Abstract
Hypoxia-inducible factor-1 (HIF-1) plays an essential role in tumor development and progression by regulating genes that are vital for proliferation, glycolysis, angiogenesis, and metastasis. To identify strategies of targeting the HIF-1 pathway, we screened a siRNA library against the entire druggable genome and a small-molecule library consisting of 691,200 compounds using a HIF-1 reporter cell line. Although the siRNA library screen failed to reveal any druggable targets, the small-molecule library screen identified a class of alkyliminophenylacetate compounds that inhibit hypoxia-induced HIF-1 reporter activity at single-digit nanomolar concentrations. These compounds were found to inhibit hypoxia but not deferoxamine-induced HIF-1α protein stabilization. Further analysis indicated that the alkyliminophenylacetate compounds likely inhibit the HIF-1 pathway through blocking the hypoxia-induced mitochondrial reactive oxygen species (ROS) production. Strikingly, all of the nonalkyliminophenylacetate HIF-1 inhibitors identified from the small-molecule library screen were also found to target mitochondria like the alkyliminophenylacetate compounds. The exclusive enrichment of mitochondria inhibitors from a library of >600,000 diverse compounds by using the HIF-1 reporter assay highlights the essential role of mitochondria in HIF-1 regulation. These results also suggest that targeting mitochondrial ROS production might be a highly effective way of blocking HIF-1 activity in tumors.
Keywords: hypoxia-inducible factor, hypoxia, reactive oxygen species, siRNA, RNAi
Hypoxia-inducible factor-1 (HIF-1) is the master regulator of cellular responses to low oxygen (1–3). It consists of a constitutively expressed β subunit and an oxygen-regulated α subunit. The proline hydroxylase domain-containing proteins (PHDs) were originally thought to be the main oxygen sensor that mediates oxygen-dependent HIF-1α degradation. Under aerobic conditions, PHDs hydroxylate HIF-1α at key proline residues by using oxygen as a substrate, which ultimately causes polyubiquitination and degradation of HIF-1α through an E3 ubiquitin ligase complex containing the von Hippel–Lindau protein (4, 5). Because oxygen is required for PHD-mediated hydroxylation, the low oxygen level under hypoxic conditions prevents HIF-1α hydroxylation, which subsequently leads to the stabilization of HIF-1α. However, several recent studies demonstrate that the hypoxia-induced production of reactive oxygen species (ROS) in mitochondria is both necessary and sufficient for hypoxia-dependent HIF-1α accumulation, suggesting that mitochondria may act as an oxygen sensor for HIF-1α regulation by generating ROS under hypoxic conditions (6–9).
HIF-1 has emerged as an attractive target for cancer therapy in the last several years. The requirement of HIF-1 for tumor growth has been examined by abrogating the HIF-1 pathway in tumors by using RNAi, small molecule inhibitors, or genetic alterations. Although the degree of tumor responses to HIF-1 inhibition varies among studies, the majority of the studies demonstrate that inhibition of HIF-1 leads to slower tumor growth in vivo (10–16). In addition to its direct impact on tumor growth, HIF-1 has also been implicated in modulating tumor responses to therapy. In particular, robust antitumor efficacy has been demonstrated by combining HIF-1 inhibition with temozolamide treatment or radiation therapy, suggesting that combining HIF-1 inhibition with other therapeutic modalities could be a promising strategy for cancer therapy (17–19).
Because of the importance of HIF-1 in tumor development and progression, a considerable amount of effort has been devoted to identify HIF-1 inhibitors for cancer therapy. Several small molecules have been reported to exhibit inhibition of the HIF-1 pathway (12, 20–26). However, these compounds often have activities other than HIF-1 inhibition, and most of them lack the desired pharmacokinetic properties or toxicity profiles required for a useful pharmaceutical agent. In this study, we carried out a small-molecule library screen aimed at identifying potent and specific HIF-1 inhibitors. In parallel, we also carried out a siRNA-based loss-of-function screen to uncover druggable targets in the HIF-1 pathway. Although the siRNA library screen failed to reveal any classic druggable targets, the small-molecule library screen identified a class of compounds that inhibit hypoxia-induced HIF-1 activation at very low concentrations.
Results
siRNA-Based Loss-of-Function Screen Failed to Reveal Classic “Druggable” Targets in the HIF-1 Pathway.
