Abstract
The environmental toxin 2,3,7,8-tetrachlorodibenzodioxin (TCDD) is a known human carcinogen; however, its precise mechanism of action remains unclear. Here we show that TCDD induces mitochondrial dysfunction, stress signaling, and tumor invasion by a mechanism similar to that described for mtDNA-depleted cells. Treatment of C2C12 cells with TCDD disrupted mitochondrial transmembrane potential in a time-dependent fashion and inhibited mitochondrial transcription and translation. TCDD also increased cytosolic [Ca2+]c and RyR1-specific Ca2+ release. These changes were associated with increased calcineurin (CnA) levels and activation of CnA-sensitive NF-κB/Rel (IκBβ-dependent) factors. Cells treated with TCDD displayed resistance to apoptosis, increased expression of the tumor marker cathepsin L, and a high degree of invasiveness as tested by the Matrigel membrane invasion assay. These effects were reversed by the CnA inhibitor FK506, and CnA mRNA silencing suggesting that TCDD triggers a signaling pathway similar to mtDNA depletion. Taken together, these results reveal that TCDD may promote tumor progression in vivo by directly targeting mitochondrial transcription and induction of mitochondrial stress signaling.
Keywords: calcineurin; mitochondrial transcription; 2,3,7,8-tetrachlorodibenzodioxin (TCDD); tumor invasion; transmembrane potential
Mitochondrial dysfunction is associated with a myriad of pathologies, including diabetes, heart disease, blindness, deafness, kidney disease, obesity, and neurodegenerative diseases, as well as aging (1). Mitochondrial dysfunction is also associated with cancer and has been reported to play a role in carcinogenesis (2–4). Diverse stimuli, including environmental toxins, drugs, ionophores, hypoxia, and mtDNA mutations/deletions are known to cause mitochondrial dysfunction (2, 4–7). Recent studies from our and others' laboratories have shown that, in a number of cell lines, mitochondrial dysfunction induced by partial depletion of mtDNA or by mitochondrial inhibitors elicits mitochondria-to-nucleus stress signaling that is propagated through activation of calcineurin (CnA) and other factors (7–11). Moreover, activation of mitochondrial stress signaling in C2C12 rhabdomyoblasts and A549 lung carcinoma cells induces invasive phenotypes that are resistant to apoptotic stimuli (8, 9, 12, 13).
Although various causes of mitochondrial dysfunction by physiological processes are relatively better understood, the environmental factors that affect mitochondrial function and lead to mtDNA mutations are much less clear. The environmental toxin 2,3,7,8-tetrachlorodibenzodioxin (TCDD), a member of a family of halogenated aromatic hydrocarbons known as dioxins, is a known carcinogenic and teratogenic agent. TCDD has deleterious effects on human as well as wildlife health. Wasting (cachexia), thymic involution, tumor promotion, hepatotoxicity, developmental toxicity, and immunosuppression are a few of the pathological effects of TCDD (14, 15). TCDD is also known to induce oxidative stress, production of superoxide and peroxide radicals, and DNA single-strand breaks (16–18). However, the cellular and molecular mechanisms of TCDD-mediated pathologies are poorly understood.
TCDD and related dioxins are well established ligands for aryl hydrocarbon receptor (AhR), which modulates transcriptional activation of many genes, including those involved in fatty acid metabolism (18), cell cycle regulation, immune response, and xenobiotic metabolism. Binding of TCDD to AhR triggers AhR nuclear translocation and its heterodimerization with AhR nuclear translocator (Arnt). The AhR–Arnt complex activates transcription by binding to dioxin-responsive elements, although some studies question the absolute requirement of AhR for transactivation of TCDD-responsive genes and hint at the existence of alternate, AhR-independent pathways (19, 20).
Here we show that exposure of C2C12 myocytes to TCDD results in inhibition of mitochondrial transcription, disruption of mitochondrial transmembrane potential (ΔΨm), and altered Ca2+ homeostasis. TCDD-treated C2C12 cells also developed resistance to apoptosis and acquired highly invasive phenotypes. Notably, these effects are dependent on CnA but not on AhR–Arnt factors. These findings suggest that TCDD may promote tumor progression by directly targeting mitochondrial function and triggering mitochondria-to-nucleus stress signaling.
Results
TCDD Induces Mitochondrial Dysfunction in C2C12 Cells.
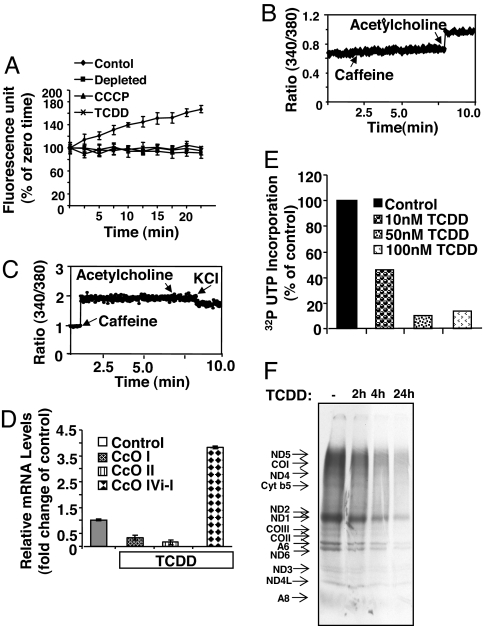
Exposure of C2C12 cells to the mitochondrial respiratory inhibitors such as carbonylcyanide m-chlorophenylhydrazone (CCCP) or depletion of mtDNA induces mitochondrial stress signaling, resulting in the activation of a number of nuclear genes and the development of invasive phenotypes (3, 7, 10). Therefore, we examined whether TCDD that induces mitochondrial damage (16) also induces stress signaling. TCDD treatment (10 nM) for 4 h resulted in a dissipation of ΔΨm as measured by MitoTracker Orange dye uptake (Fig. 1A). This effect was similar to that seen in mtDNA-depleted or CCCP-treated cells. Mitochondria from treated cells produced a higher level of reactive oxygen species [supporting information (SI) Fig. 6] possibly because of its secondary effects on membrane complexes or matrix enzymes.
