Abstract
The family of 14-3-3 proteins has emerged as critical regulators of diverse cellular responses under both physiological and pathological conditions. Here, we report an important role of 14-3-3ζ in tumorigenesis through a mechanism that involves anoikis resistance. 14-3-3ζ is up-regulated in a number of cancer types, including lung cancer. Through an RNAi approach using human lung adenocarcinoma-derived A549 cells as a model system, we have found that knockdown of a single ζ isoform of 14-3-3 is sufficient to restore the sensitivity of cancer cells to anoikis and impair their anchorage-independent growth. Enhanced anoikis appears to be mediated in part by up-regulated BH3-only proteins, Bad and Bim, coupled with decreased Mcl-1, resulting in the subsequent activation of Bax. This study suggests a model in which anchorage-independent growth of lung cancer cells requires the presence of 14-3-3ζ. This work not only reveals a critical role of 14-3-3ζ in anoikis suppression in lung cancer cells, but also identifies and validates 14-3-3ζ as a potential molecular target for anticancer therapeutic development.
Keywords: molecular target, RNAi, tumorigenesis, apoptosis, BH3-only
The 14-3-3 proteins have emerged as critical regulators of diverse cellular responses in eukaryotic organisms (see refs. 1–4 for reviews). The family of mammalian 14-3-3 proteins has seven defined isoforms: β, ε, γ, η, σ, τ, and ζ. Initially, they were described as enzyme cofactors that affect the activity of their associated tryptophan and tyrosine hydroxylase, protein kinase C, exoenzyme S ADP-ribosyltransferase, AANAT, and Raf-1 (2). The interaction of 14-3-3 with phosphorylated Raf-1 led to the discovery of 14-3-3 as the founding member of the class of phosphoserine/threonine-binding protein modules (5–9). The availability of well characterized 14-3-3 recognition motifs coupled with the use of powerful proteomics approaches has revealed an entirely new landscape in which 14-3-3 binds a variety of signaling molecules, controlling their function in response to environmental signals (9–13). In addition to protein enzymes, 14-3-3-associated client proteins include various transmembrane and nuclear receptors, adaptor proteins, and transcription factors, thus drastically expanding the functional roles of 14-3-3. Because of the primarily phosphorylation-dictated nature of 14-3-3 interactions, 14-3-3 has been tightly integrated into the central phosphor-relay signaling network that forms the core of vital signal transduction pathways. Through regulated interactions with crucial signaling mediators, 14-3-3 controls diverse cellular responses ranging from cell proliferation and differentiation to cell cycle checkpoint control and programmed cell death.
Because of its importance in the regulation of key signal transduction pathways, dysregulation of 14-3-3 has been associated with pathological consequences. In addition to their participation in various neurodegenerative disorders and inflammatory diseases, isoforms of 14-3-3 proteins have been implicated in tumorigenesis either as potential tumor suppressors or oncogenes (3, 14–16). In particular, 14-3-3σ has attracted much attention since its discovery as a p53-regulated cell cycle inhibitor (17, 18). The σ isoform plays an important role in several critical aspects of cell growth control, including mitotic translational switch (17, 19). In many cancer types, 14-3-3σ appears to be silenced by promoter methylation or degraded by an ubiquitin-mediated proteolytic pathway, which is consistent with its suggested role as a tumor suppressor (20–23). Such down-regulation of σ was correlated with oncogenesis. Interestingly, σ expression has a negative role in survival of lung cancer and colorectal carcinoma patients (24, 25). On the other hand, other 14-3-3 isoforms seem to play a role in cancer promotion, although the underlying mechanisms remain to be defined. For instance, overexpression of 14-3-3β promotes MAPK-dependent tumor formation in nude mice (26). Overexpression of 14-3-3τ leads to cell adhesion to tenascin-C and an enhanced growth rate of tumor cells (27). Thus, various 14-3-3 isoforms may offer different opportunities for the development of novel anticancer therapeutics.
Targeting 14-3-3 with a general antagonist, such as R18 or difopein, which induces a global inhibition of 14-3-3 function, has been described (28, 29). This approach suggested an essential role of the 14-3-3 family in cell survival and provided proof-of-concept that inhibition of 14-3-3 may be able to sensitize cancer cells to chemotherapeutic agents. However, it remains unknown whether an individual 14-3-3 isoform has a dominant role in promoting survival of cancer cells. Up-regulation of such an isoform would be expected to provide a survival advantage to tumor cells. At the same time, such an interdependent relationship of cancer cells on an up-regulated 14-3-3 isoform may form the basis for oncogene addiction (30), which may offer therapeutic opportunities for an isoform-specific approach. To test this hypothesis, we examined the contribution of 14-3-3ζ to oncogenesis through an RNAi approach using human A549 adenocarcinoma cells as a model system. Consistent with a previous report (31), 14-3-3ζ is indeed overexpressed in a number of lung cancer cell lines and in lung cancer tissues of patients. A549 cells are highly tumorigenic and readily form colonies in soft agar medium. Interestingly, knockdown (KD) of a single ζ isoform of 14-3-3 is sufficient to impair the anchorage-independent growth of A549 cells and restore their sensitivity to anoikis. Our study demonstrates a critical role of 14-3-3ζ in suppression of anoikis in lung cancer cells and, importantly, identifies a novel target for anticancer therapeutic intervention.