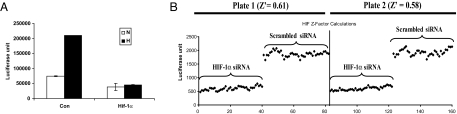
The lack of obvious druggable targets in the HIF-1 pathway has hindered the development of HIF-1-based cancer therapeutics. To identify potential druggable targets in the HIF-1 pathway, we initiated a siRNA-based loss-of-function screen using a library of siRNAs against ≈4,000 druggable targets. The aim of the screen was to identify targets that, when knocked down by siRNAs, lead to specific inhibition of the HIF-1 pathway. We have screened H1299 cells that were transiently transfected with the HIF-1 reporter using a siRNA library consisting of >500 human kinases (27). However, this transient transfection assay became prohibitively cumbersome to screen a much larger siRNA library. To increase the throughput of the HIF-1 reporter assay, we created H1299-HRE, a stable HIF-1 reporter line derived from the non-small-cell lung carcinoma cell line H1299. In this cell line, the expression of the firefly luciferase is controlled by the hypoxia-responsive element (HRE) from the enolase promoter. By using this cell line, a three- to fivefold increase of luciferase activity was observed upon hypoxia treatment. The hypoxia-dependent luciferase expression in this cell line is apparently mediated by HIF-1 because a HIF-1α siRNA abolished the induction of luciferase activity under hypoxia (Fig. 1A). By using the HIF-1α siRNA and a scrambled siRNA as positive and negative controls, Z′ factors of 0.61 and 0.58 were obtained in two independent experiments for the H1299_HRE-based 96-well reporter assay, indicating that the H1299_HRE-based assay is suitable for high-throughput screening (Fig. 1B).
Fig. 1.
siRNA library screen using the HIF-1 reporter assay. (A) The H1299_HRE cells transfected with a control siRNA (Con) or HIF-1α siRNA (Hif-1α) were subjected to hypoxia treatment and assayed for luciferase activity afterward. (B) The H1299_HRE cells were plated in 96-well plates. For each plate, half of the plate was transfected with a scramble siRNA and the other half of the plate was transfected with a HIF-1α siRNA. Forty-eight hours after transfection, the siRNA-transfected cells were subjected to hypoxia treatment for 16 h. The cells were then collected to determine the firefly luciferase activity. Z′ was calculated as Z′ = 1 − (3 σH + 3 σN)/μH − μN. In both A and B, siRNA was transfected at a final concentration of 20 nM.
Using the H1299_HRE-based reporter assay, we screened a siRNA library against ≈4,000 druggable targets using pools of four siRNAs that were designed to knock down a single target. From the primary screen and retest, 355 pools were shown to cause >40% inhibition of the HIF-1 reporter activity, resulting in an overall hit rate of 9.6%. To further test whether these hits are specific for the HIF-1 pathway, we examined the 355 confirmed hits using the dual luciferase assay in which the cells were transfected with both the HIF-1 reporter and a constitutively expressed renilla luciferase reporter. By normalizing the HIF-1-dependent firefly luciferase activity against the constitutively expressed renilla luciferase activity, pools that inhibited the HIF-1 reporter because of cytotoxicity or nonspecific transcriptional/translational inhibition were eliminated. The dual luciferase assay resulted in a significant attrition of the primary hits. Only 10 of the 355 Smartpools were found to exhibit specific inhibition of the HIF-1 reporter without significantly affecting the constitutive renilla luciferase reporter.
siRNA-mediated off-target gene silencing has been observed in many of our siRNA-library screens. In other studies, many of the top hits were invalidated as off-target hits by using multiple siRNAs against the same target (27, 28). When four individual siRNAs against each of the 10 targets from the HIF-1 reporter screen were retested, only one of the four siRNAs exhibited specific inhibition of the HIF-1 reporter. Meanwhile, 2 or more individual siRNAs from each Smartpool knocked down the target to a very high level (data not shown). The disconnection between HIF-1 reporter inhibition and the level of target knockdown suggest that the 10 pools obtained from our screen are likely off-target hits that cause HIF-1 inhibition because of off-target gene silencing triggered by one of the siRNAs in each Smartpool.
The Small-Molecule Library Screen Identified Potent HIF-1 Inhibitors.
There are several possible explanations for the failure to identify druggable targets in the HIF-1 pathway through the siRNA library screen. First, the critical intervention point in the HIF-1 pathway may not be a classic druggable target covered by our siRNA library. Another potential explanation is that a close homolog of a critical target may need to be inhibited simultaneously to block hypoxia-dependent HIF-1 activation. This could not be achieved in the siRNA-based screen because the siRNAs in our library likely only knock down one of the homologous proteins. The limitations of the siRNA-based approach can conceivably be overcome by a complimentary chemical genomics approach. A small-molecule library consists of a large number of structurally diverse compounds that may inhibit additional targets not covered by our druggable siRNA library. In addition, small molecules may not distinguish among homologous protein targets, resulting in simultaneous inhibition of many closely related proteins.
To explore the chemical genomics approach, we established a 384-well HIF-1 reporter assay using the H1299_HRE cells. A Z′ factor of 0.18 was obtained for the optimized 384-well reporter assay, indicating that the assay is suitable for high-throughput screening (data not shown). Using this assay, we screened a small-molecule library containing 691,200 compounds. From the screen, 15,756 compounds exhibited a >25% inhibition of the HIF-1 reporter activity. Among the 15,756 primary hits, 904 were confirmed by retesting. Serial dilution experiments revealed that 800 of the 904 compounds had an EC50 <10 μM on HIF-1 reporter inhibition.