Fig. 1.
TCDD disrupts ΔΨm and alters Ca2+ homeostasis in C2C12 cells. (A) Uptake of MitoTracker Orange was recorded in control (●), mtDNA-depleted (■), CCCP-treated (▴), or TCDD-treated (10 nM) (×) cultures. (B and C) Ca2+ release was measured with 1 μM Fura 2FF/FA by exposing control and TCDD-treated cells to 10 μM acetylcholine (B) and 20 mM caffeine (C), respectively. Where indicated, cultures were treated with 100 mM KCl to restore ΔΨm. (D) Mitochondrial genome-coded CcOI and CcOII and nuclear genome-coded CcOIVi1 mRNA levels were quantified by real-time PCR with β-actin gene as internal control. Treatment with 10 nM TCDD was for 4 h. (E) Level of mitochondrial transcription in isolated mitochondria. [32P]UTP incorporation was carried out as described in Methods. (F) Mitochondrial translation products were labeled with [35S]Met as described in Methods. Proteins were analyzed by SDS/PAGE followed by fluorography.
TCDD treatment also resulted in a marked increase in caffeine-mediated Ca2+ release and a concomitant reduction in acetylcholine-mediated Ca2+ release in C2C12 cells (Fig. 1 B and C). These results were similar to those obtained with mtDNA-depleted cells. Interestingly, acetylcholine-mediated Ca2+ release, signifying inositol trisphosphate channel activity, is prominent in control cells, but a markedly reduced magnitude of caffeine-mediated release (7). In contrast, cells treated with TCDD exhibited significant Ca2+ release in response to caffeine, which evokes Ca2+ release from RyR1-specific stores (Fig. 1 B and C). Addition of 100 mM KCl, which restores the transmembrane potential, reduced the level of caffeine-mediated Ca2+ release, suggesting that augmented caffeine-mediated Ca2+ release is associated with mitochondrial dysfunction and associated changes in ΔΨm.
The loss of ΔΨm and increased cytosolic [Ca2+]c observed in response to TCDD exposure suggested that mitochondria are a direct target of this toxin. Therefore, we tested the effects of TCDD treatment on mitochondrial transcription. Fig. 1D shows that mitochondrial genome-coded CcOI and CcOII mRNA levels were reduced by 60–80% in TCDD-treated cells, whereas the level of nuclear genome-coded CcOIVi1 was increased by ≈4-fold. The increase in CcOIVi1 level is consistent with our observations on mitochondrial stress-mediated activation of nuclear target genes in mtDNA-depleted C2C12 cells (7). A marked inhibition of [32P]UTP incorporation by isolated mitochondria indeed suggests a direct effect of TCDD on mitochondrial transcription (Fig. 1E). Consistent with reduced transcription, mitochondrial translation activity was reduced in a time-dependent manner after TCDD treatment (≈75% at 4 h and ≈85% at 24 h) (Fig. 1F). Additionally, we did not see any significant cell death in C2C12 or RAW 264.7 macrophage cells up to 1 μM TCDD (data not shown). Consequently, a 4-h duration of 10 nM TCDD treatment was used for all subsequent experiments. Taken together, these results demonstrate that TCDD induces mitochondrial dysfunction and increases cytosolic [Ca2+]c levels.
TCDD Activates Mitochondrial Stress Signaling in C2C12 Cells.
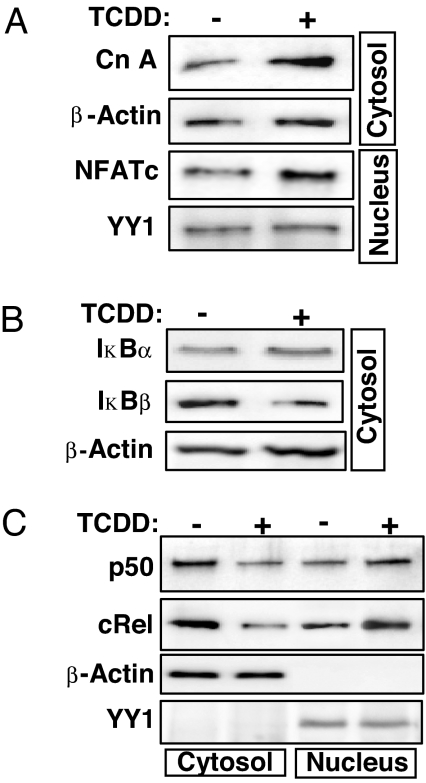
Mitochondrial stress signaling initiated by mtDNA depletion is propagated through activation of CnA and NF-κB (10). The stress-mediated NF-κB pathway is quite distinct from the canonical or noncanonical pathway and involves CnA-mediated dephosphorylation of IκBβ and activation of cRel/p50 (10). The Western blot in Fig. 2A shows that TCDD treatment caused an ≈4-fold increase in cytosolic levels of CnA, a Ca2+-dependent phosphatase. A corresponding increase in the nuclear localization of NFATc, a transcription factor known to be activated by CnA, was also observed. The cytosolic levels of IκBβ, an inhibitor of NF-κB/cRel, were significantly reduced after TCDD treatment, whereas the IκBα levels were marginally increased (Fig. 2B), suggesting suppression of the canonical NF-κB pathway. Nuclear cRel and p50 levels increased, and cytosolic cRel and p50 levels decreased in response to TCDD treatment (Fig. 2C). The role of CnA in the TCDD-induced activation pathway was further tested with the CnA inhibitor FK506 and CnA mRNA silencing by the siRNA approach. SI Fig. 7A shows that cells transfected with hairpin siRNA indeed show reduced CnA protein. In both FK506-treated CnA silenced cells cytoplasmic IκBβ level did not reduce in response to TCDD treatment. Further, the increase in nuclear cRel and p50 levels in response to TCDD was negligible or marginal in FK506-treated or CnA-silenced cells (SI Fig. 7B). Thus, TCDD, like mtDNA depletion, induces a CnA-dependent NF-κB pathway (10).