Results
14-3-3ζ Is Up-Regulated in Cancer Cells.
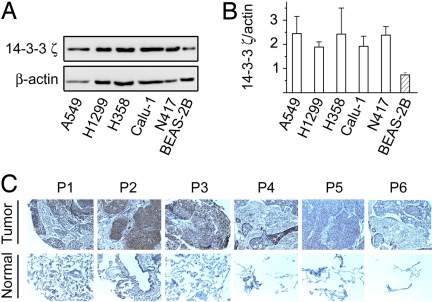
14-3-3 proteins comprise a family of seven highly homologous molecules in mammalian cells. To begin our systemic investigation of how each 14-3-3 isoform contributes to cell growth control and tumorigenesis, we examined the expression of 14-3-3ζ protein in human lung cancer cells. In comparison to its expression in the cultured human normal bronchial epithelial cell line, BEAS-2B, 14-3-3ζ appeared to be up-regulated in a panel of lung cancer cells including A549 and H358 adenocarcinoma, H1299 large cell carcinoma, Calu-1 squamous cell carcinoma, and N417 variant small cell lung cancer cells as determined by a ζ-isoform-specific antibody (Fig. 1). This result suggests that the ζ isoform of 14-3-3 is dysregulated in tumors. In support of this notion, 14-3-3ζ was found by immunohistochemistry staining to be up-regulated in four of six tumor tissues compared with normal tissues from the same patient (Fig. 1). These data raise the possibility that overexpression of 14-3-3ζ may contribute to tumorigenesis.
Fig. 1.
Expression of 14-3-3ζ isoform. (A) 14-3-3ζ expression in lung cancer cell lines. Cell lysates (10 μg) were prepared from lung cancer or control BEAS-2B cells grown in DMEM with serum and separated by SDS/PAGE (12.5%) (47). 14-3-3ζ was revealed by Western blotting with a ζ-specific antibody (Santa Cruz). (B) Summary of 14-3-3ζ expression from three experiments. The expression levels of 14-3-3ζ and β-actin were determined as in A and quantified by densitometry. Ratios of 14-3-3ζ over β-actin are expressed. (C) Expression of 14-3-3ζ in a tissue slide is shown. Immunohistochemistry was performed on a slide with paired fixed tissue samples (ABXIS) using anti-14-3-3ζ antibody (Immuno-Biological Lab). (Upper) Tissue slices from nonsmall cell lung cancer patients (P1–P6). (Lower) Normal matching tissues from the same patient. (Magnification: × 10.)
14-3-3ζ Is Required for Anchorage-Independent Growth of Tumor Cells.
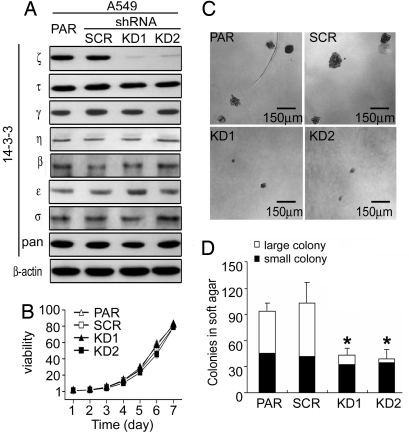
To investigate whether 14-3-3ζ has a role in cell growth control and tumorigenesis and explore the consequence of ζ isoform inhibition, we used RNAi to reduce cellular ζ levels. Two stable A549 cell lines were established expressing distinct shRNA that target two different sequences in the 3′ noncoding region of 14-3-3ζ mRNA, termed 14-3-3ζ KD1 and KD2. Compared with the parental A549 (PAR) and A549 harboring a control scrambled shRNA (SCR), KD1 and KD2 showed diminished 14-3-3ζ protein levels (Fig. 2A). Depletion of intracellular ζ showed no obvious effect on the expression of other 14-3-3 isoforms.
Fig. 2.
14-3-3ζ is required for anchorage-independent growth of A549 cells. (A) Expression of 14-3-3 isoforms in A549 parental (PAR) and scrambled (SCR) cells and A549/14-3-3ζ shRNA cells (KD1 and KD2). A549 and its derivative cell lines were grown in the presence of serum and lysed for Western blot analysis. Expression of 14-3-3 isoforms was determined by various antibodies (ζ, τ,, ε, pan from Santa Cruz; γ, η from Immuno-Biological Lab; β from GeneTex; and σ from Upstate). (B) Growth curves of A549 and its derivatives in medium with serum. (C) Colony formation of A549 and its derivative cell lines in semisolid medium. Cells on soft agar plates were grown for 3 weeks before colonies were stained and visualized microscopically. A representative view of each cell line is shown. (D) Quantification of colony formation data derived from C. Colonies were counted in a blinded fashion. Those with a diameter of >50 μm are defined as large and those <50 μm as small colonies. Results from one representative experiment are shown.