To eliminate compounds that inhibit the HIF-1 reporter activity because of cytotoxicity or nonspecific transcriptional/translational inhibition, we tested the 800 most-potent compounds in two secondary assays: (i) a dual luciferase assay in which H1299 cells were cotransfected with the HIF-1 reporter (pHRE) and the pRL-SV40 reporter and (ii) a dual luciferase assay in which H1299 cells were cotransfected with the pGL3-control and the pRL-SV40 reporter. In these assays, the pRL-SV40 and the pGL3-control reporters use the SV40 promoter to express the firefly and renilla luciferase, respectively. Because the SV40 promoter is constitutively active, the inhibitory effect of a compound on the pGL3-control or the pRL-SV40 reporter would suggest that the compound may be toxic to the cell or is a general transcriptional/translational inhibitor. Using these secondary assays, we eliminated compounds that either inhibited the pGL3-control reporter alone (likely firefly luciferase inhibitors) or inhibited both the pGL3-control and the pRL-SV40 reporters (likely cell growth inhibitors or general transcriptional/translational inhibitors). From these analyses, 250 of the 800 compounds were found to specifically inhibit the HIF-1 reporter (>50% inhibition) at 1 μM without significantly affecting the activity of the pGL3-control or the pRL-SV40 reporters (<20% inhibition).
Alkyliminophenylacetate Compounds Specifically Inhibit the HIF-1 Pathway.
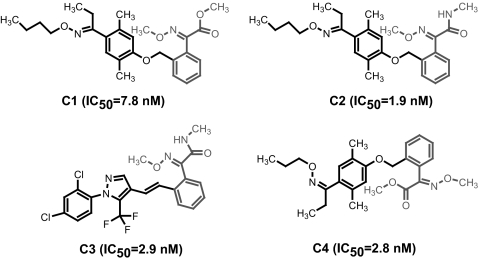
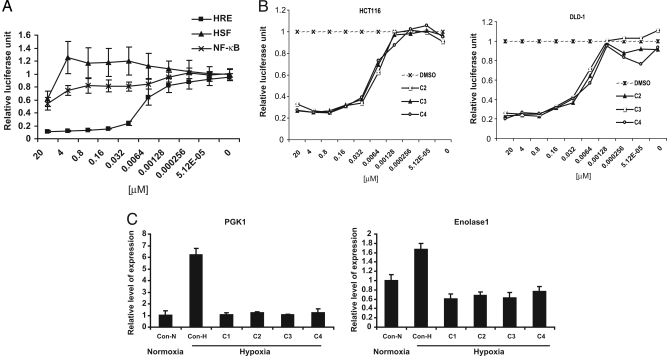
Among the 250 compounds, a group of 18 compounds share the core structure of alkyliminophenylacetate, and all of them appeared to be very potent HIF-1 inhibitors [supporting information (SI) Table 1 and Fig. 2]. Four representative compounds from this group are shown in Fig. 2. All of them exhibited an EC50 of <10 nM in the HIF-1 reporter assay. To confirm the HIF-1 specificity of the alkyliminophenylacetate compounds, we tested a representative compound from this group for its impact on luciferase reporters controlled by the heat shock transcription factor (HSF) or NF-κB. The tested compound exhibited a robust inhibition of the HIF-1 reporter activity. Meanwhile, only a marginal-to-moderate inhibition of the HSF or NF-κB reporter activity was observed for this compound (Fig. 3A). Furthermore, the alkyliminophenylacetate compounds were also found to inhibit the HIF-1 reporter activity in several other cancer cell lines, demonstrating that the inhibitory effect of these compound on the HIF-1 pathway is not restricted to a particular cancer cell line (Fig. 3B).
Fig. 2.
Small-molecule library screen using the HIF-1 reporter assay. The structures and EC50s of representative alkyliminophenylacetate compounds.
Fig. 3.
The alkyliminophenylacetate compounds are specific HIF-1 inhibitors. (A) The H1299_HRE cells, HeLa_HSP70P cells, and A549_NF-κB cells were treated with C2 by using indicated concentrations for 4 h. The cells were then treated with hypoxia, heat shock, or IL-1 to activate each reporter, respectively. At the end of each treatment, the cells were collected and assayed for luciferase activity. (B) The HCT116 and DLD-1 cells were transiently transfected with the HIF-1 reporter, pHRE, and the constitutive renilla luciferase reporter, pRL-SV40. The reporter-transfected cells were then incubated with each compound at indicated concentrations, treated with hypoxia, and assayed for luciferase activity. (C) The H1299 cells cultured at the normoxic conditions (Con-N) or the H1299 cells cultured in the presence of DMSO (Con) or 1 μM compounds (samples 1–4 represent compounds C1, C2, C3, and C4) were treated with hypoxia or DFO. The mRNA levels of PGK1 and Enolase 1 in cells were then determined by qPCR.
