SJ, Wilkinson MJ. J Mol Biol. 2000;298:833–840. doi: 10.1006/jmbi.2000.3711. [DOI] [PubMed] [Google Scholar]
- 7.Goldsbury C, Kistler J, Aebi U, Arvinte T, Cooper GJ. J Mol Biol. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]
- 8.Ban T, Hamada D, Hasegawa K, Naiki H, Goto Y. J Biol Chem. 2003;278:16462–16465. doi: 10.1074/jbc.C300049200. [DOI] [PubMed] [Google Scholar]
- 9.Ban T, Hoshino M, Takahashi S, Hamada D, Hasegawa K, Naiki H, Goto Y. J Mol Biol. 2004;344:757–767. doi: 10.1016/j.jmb.2004.09.078. [DOI] [PubMed] [Google Scholar]
- 10.Hoyer W, Cherny D, Subramaniam V, Jovin TM. J Mol Biol. 2004;340:127–139. doi: 10.1016/j.jmb.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 11.Karsai Á, Grama L, Murvai Ü, Soós K, Penke B, Kellermayer MSZ. Nanotechnology. 2007;18:345102. [Google Scholar]
- 12.Arimon M, Díez-Pérez I, Kogan MJ, Durany N, Giralt E, Sanz F, Fernández-Busquets X. FASEB J. 2005;19:1344–1346. doi: 10.1096/fj.04-3137fje. [DOI] [PubMed] [Google Scholar]
- 13.Kowalewski T, Holtzman DM. Proc Natl Acad Sci USA. 1999;96:3688–3693. doi: 10.1073/pnas.96.7.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Zhou C, Wang C, Wan L, Fang X, Bai C. Ultramicroscopy. 2003;97:73–79. doi: 10.1016/S0304-3991(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 15.Forloni G, Chiesa R, Smiroldo S, Verga L, Salmona M, Tagliavini F, Angeretti N. Neuroreport. 1993;4:523–526. doi: 10.1097/00001756-199305000-00015. [DOI] [PubMed] [Google Scholar]
- 16.Yankner BA, Duffy LK, Kirschner DA. Science. 1990;250:279–282. doi: 10.1126/science.2218531. [DOI] [PubMed] [Google Scholar]
- 17.Serpell LC. Biochim Biophys Acta. 2000;1502:16–30. doi: 10.1016/s0925-4439(00)00029-6. [DOI] [PubMed] [Google Scholar]
- 18.Tjernberg LO, Pramanik A, Björling S, Thyberg P, Thyberg J, Nordstedt C, Berndt KD, Terenius L, Rigler R. Chem Biol. 1999;6:53–62. doi: 10.1016/S1074-5521(99)80020-9. [DOI] [PubMed] [Google Scholar]
- 19.Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB. Proc Natl Acad Sci USA. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu M, Souillac PO, Ionescu-Zanetti C, Carter SA, Fink AL. J Biol Chem. 2002;277:50914–50922. doi: 10.1074/jbc.M207225200. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Darden TA, Bartolotti L, Kominos D, Pedersen LG. Biophys J. 1999;76:2871–2878. doi: 10.1016/S0006-3495(99)77442-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esler WP, Stimson ER, Jennings JM, Vinters HV, Ghilardi JR, Lee JP, Mantyh PW, Maggio JE. Biochemistry. 2000;39:6288–6295. doi: 10.1021/bi992933h. [DOI] [PubMed] [Google Scholar]
- 23.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.