Malignant transformation requires the acquisition of a number of tumor features, including increased growth rate and anchorage-independent growth potential (32). Using the defined cell lines, we aimed to test whether the ζ isoform plays any role in the process of tumorigenesis. Examination of growth rates revealed no significant difference among control and ζ KD cells (Fig. 2B). An in vitro indicator for tumorigenesis potential is the ability of transformed cells to grow in an anchorage-independent environment (33). A549 cells grew readily and formed colonies in semisolid medium without adherence to a solid substratum (Fig. 2C). Similar sizes of colonies were observed in SCR cells as in parental cells. In contrast, shRNA-mediated KD of 14-3-3ζ was sufficient to impair the anchorage-independent growth capability of A549 cells, leading to a drastic reduction in the number and size of colonies formed in soft agar medium (Fig. 2 C and D). These results revealed a novel function of 14-3-3ζ in supporting anchorage-independent growth of tumor cells. This effect is not limited to A549 cells as KD of 14-3-3ζ in H1299 lung cancer cells also led to a reduction in colonies formed in the soft agar medium [supporting information (SI) Fig. 8]. Anchorage independence is an important feature that allows tumor cells to survive under certain inappropriate host conditions, such as during invasion and metastasis. Up-regulation of 14-3-3ζ may be one of the oncogenic events that leads to anchorage-independent survival and growth of lung cancer cells.
KD of 14-3-3ζ Impairs Anoikis Avoidance of Cancer Cells.
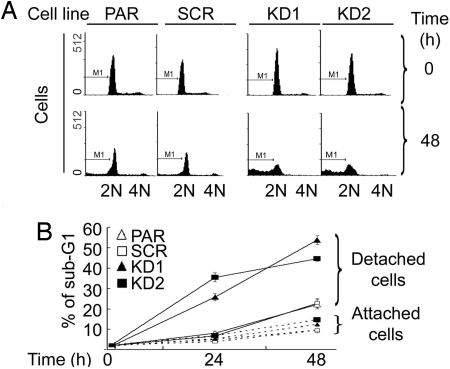
Detachment of normal epithelial cells from their substratum leads to apoptotic cell death, termed anoikis. The anchorage-independent growth of A549 cells reflects their enhanced ability to resist anoikis (34, 35). It is possible that up-regulated 14-3-3ζ provides a molecular mechanism that protects A549 cells from undergoing anoikis, leading to anchorage independence. To test this hypothesis, we compared the apoptotic tendency of A549 cells with that of 14-3-3ζ KD cells upon detachment from the plate matrix. A DNA content-based apoptosis assay was performed by using a fluorescence-activated cell analyzer. Cells with subdiploid DNA content were scored as apoptotic. As a control, a basal level of apoptosis was detected when cells were attached to culture plates (Fig. 3). No difference in apoptosis was observed in A549 cells either with or without 14-3-3ζ within the time frame tested (Fig. 3). Upon detachment from the plate matrix, PAR and SCR cells showed a slightly increased apoptotic activity, suggesting that A549 lung cancer cells are relatively resistant to anoikis. This finding is consistent with a previous report (34). On the other hand, this anoikis resistance of A549 cells was dramatically decreased by silencing of 14-3-3ζ, showing an increased population of apoptotic cells upon detachment from the matrix (Fig. 3). These data strongly support a role of up-regulated 14-3-3ζ in protecting cancer cells from anoikis.
Fig. 3.
Decreased expression of 14-3-3ζ is correlated with increased anoikis. A549 or its derivative cells were grown either under attached or detached (on polyHEMA-treated plates) conditions, and total cells were collected for DNA content analysis. (A) Representative DNA histograms of A549 cells analyzed with a flow cytometer. A549 PAR, SCR, KD1, and KD2 cells were grown in polyHEMA-coated plates and harvested at time 0 (Upper) and 48 h (Lower) after detachment for analysis. (B) Effect of 14-3-3ζ KD on sub-G1 DNA content of A549 cells. Data from a representative experiment are shown.
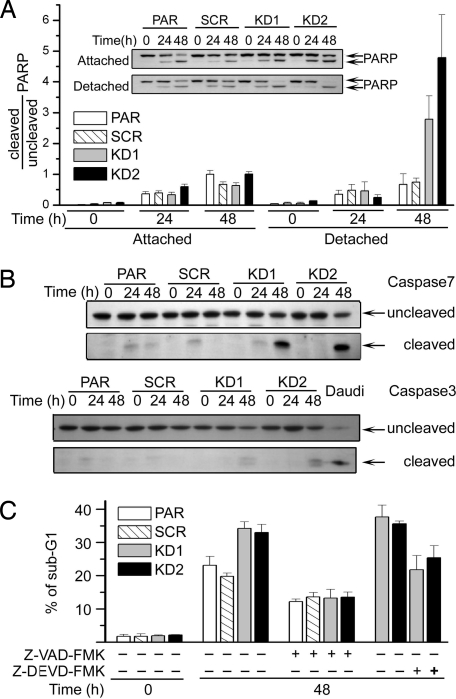
Because caspases are critical intermediates in anoikis pathways, caspase activities were used to validate the 14-3-3ζ KD effect. Executioner caspase activities were monitored by their cleavage of polyADP ribose polymerase (PARP) and caspase processing. Although cells attached to the plate matrix showed certain levels of PARP cleavage upon serum withdrawal, cells with depleted 14-3-3ζ showed almost complete PARP cleavage when detached from the matrix (Fig. 4A). With isoform-specific antibodies, fully processed caspase 7 was prominently identified in ζ KD cells undergoing anoikis over control cells (Fig. 4B). In the same sample, only trace amounts of processed caspase 3 were visible. These data suggest that 14-3-3ζ plays a role in anoikis resistance in part through suppression of caspase activation. In support of this notion, addition of a pan-caspase inhibitor, Z-VAD-FMK, or a relatively selective inhibitor of executioner caspase 3/7, Z-DEVD-FMK, abolished the ζ effect, showing decreased DNA fragmentation regardless of the 14-3-3 background (Fig. 4C). Together, these results demonstrate a critical role of 14-3-3ζ in suppression of caspase-dependent anoikis in A549 cells.