To further determine whether the inhibitory effect of the alkyliminophenylacetate compounds on the HIF-1 reporter can be extended to endogenous HIF-1 targets, we next examined several alkyliminophenylacetate compounds for their impact on hypoxia-induced expression of PGK1 and Enolase 1, two well established HIF-1 downstream targets. As reported in the literature, hypoxia treatment triggered a robust induction of PGK1 and Enolase 1 expression. However, the hypoxia-dependent expression of PGK1 and Enolase1 was abolished in cells treated with the alkyliminophenylacetate compounds, indicating that the alkyliminophenylacetate compounds can also inhibit endogenous HIF-1 targets (Fig. 3C). Taken together, these results suggest that the alkyliminophenylacetate compounds are specific and potent HIF-1 inhibitors.
Alkyliminophenylacetate Compounds Inhibit HIF-1 by Blocking Mitochondrial ROS Production.
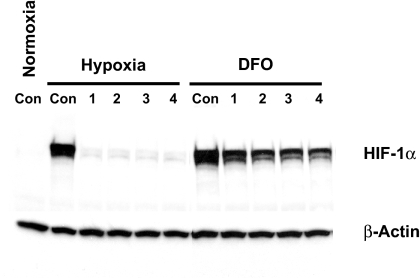
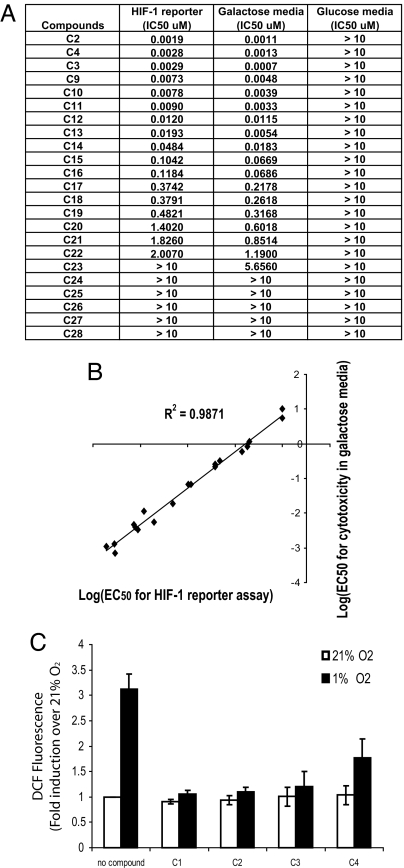
To understand the molecular mechanism by which the alkyliminophenylacetate compounds inhibit the HIF-1 pathway, we first examined the compound-treated cells for the HIF-1α protein levels before and after hypoxia or Deferoximine (DFO) treatment. Although the alkyliminophenylacetate compounds abolished hypoxia-induced HIF-1α protein accumulation, the DFO-induced HIF-1α protein accumulation was only moderately reduced (Fig. 4). The differential impact of the alkyliminophenylacetate compounds on hypoxia and DFO-induced HIF-1α accumulation resembled what was reported for ROS scavengers and mitochondrial electron transport chain inhibitors (6, 29). This prompted us to test whether the alkyliminophenylacetate compounds inhibit HIF-1 by targeting mitochondria. The ability to survive in media that are devoid of glucose but contain galactose is an indication of the mitochondrial function in cells. When cultured in galactose media, cells depend solely on the oxidative phosphorylation in mitochondria for energy, which makes the cells highly sensitive to mitochondria inhibitors (30). A number of alkyliminophenylacetate compounds were selected from Abbott's internal compound collection and tested for cytotoxicity in cells cultured in the regular glucose-containing media or the galactose-containing media. Although none of the alkyliminophenylacetate compounds triggered cell death in the regular glucose-containing media, many of them induced robust cell death in the galactose media at very low concentrations (Fig. 5A). In a parallel experiment, the same set of alkyliminophenylacetate compounds were also tested for their impact on the HIF-1 pathway, and the ability of each compound to inhibit the HIF-1 reporter was found to correlate with its ability to induce cell death in the galactose media (R2 = 0.9871) (Fig. 5 A and B). These results suggest that the alkyliminophenylacetate compounds may inhibit HIF-1 pathway by interfering with mitochondrial function.
Fig. 4.
The alkyliminophenylacetate compounds inhibit hypoxia-induced but not DFO-induced HIF-1α accumulation. The H1299 cells cultured without the presence of compounds at the normoxic conditions (Normoxia, Con) and the H1299 cells cultured in the presence of DMSO (Con) or 1 μM compounds C1, C2, C3, and C4 (1, 2, 3, and 4, respectively) were treated with hypoxia or 100 μM DFO for 4 h. The cells were then lysed and analyzed by Western blotting.
Fig. 5.