Fig. 2.
TCDD induces expression of mitochondrial stress signaling marker proteins. Subcellular fractions isolated from control and 10 nM TCDD-treated cells were analyzed by Western blotting for CnA/NFATc (A), IκBα and IκBβ (B), and NF-κB transcription factors (C). β-Actin and YY1 were used as loading controls for cytosolic and nuclear fractions, respectively.
TCDD Induces the Expression of Nuclear Genes and Promotes the Formation of Invasive Phenotypes.
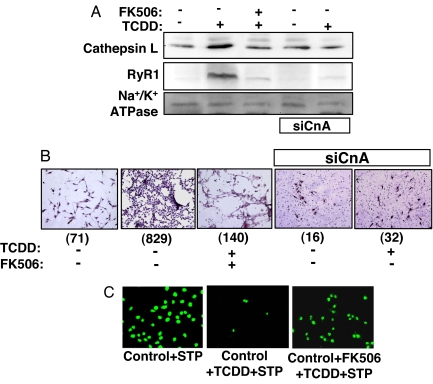
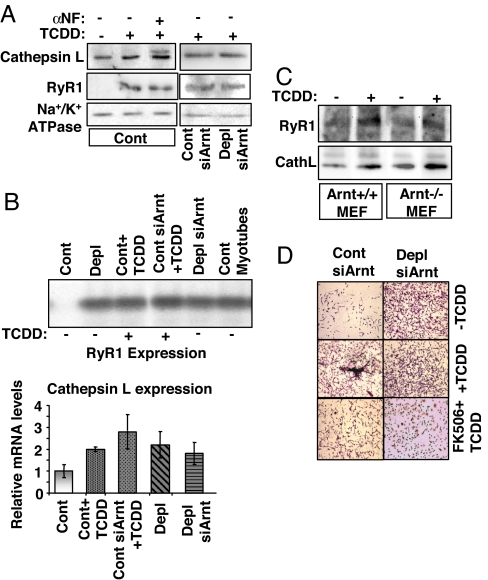
Mitochondrial stress induces the expression of a number of nuclear genes in C2C12 and A549 cells, including those involved in Ca2+ storage/release and tumorigenesis (8, 10). Western blot analysis (Fig. 3A) revealed that TCDD treatment increased levels of the Ca2+ channel receptor protein RyR1 and cathepsin L, two known markers of stress signaling (7, 8). Notably, cathepsin L is induced in many invasive tumors (21, 22). Both CnA inhibitor FK506 and CnA mRNA silencing abrogated TCDD-induced increase of both RyR1 and cathepsin L (Fig. 3A). Thus, CnA plays an important role in the TCDD-induced expression of these stress response genes.
Fig. 3.
TCDD increases cathepsin L and RyR1 levels, promotes invasive behavior, and confers resistance to apoptosis in a CnA-dependent manner. (A) Western blot analysis of cathepsin L and RyR1 levels in whole-cell lysates from control, CnA-silenced (siCnA), TCDD-treated, and TCDD + FK506-treated cells. Na+/K+-ATPase served as a loading control. (B) A Matrigel invasion assay was performed on control, CnAα-silenced (siCnAα), TCDD-treated, and TCDD + FK506-treated cells. Invaded cells were stained and viewed as described in Methods. Numbers in parentheses underneath the images indicate the number of cells in the area shown. (C) The effect of TCDD on STP-induced apoptotic cell death was assessed by TUNEL assay.
The possible role of TCDD in tumor promotion was investigated by using the Matrigel matrix invasion assay system. TCDD-treated cells displayed a high degree of invasiveness compared with that of control cells (Fig. 3B Center and Left, respectively). Moreover, this effect could be blocked by FK506 (Fig. 3B Center) or by CnA mRNA silencing (Fig. 3B Right). Although not shown, the Ca2+ chelator 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate-acetoxymethyl (BAPTA-AM) also blocked the TCDD-mediated increase of invasion in cells.
Mitochondrial stress induced by mtDNA depletion or by treatment with CCCP is known to confer resistance to staurosporine (STP)-induced apoptosis (9). TCDD treatment had similar effects on STP-induced apoptosis, as assessed by TUNEL-positive nuclei (Fig. 3C). Interestingly, inhibition of apoptosis by TCDD was fully reversed by FK506 (data not shown). These results are consistent with the hypothesis that propagation of TCDD-mediated mitochondrial stress signaling requires active CnA.
TCDD-Induced Mitochondrial Stress Signaling Occurs Independently of the Ah Receptor.