Fig. 4.
Caspase activation mediates anoikis in 14-3-3ζ KD cells. (A) Decreased 14-3-3ζ is correlated with increased caspase activity upon anoikis induction. Cells were grown either in attached or detached conditions and harvested for Western blotting with anti-PARP antibody (Inset; Cell Signaling). Ratios of cleaved to uncleaved PARP are summarized from three independent experiments. (B) Caspase 7 is activated in A549 14-3-3ζ KD1 and KD2 cells. Cells were grown on polyHEMA-treated plates and harvested for Western blotting with anti-caspase 7 and anti-caspase 3 antibodies (Cell Signaling). Lower blots show cleaved activated caspase 7 or caspase 3. (C) Caspase inhibitors decrease the level of anoikis. Cells in polyHEMA-coated plates were treated with Z-VAD-FMK or Z-DEVD-FMK before harvesting for DNA content analysis.
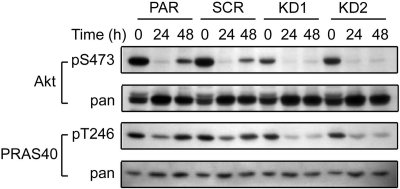
14-3-3ζ Is Involved in the Biphasic Regulation of Akt.
To understand how 14-3-3ζ deficiency affects survival signaling of lung cancer cells, we examined whether the status of the ζ isoform could alter protein kinase Src, Pyk2, and Akt/PKB activity. Src and Pyk2 are known to mediate antianoikis signaling in A549 cells (34). However, silencing of 14-3-3ζ had no effect on Src and Pyk2 activity during anoikis (SI Fig. 9). Akt plays a major role in cell survival by phosphorylating a number of key proapoptotic 14-3-3 client proteins, including Bad (36). Cellular Akt activity was assessed by the phosphorylation status of Akt at S473 and that of its substrate PRAS40 at T246. Cells with 14-3-3ζ KD showed no difference in Akt activity from PAR and SCR control cells under attached conditions (data not shown). Interestingly, when A549PAR and SCR control cells were grown in poly (2-hydroxyethyl-methacrylate) (polyHEMA)-treated culture plates, Akt exhibited a biphasic activity. pS473-Akt decreased at 24 h and recovered at 48 h, consistent with a previous report (37). It is possible that the recovered Akt activity represents a cellular protective feedback mechanism to prolong survival. In contrast, 14-3-3ζ KD cells failed to show recovery of Akt activity after its initial decline (Fig. 5). Thus, 14-3-3ζ seems to be required for the protective feedback mechanism of Akt upon prolonged cell detachment from matrix. However, this ζ-regulated Akt activity appears to be insufficient to explain the anoikis suppression function of ζ as seen in Fig. 3. Failed Akt reactivation at this late stage unlikely contributes to the initially increased anoikis response in 14-3-3ζ-defective cells.
Fig. 5.
Decreased 14-3-3ζ is associated with reduced Akt/PKB activity. A549 or its derivative cells were grown on polyHEMA-coated plates in medium without serum. At the indicated times, cells were collected and lysed for Western blotting with the antibodies indicated.
Dysregulation of Bcl-2 Family Proteins Is Associated with 14-3-3ζ Deficiency.