The alkyliminophenylacetate compounds likely inhibit HIF-1 by blocking mitochondrial ROS production. (A) The EC50s of the alkyliminophenylacetate compounds for the HIF-1 reporter inhibition and the cytotoxicity in the galactose and glucose media. (B) The log(EC50) of each compound for the HIF-1 reporter inhibition (x axis) and the cytotoxicity in the galactose media (y axis) was plotted. (C) The H1299 cells cultured under nomoxic or hypoxic conditions were either untreated or treated with various alkyliminophenylacetate compounds. The ROS levels in cells were determined by quantifying the DCF fluorescence.
It has been suggested that the ROS produced in complex III of the mitochondrial electron transport chain is responsible for HIF-1α stabilization under hypoxia (7–9, 31). To further define the mechanism of action for the alkyliminophenylacetate compounds, we examined whether the alkyliminophenylacetate compounds can inhibit ROS production under hypoxic conditions using a cell-permeable fluorescent indicator of ROS levels [2,7-dichlorodihydrofluorescein diacetate (DCF)]. In control cells, hypoxia treatment triggered a 3-fold increase of the DCF fluorescent signal, indicating that ROS increases under hypoxia. However, treating the cells with the alkyliminophenylacetate compounds severely impaired the hypoxia-dependent ROS production (Fig. 5C), suggesting that the alkyliminophenylacetate compounds may inhibit HIF-1 by blocking mitochondrial ROS production.
Inhibition of Mitochondrial ROS Production Is a Common Mechanism Used by Structurally Diverse HIF-1 Inhibitors.
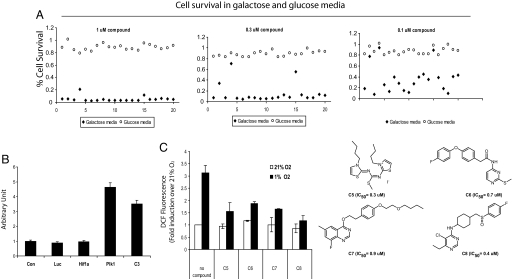
Upon confirming the effect of the alkyliminophenylacetate compounds on mitochondrial ROS production, we tested whether this is a common mechanism used by the HIF-1 inhibitors obtained in our chemical genomics screen. To identify compounds that affect the mitochondrial function, we examined 200 HIF-1 inhibitors, including both the alkyliminophenylacetate compounds and the nonalkyliminophenylacetate compounds, for their ability to trigger cell death in galactose media. Surprisingly, all HIF-1 inhibitors tested, including all of the nonalkyliminophenylacetate compounds, were found to induce robust cell death at very low concentrations in cells cultured in galactose media. As a control, the same set of compounds did not induce cell death in cells cultured in standard glucose-containing media (Fig. 6A and data not shown). To exclude the possibility that inhibiting HIF-1 by itself causes cell death in galactose media, we examined the viability of cells in galactose media upon siRNA-mediated HIF-1α knockdown. In these experiments, cells that were transfected with a validated potent HIF-1α siRNA were found to exhibit a similar degree of viability in galactose media compared with cells transfected with a control siRNA or a siRNA against luciferase (Fig. 6B) (27). As controls, knockdown of the mitotic kinase PLK1 or treating the cells with an alkyliminophenylacetate compound triggered robust cell death, indicating that siRNAs were efficiently transfected under our experimental conditions, and the cells remain responsive to death stimuli. These results indicate that the inhibition of HIF-1 is not the cause of cell death in the galactose media. Therefore, the cytotoxic effect of the HIF-1 inhibitors is likely due to the ability of these compounds to interfere with mitochondrial function. To further define whether the nonalkyliminophenylacetate HIF-1 inhibitors also prevent ROS production under hypoxia, we selected several nonalkyliminophenylacetate HIF-1 inhibitors with diverse structures, and tested these compounds for their impact on hypoxia-induced ROS production. Similar to what was observed with the alkyliminophenylacetate compounds, all of the nonalkyliminophenylacetate HIF-1 inhibitors were also found to block the hypoxia-induced ROS production (Fig. 6C). The decrease in DCF fluorescence during hypoxia by these compounds is not due to interference of the assay with our compounds because neither the alkyliminophenylacetate nor the nonalkyliminophenylacetate compounds decreased DCFH oxidation to DCF in cultured cells exposed to glucose oxidase, a well established generator of H2O2 (SI Fig. 7). In addition, the HIF-1b null cells were found to maintain the ability to generate ROS under hypoxia, suggesting that the reduction of ROS in compound-treated cells is not a consequence of inhibiting HIF-1 by these compounds (SI Fig. 8). Furthermore, the administration of a mitochondria-targeted ROS scavenger, MitoQ, was found to prevent hypoxia-induced ROS generation and HIF-1α stabilization but not HIF-1α stabilization induced by prolyl hydroxylase inhibitor DMOG (SI Fig. 9 A and B). In contrast to the HIF-1 inhibitors identified in this study, MitoQ did not inhibit respiration or diminish survival in the galactose media (SI Fig. 9 C and D). Taken together, these results suggest that inhibition of hypoxia-dependent mitochondrial ROS production appears to be the common mechanism used by the HIF-1 inhibitors identified from our screen.