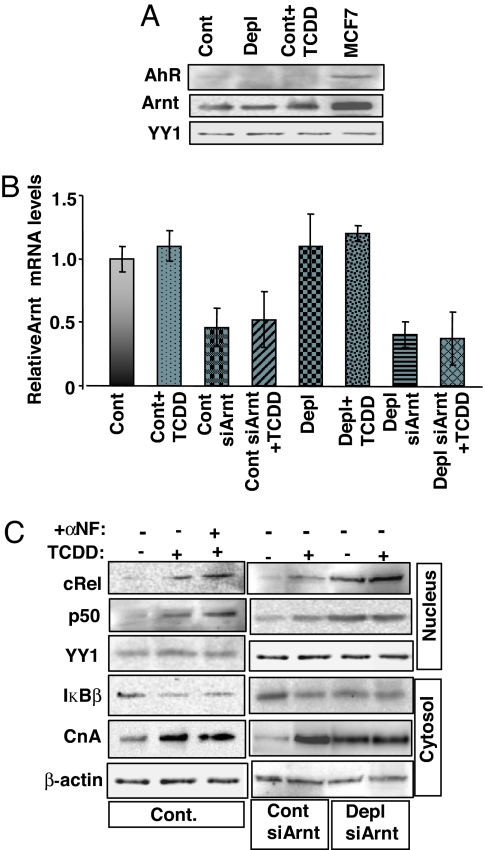
A well established mechanism of TCDD action is AhR–Arnt-mediated transcription activation. Therefore, we examined whether TCDD-induced mitochondrial stress signaling in C2C12 cells is propagated through AhR–Arnt or through Ca2+-activated CnA. As shown in Fig. 4A Top, nuclear AhR protein levels were undetectable in control C2C12 cells as also in TCDD-treated and mtDNA-depleted C2C12 cells. Although nuclear Arnt levels could be detected in these cells, no major changes in the levels were observed in response to TCDD exposure (Fig. 4A Middle). MCF7 cell lysates, which were used as a positive control, showed an abundance of both AhR and Arnt proteins.
Fig. 4.
TCDD-induced mitochondrial stress signaling is unaffected by AhR inhibition and Arnt mRNA silencing. (A) Nuclear fractions isolated from control (Cont), mtDNA-depleted (Depl), and TCDD-treated cells were subjected to Western blot analysis of AhR and Arnt. MCF7 nuclear fractions were included as a positive control. YY1 was used as a loading control. (B) Arnt mRNA levels were assessed by real-time PCR in control cells, TCDD-treated cells, mtDNA-depleted cells, Arnt-silenced control cells, and Arnt-silenced mtDNA-depleted cells. (C) Effect of α-NF, an AhR antagonist, and Arnt mRNA silencing on the induction of mitochondrial stress signaling markers by TCDD. Levels of the indicated factors were assessed by Western blot analysis. YY1 and β-actin were used as loading controls for nuclear and cytosolic fractions, respectively.
The AhR-heterodimerizing partner, Arnt, modulates xenobiotic agent- and hypoxia-responsive transcription factor (23) by functionally heterodimerizing with other non-AhR factors. Therefore, to verify whether other Arnt-dimerizing factors are involved in TCDD-mediated mitochondrial stress signaling, we tested the effect of α-naphthoflavone (α-NF), a known antagonist of AhR, as well as siRNA-mediated Arnt knockdown in C2C12 cells. As shown in Fig. 4B, Arnt mRNA levels were significantly reduced in both control and mtDNA-depleted cells stably expressing siArnt vector (siArnt cells). However, TCDD had no effect on Arnt mRNA levels in any of these cells. Target proteins of mitochondrial stress signaling were analyzed by Western blotting, and the results are presented in Fig. 4C. Treatment with α-NF did not affect TCDD-induced expression of target proteins. In the same way, TCDD did not elicit any changes in target protein levels in siArnt control and siArnt mtDNA-depleted cells. Levels of nuclear cRel and p50 as well as of cytosolic CnA were roughly equal in control cells and siArnt cells treated with TCDD. At the same time, the cytosolic levels of NF-κB inhibitor IκBβ were reduced after these treatments.
Because C2C12 cells lack detectable AhR, we used Arnt+/+ mouse embryonic fibroblasts (MEFs) and Arnt−/− MEFs to test the TCDD-mediated stress signaling (SI Fig. 8). Immunoblot results show that a TCDD-induced increase in CnA (SI Fig. 8A) and CnA-dependent NF-κB activation (SI Fig. 8B) occurred normally in Arnt−/− cells, suggesting that the signaling proceeds normally in the absence of the AhR pathway.
The protein levels of RyR1 and cathepsin L, two known targets of mitochondrial stress signaling (7, 8, 10, 12), were increased in cells treated with TCDD (Fig. 5A). However, α-NF or Arnt knockdown had no apparent effect on TCDD-mediated increases in protein levels in C2C12 cells. We also tested Arnt−/− MEFs (Fig. 5B) and found that the TCDD-mediated increase in the levels of RyR1 and cathepsin L proteins was not affected. These results suggest that the observed induction may not involve the AhR–Arnt pathway. RyR1 mRNA levels were increased in mtDNA-depleted cells and in cells treated with TCDD (Fig. 5C Upper). In both cases, Arnt mRNA silencing did not alter RyR1 mRNA levels. Cathepsin L mRNA levels showed a similar pattern in that Arnt mRNA silencing had no significant effect on TCDD-mediated induction of mRNA (Fig. 5C Lower). Similarly, Arnt silencing had no effect on TCDD-mediated increases in cellular invasiveness (Fig. 5D). The invasive behavior of mtDNA-depleted cells was also not decreased by silencing of Arnt mRNA, whereas FK506 treatment inhibited the invasiveness in both control and mtDNA-depleted siArnt cells. These findings confirm that TCDD-mediated transcriptional activation of target genes occurs through an AhR–Arnt-independent pathway. In contrast, transcriptional up-regulation of marker genes and increased tumor invasiveness were sensitive to FK506 and CnA mRNA silencing, suggesting the involvement of the CnA pathway.