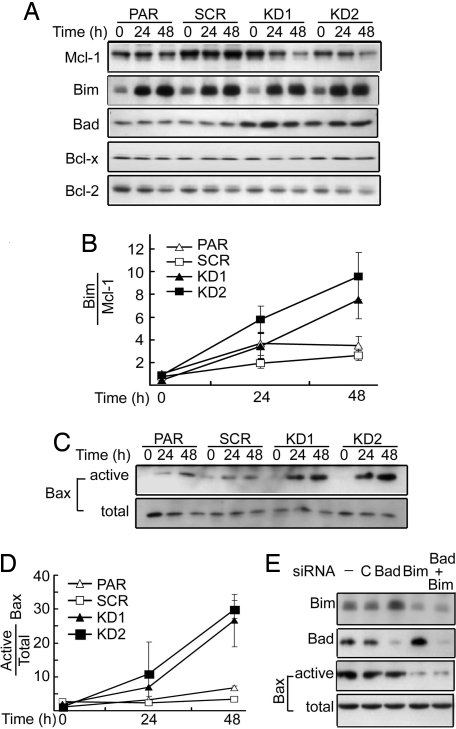
To identify the molecular mechanism that underlies the enhanced anoikis in 14-3-3ζ-deficient cells, we focused on the Bcl-2 family of apoptosis regulators that integrate cellular survival and apoptosis signals through their action on mitochondria (38). The prosurvival members of the Bcl-2 family, Bcl-2, Bcl-xL, and Mcl-1, have been shown to restrain the proapoptotic activity of Bax and Bak, which function to induce mitochondrial outer membrane permeability and the release of apoptogenic factors. The BH3-only proteins sense cellular stress signals and antagonize the function of prosurvival Bcl-2 members that guard Bax and Bak. We reasoned that 14-3-3ζ levels might control the status of BH3-only proteins in A549 cells, which, in turn, determine cancer cell susceptibility to anoikis. To test this model, we monitored the expression levels of both broadly acting BH3-only proteins (Bid, Puma, and Bim) and selectively acting BH3-only proteins (Bad and Noxa) (39). Among BH3-only proteins tested, Puma, Noxa, and Bid showed no change in their expression levels upon cell detachment regardless of 14-3-3ζ status (data not shown). Strikingly, levels of Bim increased with time when cells were grown in polyHEMA-coated plates (Fig. 6A). However, the ζ status of A549 cells did not show any effect on Bim expression, with a similar trend of Bim increase upon cell detachment seen in both PAR and 14-3-3ζ KD cells. The change in Bim level per se seems insufficient to explain the enhanced anoikis in ζ KD cells. On the other hand, the expression level of the proapoptotic protein Bad was significantly increased in 14-3-3ζ KD1 and KD2 cells. It is possible that Bad coordinates with Bim to enhance anoikis in cells with decreased 14-3-3ζ.
Fig. 6.
Altered expression of Bcl-2 family members in 14-3-3ζ KD cells. (A) A549 cells and its derivatives were serum-starved under detached conditions and harvested for Western blot analysis of Bim and Bad, Bcl-2, Bcl-xL, and Mcl-1. (B) Quantification of ratio of Bim to Mcl-1 from three experiments as in A. (C) Detection of active Bax. Cells were detached from matrix and harvested at the indicated times. Active Bax was immunoprecipitated with an active-conformation-specific mAb (6A7) and revealed by Western blotting with an anti-Bax polyclonal antibody. (Upper) The conformationally changed Bax. (Lower) Total Bax in the lysate. (D) Quantification of results from C. (E) KD of Bim abolished detachment-induced Bax activation. A549 KD2 cells were transfected with siRNA for Bad, Bim, Bad/Bim, or control from Dharmacon, and expression of Bad and Bim was revealed by Western blotting. Activated Bax at 48 h after detachment was detected as in C.
The BH3-only proteins control apoptosis sensitivity by engaging prosurvival members of the Bcl-2 family. Therefore, it is the relative ratio of BH3-only over prosurvival Bcl-2 members that impacts the outcome of stress-induced apoptosis (38). Thus, we examined the status of prosurvival Bcl-2 members in anoikis in relation to 14-3-3ζ status. No significant change in Bcl-2 or Bcl-xL was observed in all four cell lines tested under anoikis conditions (Fig. 6A). In contrast, Mcl-1 appeared to decrease with time upon cell detachment in 14-3-3ζ KD1 and KD2 cells versus parental control cells (Fig. 6 A and B). Decreased Mcl-1 and increased Bad in 14-3-3ζ shRNA-treated stable cell lines along with anoikis-induced Bim may tip the balance in A549 cells toward enhanced apoptotic potential.
The above model predicts a decreased threshold for the induction of active Bax when 14-3-3ζ is reduced. To test this notion, we monitored conformational change of Bax as an indicator of its activation (40). Upon cell detachment, conformationally changed active Bax was slightly increased in PAR and SCR control cells (Fig. 6 C and D). On the other hand, both KD1 and KD2 cells exhibited a drastically increased population of cells with activated Bax, whereas total Bax levels remained the same. These data together suggest a vital role of the ζ isoform of 14-3-3 in suppression of mitochondria-mediated anoikis in cancer cells. To further define a role of Bad and/or Bim in mediating anoikis in KD1/KD2 cells, a siRNA approach was used to knock down Bad and Bim (Fig. 6E). However, Bad siRNA showed no effect on anoikis. On the other hand, down-regulation of Bim, either alone or in combination with Bad, significantly decreased detachment-induced Bax activation in 14-3-3ζ KD cells. These data support the model that Bim plays an intimate role in transmitting enhanced anoikis signaling in 14-3-3ζ-deficient A549 cells.