Fig. 6.
Inhibiting mitochondrial ROS generation is a common mechanism used by the HIF-1 inhibitors identified from our chemical genomic screening. (A) The H1299_HRE cells were cultured in the glucose or galactose media in the presence of 1 μM (Left), 0.3 μM (Center), or 0.1 μM (Right) concentrations of different HIF-1 inhibitors. The cell viability was determined 72 h after compound treatment, and the percent cell survival was calculated as the percentage of viable cells in the compound-treated samples versus the DMSO-treated control sample. Results from a subset (19 compounds) of the 200 HIF-1 inhibitors tested were shown here. (B) The H1299_HRE cells cultured in the galactose media were transfected with different siRNAs or treated with 1 μM C3, and cell death was determined 72 h after siRNA transfection or compound treatment by using the Toxilight assay. (C) The H1299 cells cultured under nomoxic or hypoxic conditions were either untreated or treated with various compounds. The ROS levels in cells were determined by quantifying the DCF fluorescence.
Discussion
HIF-1 activation under hypoxic conditions is a multistep process involving the stabilization of HIF-1α protein, dimerization of the α and β subunits, translocation to the nucleus, binding to the HIF-1 responsive promoters, and the formation of active transcriptional complexes with accessory proteins such as p300/CBP. Because all of these steps could be potential targets of HIF-1 specific inhibitors, it is surprising that all of the HIF-1 inhibitors identified from a very large and highly diverse small-molecule library targeted the mitochondria. The involvement of mitochondria in HIF-1 regulation was proposed many years ago. However, the essential role of mitochondria as an oxygen sensor upstream of PHDs is just beginning to be appreciated. Several recent studies indicated that ROS produced from complex III of the mitochondrial electron transport chain is responsible for hypoxia-dependent stabilization of HIF-1α, suggesting that mitochondria may act as an oxygen sensor by increasing ROS generation under low-oxygen conditions. In this study, all of the HIF-1 inhibitors tested were found to perturb mitochondrial function, and many structurally unrelated HIF-1 inhibitors were found to inhibit hypoxia-dependent ROS production. The fact that the HIF-1 inhibitors identified in a screen of >600,000 compounds are all mitochondria inhibitors strongly supports the notion that mitochondria play a central role in regulating HIF-1 activity. It is noteworthy that the HIF-1 inhibitors identified from our screen are extremely potent. Many of the alkyliminophenylacetate compounds have EC50s in the single-digit nanomolar range. This is in sharp contrast to many micromolar HIF-1 inhibitors reported in the literature that work through nonmitochondrial mechanisms. These results suggest that HIF-1 activity may be highly sensitive to the perturbation of mitochondrial ROS production, making the inhibition of hypoxia-dependent ROS generation an attractive strategy to block the HIF-1 pathway.
The molecular targets of the HIF-1 inhibitors identified from our screen remain to be defined. A structure-based patent search revealed that the alkyliminophenylacetate compounds contain the toxophores of the strobilurin fungicides (32, 33). Azoxystrobin (Syngenta), Kresoxim-Methyl (BASF), and Metominnostrobin (Shionogi) are marketed strobilurin-type fungicides with the representative toxophores methyl-methoxyacrylate, methyl-methoxyiminoacetate, and N-methyl-methoxyiminoacetamide, respectively. Strobilurins are well established inhibitors of complex III, where ROS was produced at the Qo site. The Qo site of complex III is intact in the cytochrome b null cells, and cells that do not contain cytochrome b are still able to generate ROS (34). Strobilurin inhibits electron transfer in the cytochrome bc1 complex of the mitochondrial respiratory chain before cytochrome b, resulting in the preventing cytochrome b reduction and thereby attenuating ROS production (35, 36). A recent study indicates that Qo site inhibitors of complex III (myxothiazol, stigmatellin) are most effective in preventing hypoxia-induced ROS production and HIF-1 activation. By contrast, antimycin, the complex III inhibitor that does not inhibit at the Qo site, is not effective at suppressing ROS generation or HIF activation (34, 37). Because all alkyliminophenylacetate class of HIF-1 inhibitors contain one of the strobilurin toxophores, we suspect that the alkyliminophenylacetate compounds target the Qo site of complex III, resulting in the inhibition of mitochondrial electron transfer in complex III (SI Fig. 10). This proposed activity is consistent with the two distinct cellular activities of the alkyliminophenylacetate. On one hand, inhibiting complex III blocks oxidative phosphorylation, making cells highly dependent on glycolysis. Therefore, when glycolysis was blocked in cells cultured in galactose-containing media, alkyiminopheylacetates treatment caused dramatic cell death. On the other hand, inhibiting complex III blocks ROS production under hypoxia, which resulted in the inhibition of HIF-1 activity by alkyiminophenylacetates. For the variety of nonalkyliminophenylacetate compounds, given their abilities to induce cell death in the galactose media, they likely block mitochondrial respiration as the alkyliminophenylacetate compounds. However, the exact targets for these compounds are unknown.