Fig. 5.
TCDD-induced expression of marker genes in Arnt mRNA-silenced cells. (A) Cathepsin L and RyR1 protein levels in postnuclear fractions were analyzed by Western blotting. (B) immunoblot of cathepsin L and RyR1 levels in Arnt+/+ and Arnt−/− MEFs as in A. (C) Levels of RyR1 mRNA (Upper) and cathepsin L mRNA (CathL; Lower) were assessed by RT-PCR amplification followed by Southern blot hybridization and real-time PCR, respectively. β-Actin served as an internal control. (D) Control cells, TCDD-treated cells, mtDNA-depleted cells, Arnt silenced control cells, and Arnt-silenced mtDNA-depleted cells were evaluated for invasiveness in response to TCDD treatment by using a Matrigel invasion assay.
Discussion
Previously we have shown that mitochondrial dysfunction or disruption of ΔΨm, either by mtDNA depletion or treatment with respiratory inhibitors (CCCP, rotenone, and antimycin) alters intracellular Ca2+ homeostasis and induces stress signaling (7, 10, 13). The protein phosphatase, CnA, and PKC (13) are two important intermediates in this pathway. Mitochondrial stress signaling also confers resistance to apoptosis and enhances invasiveness. In this work, we show that TCDD treatment induces mitochondrial dysfunction as seen by disruption of ΔΨm and initiates mitochondrial stress signaling, similar to that triggered by mtDNA depletion and CCCP treatment (7, 10, 13). The signaling cascade appears to be independent of AhR.
TCDD appears to elicit its effect by interfering with mitochondrial transcription as seen by reduced runoff transcription by isolated mitochondria and markedly reduced mitochondria encoded mRNAs (Fig. 1 D and E). We were unable to detect any reduction in mtDNA content or DNA chain breaks under the present conditions (results not shown). As reported by others, we also saw an increased reactive oxygen species production (SI Fig. 6), which may be the result of its effects on mitochondrial membrane complexes or matrix enzymes. However, we do not rule out other possibilities.
Numerous studies suggest that TCDD blocks cell death processes in cancer cells, thereby promoting tumor development (24, 25). However, the precise mechanism by which TCDD inhibits apoptosis remains unclear. We show that exposure to TCDD leads to formation of invasive phenotypes, which is accompanied by resistance to apoptosis (3, 8, 13). Our finding that TCDD-induced activation of cRel/p50, up-regulation of cathepsin L and RyR1, acquisition of invasive phenotypes, and resistance to apoptosis are all sensitive to FK506, and CnA mRNA silencing suggests a direct involvement of CnA in these processes.
In mammary epithelial cells, TCDD is known to trigger EGFR signaling by induction of TGFα mRNA and protein (26, 27). Studies using HeLa cells and MEFs have shown that TCDD suppresses the checkpoint protein MAD2 or induces MAPKs in an AhR-independent manner (28, 29). Another study (30) employing AhR−/− mice demonstrated that TCDD inhibits cell death in response to DNA-damaging agents by activating MDM2 and inducing degradation of p53 in rat liver in an AhR-dependent manner. TCDD is also known to induce PKC and ERK in an AhR-independent manner (19, 29). PKC alters the recruitment and binding ability of a number of transcriptional coactivators (25, 31, 32). cAMP response element-binding (CREB)-binding protein/protein 300 (CBP/p300) is one such coactivator that can bind to the transactivation domain of Arnt both in vitro and in vivo (33). Mitochondrial stress signaling induced by mtDNA depletion has been shown to activate a number of factors, including CREB (ref. 11). Our unpublished results suggest that CREB participates with cRel/p50 proteins in the transcription activation of mitochondrial stress target genes, RyR1 and cathepsin L. In this respect, our results are consistent with these various observations and suggest the existence of an AhR-independent mechanism of TCDD-induced signaling cascade.
Our results reveal that TCDD triggers activation of IκBβ-associated NF-κB/cRel/p50. Interestingly, TCDD has been shown to modulate the expression of genes encoding IL-1β, TGFα, TGFβ, EGFR, ER, c-Fos, and c-Jun by inducing the binding of many NF-κB/Rel proteins to the κB site that overlaps with the dioxin-responsive element-like site (34–39). The fact that TCDD-induced expression of nuclear target genes observed in the present work was highly sensitive to FK506 treatment and CnA mRNA silencing suggests that CnA is involved in these transcriptional responses and in the activation of NF-κB. In keeping with this possibility it has been reported that TCDD activates NF-κB by AhR-independent mechanisms (40, 41).
TCDD has been reported to increase cytosolic Ca2+ and also cause activation of PKC, PKA, and certain MAPKs (19, 42–44). In this regard, our data provide a mechanistic insight on how TCDD induces a change in Ca2+ homeostasis through its direct action on mitochondrial function causing disruption of ΔΨm and culminating with the activation of mitochondria-to-nucleus stress signaling. In previous studies we have shown that mitochondrial stress signaling also causes the activation of PKA and PKC pathways (3, 13). Results presented in this work are consistent with the hypothesis that Ca2+-dependent activation/promotion of tumor invasion in nontumorigenic C2C12 cells involves mitochondrial stress signaling that is propagated through CnA-mediated activation of NF-κB and leads to the transcription activation of nuclear genes such as cathepsin L. Our observation that TCDD induces invasive phenotypes in otherwise noninvasive C2C12 rhabdomyoblasts is of direct significance in understanding mechanisms by which a large family of polychlorinated biphenyls cause cancer in humans and animals.