Discussion
One of the most important oncogenic properties of cancer cells is their ability to survive and grow in the absence of anchorage to the extracellular matrix (32, 35). Unlike normal cells in which the anoikis program is activated after loss of adhesion to substratum, many cancer cells develop mechanisms that lead to anoikis resistance. Such breakdown of anoikis control has been shown to contribute significantly to the malignancy of many solid tumors, including lung cancer (35). Thus, identification of molecular events that control anoikis in cancer cells has significant therapeutic implications. Here, we identify a particular isoform of 14-3-3, ζ, as a critical suppressor of anoikis in lung cancer cells. KD of ζ restores the sensitivity of A549 cancer cells to anoikis and inhibits their anchorage-independent growth. This effect is mediated in part by dysregulated BH3-only protein function, leading to a lowered threshold for the activation of Bax. Our work not only reveals an important role of 14-3-3ζ in the suppression of anoikis, but also validates 14-3-3ζ as a potential molecular target for the development of anticancer agents. This 14-3-3ζ-targeted strategy is supported by recent clinical data that associate 14-3-3ζ expression with advanced disease grade and poor survival outcome of lung cancer patients (41).
Among BH3-only proteins, Bim was shown to mediate anoikis in mammary epithelial cells (42). Our results indicate a critical role of Bim in mediating anoikis in lung cancer cells. Although Bad did not further increase with time upon cell detachment, an increase in Bad basal level in 14-3-3ζ-deficient cells may enhance cell susceptibility to anoikis. Interestingly, increased Bim levels upon cell detachment were associated with decreased Mcl-1 in 14-3-3ζ KD cells. Bim functions in part by inhibiting Mcl-1 (39). Taken together, matrix detachment induced a significantly up-regulated ratio of Bim over Mcl-1 in ζ-reduced cells, leading to an amplified Bim proapoptotic effect (Fig. 6). Neutralization of both classes of Bcl-2/Bcl-xL and Mcl-1 by up-regulated Bad and Bim may account for enhanced Bax activation, resulting in a potent anoikis response.
There are seven known isoforms in the mammalian 14-3-3 family. KD of ζ appears to be sufficient to give rise to a significant phenotype, anoikis restoration, in A549 cells, suggesting a unique function of ζ that other isoforms cannot replace. These results also point to the possibility that up-regulated 14-3-3ζ may be part of the oncogene addiction machinery that A549 lung cancer cells rely on for survival (30). The gained ability to resist anoikis allows cancer cells to invade and metastasize, which is often fatal to patients (Fig. 7). This 14-3-3ζ effect is not limited to A549 cells. Further shRNA studies with H1299 lung cancer cells also show a role of 14-3-3ζ in anoikis resistance, albeit with a delayed anoikis response (SI Fig. 8). This notion offers an attractive opportunity for therapeutic development by inhibiting 14-3-3ζ to sensitize lung cancers to anoikis. Beyond lung cancer cells, KD of 14-3-3ζ has been shown to promote oncogenic properties of HaCaT cells, with increased sensitivity to UV-induced apoptosis and cell adhesion (43).
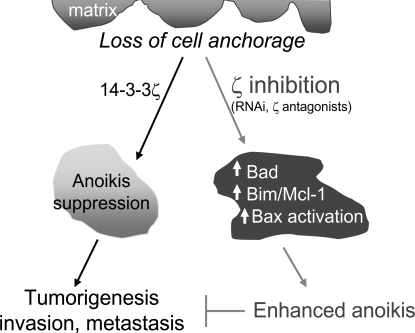
Fig. 7.
Working model for a critical role of 14-3-3ζ in anoikis suppression and tumorigenesis. In A549 lung cancer cells, 14-3-3ζ suppresses anoikis upon cell detachment from the matrix, which contributes to tumorigenesis, invasion, and metastasis. Inhibition of ζ through either RNAi or a ζ antagonist up-regulates Bad, Bim/Mcl-1, leading to Bax activation and the restoration of the anoikis program. Enhanced anoikis suppresses tumorigenesis.
The 14-3-3ζ KD effect could be achieved by a general 14-3-3 antagonist, like R18 or difopein, which competitively binds to the amphipathic groove of 14-3-3, or preferably by 14-3-3ζ-selective agents (28, 44). Although a global 14-3-3 inhibitor may be able to achieve a desired effect on multiple pathways important for selected disease processes, small-molecule inhibitors are preferred for this purpose given the ability to fine tune their use. On the other hand, a 14-3-3ζ-selective agent may offer additional advantages such as selectivity for tumors with 14-3-3ζ dependency. Developing selective 14-3-3ζ-targeted agents is more challenging and could involve the disruption of 14-3-3ζ–specific protein–protein interactions or ζ-regulated pathways. Alternatively, dissection of transcription mechanisms that control the specific expression of the ζ isoform may offer additional opportunities for ζ-specific inhibition. Because 14-3-3ζ is up-regulated in many other cancers, such as oral and stomach cancers (45, 46), development of small-molecule inhibitors of 14-3-3ζ may find broad therapeutic applications.
Materials and Methods
Cell Culture and Growth Conditions.
Attached cells were grown in monolayer culture in RPMI medium 1640 with or without FBS (10%). Detached cells were grown in six-well plates precoated with polyHEMA (1.2%). Cells were cultured at 37°C in a humidified incubator with CO2 (5%) and air (95%).
Immunological Analysis.