It is somewhat surprising that the large-scale siRNA library screen failed to identify any classic druggable targets in the HIF-1 pathway, given that a number of signaling pathways/molecules (e.g., PI3 kinase and HSP90) have been implicated in HIF-1 regulation. To investigate the reasons for the lack of targets from the siRNA library screen, we first examined the siRNAs against several PI3K members for their ability to knock down the respective targets. Although multiple siRNA were found to knock down PI3K catalytic subunits p110α and p110β, these siRNAs did not inhibit the HIF-1 reporter activity, suggesting that knockdown of p110α or p110β is not sufficient to affect HIF-1 activity (SI Fig. 11 A and B). Furthermore, PI3K inhibitors wortmannin and LY294002, which are expected to inhibit the activity of all forms of PI3K, failed to block HIF-1 activation in our reporter cell line (SI Fig. 11C). These results suggest that PI3K may not be involved in HIF-1 activation in the HIF-1 reporter cell line used in our study, which explains why PI3K was not identified from the siRNA screen. On the other hand, geldanamycin, the small-molecule inhibitor of HSP90, was found to inhibit HIF-1 reporter activity in our assay, suggesting that the failure to identify HSP90 from the siRNA library screen could be due to technical reasons such as insufficient target knockdown or redundancy (SI Fig. 11D). Taken together, the inability to identify targets by siRNA library screen could be attributed to both technical and biological reasons.
Although mitochondria inhibitors such as rotenone and myxothiazol are very toxic, the complex III inhibitor strobilurin appeared to be considerably less toxic to mice. In particular, the strobilurin type of fungicides have gone through stringent toxicity scrutiny. In a chronic toxicity study in rats using Kresoxim-Methyl, oral dosing up to 500 mg/kg/day for prolonged period did not produce any toxic side effects (U.S. Environmental Protection Agency, Pesticide Fact Sheet for Kresoxim-methyl). Therefore, it remains to be determined whether the alkyliminophenylacetate class of compounds could be tolerated in animal models at doses that inhibit HIF-1. Instead of inhibiting the electron-transport chain, the use of antioxidants is an attractive alternative for inhibiting hypoxia-dependent ROS production. This strategy allows the inhibition of ROS signal without interfering with mitochondrial respiration, thereby minimizing the toxicity concerns. Mitochondria-targeted ubiquinone such as MitoQ has been shown to inhibit HIF-1 in vitro and be safe and effective in vivo via oral dosing to protect cardiac ischemia–reperfusion injury (38, 39). It will be interesting to determine whether similar reagents can be developed as HIF-1 inhibitors for cancer therapy.
Materials and Methods
The H1299_HRE cells were incubated with 1 μM concentrations of each compound and treated with hypoxia for 18 h. Luciferase activity was subsequently determined by using the Steady-Glo Luciferase Assay System (Promega). More detailed methods are available in SI Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We are grateful to M. Celeste Simon (University of Pennsylvania School of Medicine) for ARNT null MEFs. This work was supported in part by National Institutes of Health Grants R01 CA123067-01 and R01 GM060472-09 (to N.S.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706585104/DC1.