Methods
Cell Culture.
C2C12 skeletal myoblasts were grown in high-glucose DMEM containing 10% FBS and 0.1% gentamicin. Depletion of mtDNA was carried out by ethidium bromide treatment (100 ng/ml) for ≈70 passages as described previously (7). Clones containing mtDNA contents reduced by 40–80% were selected and grown in the presence of 1 mM sodium pyruvate and 50 μg/ml uridine. Clones were subsequently divided into aliquots and frozen in liquid N2.
Isolation of Subcellular Fractions and Western Blot Analyses.
Cells were homogenized in buffer containing 0.3 M sucrose, 10 mM Tris·HCl (pH 8.0), 10 mM NaCl, 3 mM MgCl2, protease inhibitors (i.e., 0.2 mM EDTA, 1 mM PMSF, and 50 μg/ml leupeptin, aprotinin, chymostatin, pepstatin, and antipain), and phosphatase inhibitors (i.e., 1 mM NaVO4, 100 μM molybdic acid, and 10 mM NaF). Subcellular fractions were isolated as described elsewhere (7). Proteins were resolved on polyacrylamide gels and subjected to Western blot analysis using appropriate antibodies (1:1,500 dilutions). Western blots were developed by using the SuperSignal West femto maximum sensitivity substrate kit (Pierce, Rockford, IL) and were imaged and quantified with a VersaDoc imaging system (Bio-Rad, Hercules, CA).
Measurement of ΔΨm.
The ΔΨm was measured spectrofluorometrically by using MitoTracker Orange (Invitrogen, Carlsbad, CA) CM-H2 TMRos as described previously (8). Each assay was carried out with 1.5 × 106 cells suspended in 1 ml of extracellular medium. MitoTracker Orange CM-H2 TMRos (50 nM/6 × 106 cells) was added directly to the cell suspension in the cuvette. The rate of dye uptake, a measure of ΔΨm, was monitored by using a Delta RAM spectrofluorometer (Photon Technology International) at excitation and detection wavelengths of 525 nm and 575 nm, respectively. All data were recorded as fluorescence units per minute.
Calcium Release Assay.
Calcium release was measured essentially as described previously (8). Briefly, cells were harvested and washed with ice-cold extracellular medium containing 120 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 0.2 mM MgCl2, 0.1 mM EGTA, and 20 mM Hepes/Tris (pH 7.2). Nonpermeabilized cells were suspended in buffer containing 20 mM Hepes/Tris (pH 7.2), 120 mM NaCl, 5 mM KCl, 1 mM KH2PO4. All buffers and solutions were passed through a Chelex column (Sigma) to remove residual Ca2+. For each assay, 1.5 × 106 cells were loaded with membrane-permeable Fura 2FF/FA (1 μM) for 20 min at room temperature. Ca2+ release in response to 20 mM caffeine and 10 μM acetylcholine was measured as fluorescence units recorded at excitation 340/380 nm and emission at 510 nm. Calibration of the Fura 2FF/FA signal was carried out in a calibration buffer containing 10 mM EGTA/Tris/Hepes (pH 8.5) and 5 mM CaCl2, according to the protocol provided with the Delta RAM.
Mitochondrial Protein Synthesis and Transcription.
Cells were incubated in 200 μg/ml cycloheximide for 10 min. Labeling was carried out with [35S]Met (100 μCi per plate) for 6 h. Cells were washed and homogenized in mitochondrial isolation buffer with a glass homogenizer, and mitochondria were isolated by differential centrifugation as described above. Mitochondrial protein (50 μg) was separated on a SDS/15–20% gradient polyacrylamide gel and imaged by using a Bio-Rad GS-525 molecular imager. For in vitro transcription, isolated mitochondria (2.5 mg/ml) were incubated with [32P]UTP in an energy-regenerating system (45) supplemented with dNTPs, amino acid mix, and substrate mix containing succinate, malate, pyruvate and isocitrate (2 mM each).
Matrigel Invasion Assay.
In vitro invasion assays were carried out as described previously (8). The Matrigel invasion chambers were prepared at 1:3 dilution of Matrigel (Becton Dickinson) according to manufacturer's protocol. Approximately 4 × 104 cells were suspended in 500 μl of growth medium and layered on top of the Matrigel layer. Invading cells were stained with Meyer's hematoxylin and observed under brightfield microscope.
TUNEL Assay.
Cells were grown on lysine-coated glass coverslips in 6-well plates and treated with 2 μM STP for 4 h to induce apoptosis (9). Nuclear DNA breaks were measured by using ApopTag Fluorescein from the in situ apoptosis detection kit (Intergen Company). The coverslips were mounted on slides with mounting medium from the Prolong antifade kit (Molecular Probes). Cells were imaged through an Olympus BX61 fluorescence microscope at Ex/Em-496/518.
RT-PCR and Southern Hybridization.
Total cellular RNA (2.5 μg) was reverse-transcribed with appropriate reverse primers and amplified with a commercially available RT-PCR kit (PerkinElmer) and RyR1-specific forward and reverse primers described before (7). Amplicons were separated on 1.2% agarose gel and transferred onto Zeta probe-GT membrane (Bio-Rad) for Southern hybridization. The mouse RyR1 DNA fragment was used as a probe. Hybridization methods were identical to those described previously (7).
Real-Time PCR.