Cell lysate preparation and Western blotting with enhanced chemiluminescence system were performed essentially as described (28).
Generation of 14-3-3ζ KD Cell Lines.
Recombinant retroviruses carrying 14-3-3ζ-KD shRNA were prepared with pSilencer 5.1 (Ambion). Two distinct sequences for 14-3-3ζ were selected: ACGGTTCACATTCCATTAT and ATAGTTAACAGGGAAATAA. To generate 14-3-3ζ-silenced stable cell lines, infected cells were selected in the presence of puromycin (1.25 μg/ml). Drug-resistant clones were collected, pooled, and expanded. The 14-3-3ζ level was verified for each experiment.
Soft Agar Colony Formation Assay.
Cells (1 × 103) were resuspended in RPMI medium 1640 (1.0 ml with 20% FBS and 0.33% agar) and plated over a layer of solidified RPMI medium 1640/20% FBS/0.66% agar (2.0 ml). Plates were incubated at 37°C, and colonies were stained with crystal violet (0.005%; Sigma–Aldrich) and scored in a blinded fashion.
Anoikis and Cell Death Assays.
For anoikis assays, cells were seeded on culture plates pretreated with polyHEMA (1.2%). For DNA content analysis, both attached and detached cells were collected, fixed, stained with propidium iodide, and analyzed by a FACScan cytometer equipped with Cell Quest software (BD Biosciences). Apoptosis marker analysis including caspase and Bax activation was detected by Western blotting. To detect conformational change of Bax, cells were lysed in CHAPS buffer and immunoprecipitated with anti-Bax mAb (6A7). Active Bax was detected with polyclonal anti-Bax antibody (40).
Supplementary Material
ACKNOWLEDGMENTS.
We thank members of H.F.'s laboratory for helpful discussions and Anthea Hammond for editing. This work was supported in part by National Institutes of Health Grants R01 GM53165 (to H.F.) and P01 CA116676 (to F.R.K., H.F., and S.S.), Emory University Research Committee, and Golfers Against Cancer. F.R.K., S.S., and H.F. are Georgia Cancer Coalition Distinguished Scholars. H.F. is a Georgia Research Alliance Distinguished Investigator. Y.D. is an Emory Drug Development and Pharmacogenomics Academy Fellow.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710905105/DC1.
References
- 1.Fu H, Subramanian R-R, Masters S-C. 14-3-3 proteins: Structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Aitken A. 14-3-3 proteins: A historic overview. Semin Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Wilker E, Yaffe M-B. 14-3-3 Proteins: A focus on cancer and human disease. J Mol Cell Cardiol. 2004;37:633–642. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 4.Muslin A-J, Lau J-M. Differential functions of 14-3-3 isoforms in vertebrate development. Curr Top Dev Biol. 2005;65:211–228. doi: 10.1016/S0070-2153(04)65008-3. [DOI] [PubMed] [Google Scholar]
- 5.Fantl W-J, et al. Activation of Raf-1 by 14-3-3 proteins. Nature. 1994;371:612–614. doi: 10.1038/371612a0. [DOI] [PubMed] [Google Scholar]
- 6.Fu H, et al. Interaction of the protein kinase Raf-1 with 14-3-3 proteins. Science. 1994;266:126–129. doi: 10.1126/science.7939632. [DOI] [PubMed] [Google Scholar]
- 7.Irie K, et al. Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science. 1994;265:1716–1719. doi: 10.1126/science.8085159. [DOI] [PubMed] [Google Scholar]
- 8.Michaud N-R, Fabian J-R, Mathes K-D, Morrison D-K. 14-3-3 is not essential for Raf-1 function: Identification of Raf-1 proteins that are biologically activated in a 14-3-3- and Ras-independent manner. Mol Cell Biol. 1995;15:3390–3397. doi: 10.1128/mcb.15.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muslin A-J, Tanner J-W, Allen P-M, Shaw A-S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 10.Yaffe M-B, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 11.Pozuelo R-M, et al. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation, and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meek S-E, Lane W-S, Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J Biol Chem. 2004;279:32046–32054. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 13.Jin J, et al. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 14.Porter G-W, Khuri F-R, Fu H. Dynamic 14-3-3/client protein interactions integrate survival and apoptotic pathways. Semin Cancer Biol. 2006;16:193–202. doi: 10.1016/j.semcancer.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Toyo-oka K, et al. 14-3-3epsilon is important for neuronal migration by binding to NUDEL: A molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- 16.Kilani R-T, et al. Detection of high levels of 2 specific isoforms of 14-3-3 proteins in synovial fluid from patients with joint inflammation. J Rheumatol. 2007;34:1650–1657. [PubMed] [Google Scholar]