References
- 1.Semenza GL. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 2.Wenger RH. FASEB J. 2002;16:1151–1162. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]
- 3.Poellinger L, Johnson RS. Curr Opin Genet Dev. 2004;14:81–85. doi: 10.1016/j.gde.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Yang H, Kaelin WG. Cell Growth Differ. 2001;12:447–455. [PubMed] [Google Scholar]
- 5.Semenza GL. Cell. 2001;107:1–3. doi: 10.1016/s0092-8674(01)00518-9. [DOI] [PubMed] [Google Scholar]
- 6.Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Proc Natl Acad Sci USA. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunelle JK, Bell EL, Quesada NM, Vercauteren K, Tiranti V, Zeviani M, Scarpulla RC, Chandel NS. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Mansfield KD, Guzy RD, Pan Y, Young RM, Cash TP, Schumacker PT, Simon MC. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Keshert E. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 11.Kung AL, Wang S, Klco JM, Kaelin WG, Livingston DM. Nat Med. 2000;6:1335–1340. doi: 10.1038/82146. [DOI] [PubMed] [Google Scholar]
- 12.Kung AL, Zabludoff SD, France DS, Freedman SJ, Wood AW, Livingston DM. Cancer Cell. 2004;6:33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Rapisarda A, Zalek J, Hollingshead M, Braunshweig T, Uranchimeg B, Shoemaker RH, Melillo G. Cancer Res. 2004;64:6845–6848. doi: 10.1158/0008-5472.CAN-04-2116. [DOI] [PubMed] [Google Scholar]
- 14.Ryan HE, Lo J, Johnson RS. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- 16.Li L, Lin X, Staver M, Shoemaker A, Semizarov D, Fesik SW, Shen Y. Cancer Res. 2005;65:7249–7258. doi: 10.1158/0008-5472.CAN-04-4426. [DOI] [PubMed] [Google Scholar]
- 17.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Cancer Cell. 2004;5:429–441. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 18.Moeller BJ, Dreher MR, Rabbani ZN, Schroeder T, Cao Y, Li CY, Dewhirst MW. Cancer Cell. 2005;8:99–110. doi: 10.1016/j.ccr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Lin X, Shoemaker AR, Albert DH, Fesik SW, Shen Y. Clin Cancer Res. 2006;12:4747–4754. doi: 10.1158/1078-0432.CCR-05-2842. [DOI] [PubMed] [Google Scholar]
- 20.Rapisarda A, Uranchimeg B, Scudiero DA, Selby M, Sausville EA, Shoemaker RH, Melillo G. Cancer Res. 2002;62:4316–4324. [PubMed] [Google Scholar]
- 21.Tan C, de Noronha RG, Roecker AJ, Pyrzynska B, Khwaja F, Nicolaou KC, Van Meir EG. Cancer Res. 2005;65:605–612. [PubMed] [Google Scholar]
- 22.Mabjeesh NJ, Escuin D, laVallee TM, Pribluda VS, Simons JW, Giannakakou P. Cancer Cell. 2003;3:363–375. doi: 10.1016/s1535-6108(03)00077-1. [DOI] [PubMed] [Google Scholar]
- 23.Kong X, Lin Z, Liang D, Fath D, Sang N, Caro J. Mol Cell Biol. 2006;26:2019–2028. doi: 10.1128/MCB.26.6.2019-2028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong D, Park EJ, Stephen AG, Calvani M, Cardellina JH, Monks A, Fisher RJ, Shomaker RH, Melillo G. Cancer Res. 2005;65:9047–9055. doi: 10.1158/0008-5472.CAN-05-1235. [DOI] [PubMed] [Google Scholar]
- 25.Chau NM, Rogers P, Aherne W, Carroll V, Collins I, McDonald E, Workman P, Ashcroft M. Cancer Res. 2005;65:4918–4928. doi: 10.1158/0008-5472.CAN-04-4453. [DOI] [PubMed] [Google Scholar]
- 26.Welsh S, Williams R, Kirkpatrick L, Paine-Murrieta G, Powis G. Mol Cancer Ther. 2004;3:233–244. [PubMed] [Google Scholar]
- 27.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin X, Morgan-Lappe S, Huang X, Li L, Zakula DM, Vernetti LA, Fesik SW, Shen Y. Oncogene. 2006;26:3972–3979. doi: 10.1038/sj.onc.1210166. [DOI] [PubMed] [Google Scholar]
- 29.Hagen T, Taylor CT, Lam F, Moncada S. Science. 2003;302:1975–1978. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- 30.Whitfield CD, Bostedor R, Goodrum D, Haak M, Chu EH. J Biol Chem. 1981;256:6651–6656. [PubMed] [Google Scholar]
- 31.Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT. J Biol Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 32.Bartlett DW, Clough JM, Godwin JR, Hall AA, Hamer M, Parr-Dobrzanski B. Pest Manag Sci. 2002;58:649–662. doi: 10.1002/ps.520. [DOI] [PubMed] [Google Scholar]
- 33.Balba H. J Environ Sci Health B. 2007;42:441–451. doi: 10.1080/03601230701316465. [DOI] [PubMed] [Google Scholar]
- 34.Bell EL, Eisenbart J, Moraes CT, Murphy M, Budinger S, Chandel NS. J Cell Biol. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller FL, Roberts AG, Bowman MK, Kramer DM. Biochemistry. 2003;42:6493–6499. doi: 10.1021/bi0342160. [DOI] [PubMed] [Google Scholar]
- 36.Von Jagow G, Gribble GW, Trumpower BL. Biochemistry. 1986;25:775–780. doi: 10.1021/bi00352a006. [DOI] [PubMed] [Google Scholar]
- 37.Kim H, Yu CA, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Proc Natl Acad Sci USA. 1998;95:8026–8033. doi: 10.1073/pnas.95.14.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanjuan-Pla A, Cervera AM, Apostolova N, Garcia-Bou R, Victor VM, Murphy MP, McCreath KJ. FEBS Lett. 2005;579:2669–2674. doi: 10.1016/j.febslet.2005.03.088. [DOI] [PubMed] [Google Scholar]
- 39.Adlam VJ, Harrison JC, Porteous CM, James AM, Smith RA, Murphy MP, Sammut IA. FASEB J. 2005;19:1088–1095. doi: 10.1096/fj.05-3718com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.