Total RNA (5 μg) from TCDD-treated and untreated cells was reverse-transcribed by using the high-capacity cDNA archive kit (Applied Biosystems). Primers were designed by using Primer Express software (Applied Biosystems). Real-time amplifications for CcOI, CcOII, CcOIVi1, Arnt, and cathepsin L were performed in an Applied Biosystems 7300 real-time PCR machine by using SYBR Green Master mix (Applied Biosystems). Each 25-μl reaction contained 25 ng of cDNA and 200 nM forward and reverse primers. Two-step RT-PCR was carried for 40 cycles. Data were analyzed by using Applied Biosystems RQ analysis software. β-Actin served as an internal control. Target gene expression was presented as fold increase over control levels.
siRNA-Mediated Gene Knockdown.
A 19-nucleotide hairpin oligonucleotide (5′-CTG GCA ACA CAT CTA CTG A-3′) with a 6-nucleotide spacer was cloned into BamHI and HindIII sites of pSilencer 2.1 U6-neo vector. Control and mtDNA-depleted cells grown in 6-well plates were transfected with 2 μg of siArnt construct or scrambled DNA sequence by using FuGENE6 transfection reagent. Isoform-specific Silencer Predesigned siRNA for CnA mRNA was purchased from Ambion. The control cells were transfected by reverse-transfection with siPORT NeoFX transfection reagent (Ambion) according to the manufacturer's instructions. Transfected cells were grown in medium containing 90% DMEM, 10% FBS, 0.1% gentamicin, and G418 (800 μg/ml) to selecting positive siArnt clones. Cells transfected with siRNA for CnAα were grown for 48 h and used for invasion assay. Arnt protein and mRNA levels in isolated clones were determined by Western blotting and real-time PCR, respectively.
Supplementary Material
ACKNOWLEDGMENTS.
We thank the National Cancer Institute chemical repository (Bethesda, MD) for supplying TCDD. This work was supported by National Institutes of Health Grant CA-22762.
Abbreviations
- AhR
aryl hydrocarbon receptor
- α-NF
α-naphthoflavone
- Arnt
AhR nuclear translocator
- CCCP
carbonylcyanide m-chlorophenylhydrazone
- CnA
calcineurin
- CREB
cAMP response element-binding
- ΔΨm
mitochondrial transmembrane potential
- MEF
mouse embryonic fibroblast
- siArnt
Arnt-silenced
- STP
staurosporine
- TCDD
2,3,7,8-tetrachlorodibenzodioxin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706183104/DC1.
References
- 1.Wallace DC. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh KK, Russell J, Sigala B, Zhang Y, Williams J, Keshav KF. Oncogene. 1999;18:6641–6646. doi: 10.1038/sj.onc.1203056. [DOI] [PubMed] [Google Scholar]
- 3.Amuthan G, Biswas G, Zhang SY, Klein-Szanto A, Vijayasarathy C, Avadhani NG. EMBO J. 2001;20:1910–1920. doi: 10.1093/emboj/20.8.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandy B, Davison AJ. Free Radic Biol Med. 1990;8:523–539. doi: 10.1016/0891-5849(90)90152-9. [DOI] [PubMed] [Google Scholar]
- 5.Niranjan BG, Bhat NK, Avadhani NG. Science. 1982;215:73–75. doi: 10.1126/science.6797067. [DOI] [PubMed] [Google Scholar]
- 6.Grossman LI. Environ Mol Mutagen. 1995;25(Suppl 26):30–37. doi: 10.1002/em.2850250607. [DOI] [PubMed] [Google Scholar]
- 7.Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amuthan G, Biswas G, Ananadatheerthavarada HK, Vijayasarathy C, Shephard HM, Avadhani NG. Oncogene. 2002;21:7839–7849. doi: 10.1038/sj.onc.1205983. [DOI] [PubMed] [Google Scholar]
- 9.Biswas G, Anandatheerthavarada HK, Avadhani NG. Cell Death Differ. 2005;12:266–278. doi: 10.1038/sj.cdd.4401553. [DOI] [PubMed] [Google Scholar]
- 10.Biswas G, Anandatheerthavarada HK, Zaidi M, Avadhani NG. J Cell Biol. 2003;161:507–519. doi: 10.1083/jcb.200211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnould T, Vankoningsloo S, Renard P, Houbion A, Ninane N, Demazy C, Remacle J, Raes M. EMBO J. 2002;21:53–63. doi: 10.1093/emboj/21.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biswas G, Guha M, Avadhani NG. Gene. 2005;354:132–139. doi: 10.1016/j.gene.2005.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butow RA, Avadhani NG. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 14.Poland A, Knutson JC. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- 15.Davis D, Safe S. Toxicology. 1990;63:97–111. doi: 10.1016/0300-483x(90)90072-o. [DOI] [PubMed] [Google Scholar]
- 16.Shen D, Dalton TP, Nebert DW, Shertzer HG. J Biol Chem. 2005;280:25305–25312. doi: 10.1074/jbc.M500095200. [DOI] [PubMed] [Google Scholar]