- 17.Hermeking H. 14-3-3 proteins and cancer biology. Semin Cancer Biol. 2006;16:161. doi: 10.1016/j.semcancer.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Hermeking H, et al. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 19.Wilker E-W, et al. 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446:329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- 20.Iwata N, et al. Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene. 2000;19:5298–5302. doi: 10.1038/sj.onc.1203898. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki H, et al. Inactivation of the 14-3-3 sigma gene is associated with 5′ CpG island hypermethylation in human cancers. Cancer Res. 2000;60:4353–4357. [PubMed] [Google Scholar]
- 22.Ferguson AT, et al. High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci USA. 2000;97:6049–6054. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urano T, et al. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumor growth. Nature. 2002;417:871–875. doi: 10.1038/nature00826. [DOI] [PubMed] [Google Scholar]
- 24.Perathoner A, et al. 14-3-3sigma expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Clin Cancer Res. 2005;11:3274–3279. doi: 10.1158/1078-0432.CCR-04-2207. [DOI] [PubMed] [Google Scholar]
- 25.Ramirez J-L, et al. 14-3-3sigma methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival: The Spanish Lung Cancer Group. J Clin Oncol. 2005;23:9105–9112. doi: 10.1200/JCO.2005.02.2905. [DOI] [PubMed] [Google Scholar]
- 26.Takihara Y, Matsuda Y, Hara J. Role of the beta isoform of 14-3-3 proteins in cellular proliferation and oncogenic transformation. Carcinogenesis. 2000;21:2073–2077. doi: 10.1093/carcin/21.11.2073. [DOI] [PubMed] [Google Scholar]
- 27.Martin D, Brown-Luedi M, Chiquet-Ehrismann R. Tenascin-C signaling through induction of 14-3-3 tau. J Cell Biol. 2003;160:171–175. doi: 10.1083/jcb.200206109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masters S-C, Fu H. 14-3-3 proteins mediate an essential anti-apoptotic signal. J Biol Chem. 2001;276:45193–45200. doi: 10.1074/jbc.M105971200. [DOI] [PubMed] [Google Scholar]
- 29.Wang B, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry. 1999;38:12499–12504. doi: 10.1021/bi991353h. [DOI] [PubMed] [Google Scholar]
- 30.Weinstein I-B. Cancer: Addiction to oncogenes–the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 31.Qi W, Liu X, Qiao D, Martinez J-D. Isoform-specific expression of 14-3-3 proteins in human lung cancer tissues. Int J Cancer. 2005;113:359–363. doi: 10.1002/ijc.20492. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D, Weinberg R-A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 33.Freedman V-H, Shin S-I. Cellular tumorigenicity in nude mice: Correlation with cell growth in semi-solid medium. Cell. 1974;3:355–359. doi: 10.1016/0092-8674(74)90050-6. [DOI] [PubMed] [Google Scholar]
- 34.Wei L, Yang Y, Zhang X, Yu Q. Altered regulation of Src upon cell detachment protects human lung adenocarcinoma cells from anoikis. Oncogene. 2004;23:9052–9061. doi: 10.1038/sj.onc.1208091. [DOI] [PubMed] [Google Scholar]
- 35.Frisch S-M, Screaton R-A. Anoikis mechanisms. Curr Opin Cell Biol. 2001;13:555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 36.Manning B-D, Cantley L-C. AKT/PKB signaling: Navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kodama K, et al. Laminin 5 expression protects against anoikis at aerogenous spread and lepidic growth of human lung adenocarcinoma. Int J Cancer. 2005;116:876–884. doi: 10.1002/ijc.21136. [DOI] [PubMed] [Google Scholar]
- 38.Danial N-N, Korsmeyer S-J. Cell death: Critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 39.Chen L, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 40.Yamaguchi H, et al. Epothilone B analogue (BMS-247550)-mediated cytotoxicity through induction of Bax conformational change in human breast cancer cells. Cancer Res. 2002;62:466–471. [PubMed] [Google Scholar]
- 41.Fan T, et al. Up-regulation of 14-3-3zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 2007;67:7901–7906. doi: 10.1158/0008-5472.CAN-07-0090. [DOI] [PubMed] [Google Scholar]
- 42.Reginato M-J, et al. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5:733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- 43.Niemantsverdriet M, Wagner K, Visser M, Backendorf C. Cellular functions of 14-3-3zeta in apoptosis and cell adhesion emphasize its oncogenic character. Oncogene. 2007 doi: 10.1038/sj.onc.1210742. [DOI] [PubMed] [Google Scholar]
- 44.Du Y, Masters S-C, Khuri F-R, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay. J Biomol Screen. 2006;11:269–276. doi: 10.1177/1087057105284862. [DOI] [PubMed] [Google Scholar]
- 45.Arora S, Matta A, Shukla N-K, Deo S-V, Ralhan R. Identification of differentially expressed genes in oral squamous cell carcinoma. Mol Carcinog. 2005;42:97–108. doi: 10.1002/mc.20048. [DOI] [PubMed] [Google Scholar]
- 46.Jang J-S, Cho H-Y, Lee Y-J, Ha W-S, Kim H-W. The differential proteome profile of stomach cancer: Identification of the biomarker candidates. Oncol Res. 2004;14:491–499. doi: 10.3727/0965040042380441. [DOI] [PubMed] [Google Scholar]
- 47.Sun S-Y, et al. Differential responses of normal, premalignant, and malignant human bronchial epithelial cells to receptor-selective retinoids. Clin Cancer Res. 1999;5:431–437. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.