- 17.Senft AP, Dalton TP, Nebert DW, Genter MB, Puga A, Hutchinson RJ, Kerzee JK, Uno S, Shertzer HG. Free Radic Biol Med. 2002;33:1268–1278. doi: 10.1016/s0891-5849(02)01014-6. [DOI] [PubMed] [Google Scholar]
- 18.Hassoun EA, Wang H, Abushaban A, Stohs SJ. J Toxicol Environ Health. 2002;65:825–842. doi: 10.1080/00984100290071054. [DOI] [PubMed] [Google Scholar]
- 19.Long WP, Perdew GH. Arch Biochem Biophys. 1999;371:246–259. doi: 10.1006/abbi.1999.1452. [DOI] [PubMed] [Google Scholar]
- 20.Reiners JJ, Jr, Clift RE. J Biol Chem. 1999;274:2502–2510. doi: 10.1074/jbc.274.4.2502. [DOI] [PubMed] [Google Scholar]
- 21.Denhardt DT, Greenberg AH, Egan SE, Hamilton RT, Wright JA. Oncogene. 1987;2:55–59. [PubMed] [Google Scholar]
- 22.Kirschke H, Eerola R, Hopsu-Havu VK, Bromme D, Vuorio E. Eur J Cancer. 2000;36:787–795. doi: 10.1016/s0959-8049(00)00014-9. [DOI] [PubMed] [Google Scholar]
- 23.Long WP, Chen X, Perdew GH. J Biol Chem. 1999;274:12391–12400. doi: 10.1074/jbc.274.18.12391. [DOI] [PubMed] [Google Scholar]
- 24.Puga A, Barnes SJ, Chang C, Zhu H, Nephew KP, Khan SA, Shertzer HG. Biochem Pharmacol. 2000;59:997–1005. doi: 10.1016/s0006-2952(99)00406-2. [DOI] [PubMed] [Google Scholar]
- 25.Stinchcombe S, Buchmann A, Bock KW, Schwarz M. Carcinogenesis. 1995;16:1271–1275. doi: 10.1093/carcin/16.6.1271. [DOI] [PubMed] [Google Scholar]
- 26.Davis JW, II, Lauer FT, Burdick AD, Hudson LG, Burchiel SW. Cancer Res. 2001;61:3314–3320. [PubMed] [Google Scholar]
- 27.Davis JW, II, Melendez K, Salas VM, Lauer FT, Burchiel SW. Carcinogenesis. 2000;21:881–886. doi: 10.1093/carcin/21.5.881. [DOI] [PubMed] [Google Scholar]
- 28.Oikawa K, Ohbayashi T, Mimura J, Iwata R, Kameta A, Evine K, Iwaya K, Fujii-Kuriyama Y, Kuroda M, Mukai K. Cancer Res. 2001;61:5707–5709. [PubMed] [Google Scholar]
- 29.Tan Z, Chang X, Puga A, Xia Y. Biochem Pharmacol. 2002;64:771–780. doi: 10.1016/s0006-2952(02)01138-3. [DOI] [PubMed] [Google Scholar]
- 30.Paajarvi G, Viluksela M, Pohjanvirta R, Stenius U, Hogberg J. Carcinogenesis. 2005;26:201–208. doi: 10.1093/carcin/bgh289. [DOI] [PubMed] [Google Scholar]
- 31.Long WP, Pray-Grant M, Tsai JC, Perdew GH. Mol Pharmacol. 1998;53:691–700. doi: 10.1124/mol.53.4.691. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y-H, Tukey RH. J Biol Chem. 1996;271:26261–26266. doi: 10.1074/jbc.271.42.26261. [DOI] [PubMed] [Google Scholar]
- 33.Kallio PJ, Okamoto K, O'Brien S, Carrero P, Makino Y, Tanaka H, Poellinger L. EMBO J. 1998;17:6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Astroff B, Rowlands C, Dickerson R, Safe S. Mol Cell Endocrinol. 1990;72:247–252. doi: 10.1016/0303-7207(90)90149-3. [DOI] [PubMed] [Google Scholar]
- 35.Gaido KW, Maness SC, Leonard LS, Greenlee WF. J Biol Chem. 1992;267:24591–24595. [PubMed] [Google Scholar]
- 36.Sutter TR, Guzman K, Dold KM, Greenlee WF. Science. 1991;254:415–418. doi: 10.1126/science.1925598. [DOI] [PubMed] [Google Scholar]
- 37.Puga A, Nebert D, Carrier F. DNA Cell Biol. 1992;11:269–281. doi: 10.1089/dna.1992.11.269. [DOI] [PubMed] [Google Scholar]
- 38.Silverstone AE, Frazier DE, Jr, Fiore NC, Soults JA, Gasiewicz TA. Toxicol Appl Pharmacol. 1994;126:248–259. doi: 10.1006/taap.1994.1114. [DOI] [PubMed] [Google Scholar]
- 39.Lu Y, Wang X, Safe S. Toxicol Appl Pharmacol. 1994;127:1–8. doi: 10.1006/taap.1994.1132. [DOI] [PubMed] [Google Scholar]
- 40.Sulentic CEW, Holsapple MP, Kaminski ME. J Pharmacol Exp Ther. 2000;295:705–716. [PubMed] [Google Scholar]
- 41.Ashida H, Matsumura F. J Biochem Mol Toxicol. 1998;12:191–204. doi: 10.1002/(sici)1099-0461(1998)12:4<191::aid-jbt1>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 42.Matsumura F. Biochem Pharmacol. 2003;66:527–540. doi: 10.1016/s0006-2952(03)00157-6. [DOI] [PubMed] [Google Scholar]
- 43.Weiss C, Faust D, Durk H, Kolluri SK, Pelzer A, Schneider S, Dietrich C, Oesch F, Gottlicher M. Oncogene. 2005;24:4975–4983. doi: 10.1038/sj.onc.1208679. [DOI] [PubMed] [Google Scholar]
- 44.Vogel CF, Sciullo E, Park S, Liedtke C, Trautwein C, Matsumura F. J Biol Chem. 2004;279:8886–8894. doi: 10.1074/jbc.M310190200. [DOI] [PubMed] [Google Scholar]
- 45.Kantharaj GR, Bhat KS, Avadhani NG. Biochemistry. 1983;22:3151–3156. doi: 10.1021/bi00282a018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.