Abstract
The Formin proteins are central players in mediating cytoskeletal reorganization and are epistatically positioned in a pathway downstream of Rho activation. These proteins exist in the cytoplasm in an autoinhibited state, which is mediated by intramolecular interactions between the amino-terminal GTPase binding domain (GBD) that encompasses the diaphanous inhibitory domain (DID) and the carboxyl-terminal diaphanous autoregulatory domain (DAD). It has been proposed that the binding of Rho within the GBD releases this molecule from autoinhibition by disrupting the DID/DAD interactions. Here we report that Daam1 is not significantly activated by Rho binding but rather by its interaction with Dishevelled (Dvl). Removal of the DAD domain disrupts interactions between Dvl and Daam1, and the binding of Dvl to Daam1 disrupts the interaction between the GBD and DAD that mediates Daam1 autoinhibition. Mutations within or removal of the DAD converts Daam1 into an active protein that can induce Rho activation. We further demonstrate that Dvl synergizes with Daam1 to regulate gastrulation during Xenopus embryogenesis and that expression of activated Daam1 can rescue impaired convergent extension movements resulting from deregulated noncanonical Wnt signaling. Our studies together define the importance of a carboxyl-terminal binding partner, Dvl, that leads to the activation of Daam1.
Keywords: Dishevelled, Wnt, Rho
Directional cell migration is required for the development of an organism with proper polarity including dorsoventral, anterior–posterior, and left–right symmetry. Examples of these cell movements include those of gastrulation and neural fold closure. These cell movements are tightly regulated by secreted ligands (1, 2). One of these signaling pathways required for cell movements is the noncanonical Wnt pathway (3–5).
Noncanonical Wnt signaling, also termed the planar cell polarity pathway, regulates cell movements through modification of the actin cytoskeleton (1, 3, 4, 6). A number of molecular components for this pathway have been identified including Wnt11, Fz, Dvl, Daam1, Rho, Rac, JNK, Strabismus, and Prickle (reviewed in ref. 5). For noncanonical Wnt signaling, the binding of Wnt to the Frizzled (Fz) receptor stimulates a signal that is transduced to the cytoplasmic phosphoprotein Dishevelled (Dvl). At the level of Dvl, two independent and parallel pathways lead to the activation of the small GTPases Rho and Rac. The first pathway signaling to the small GTPase Rho occurs through the molecule Daam1 (7). This Rho pathway leads to the activation of the Rho-associated kinase Rock and mediates cytoskeletal reorganization (5, 8). The second activates another small GTPase of the Rho-family, Rac, which in turns stimulates JNK activity (9–11). Daam1 is a Formin protein family and has been shown to regulate gastrulation; however, how Daam1 is activated for its function remains unknown.
The Formin proteins are central players in regulating cytoskeletal reorganization in mammalian cells (12). The Formin proteins contain three major domains termed the GTPase binding domain (GBD), Formin homology 1 (FH1) domain, and Formin homology 2 (FH2) domain (13). These proteins are proposed to exist in the cytoplasm in an autoinhibited state, which is mediated by a domain termed the diaphanous autoinhibitory domain (DAD) (12). This DAD found in the carboxyl terminus mediates interaction with the amino terminus of the protein and serves to “lock” the protein in a folded or closed conformation (12). It is proposed that Rho activation allows for Rho-GTP to bind to the GBD and release this molecule from autoinhibition. The FH1 and FH2 can then bind to effectors to mediate effects on the cytoskeleton. Intriguingly, the FH2 domain has recently been shown to be capable of nucleating actin filaments by itself in vitro, suggesting a complex interplay between the FH1 and FH2 domains along with their effectors for actin polymerization (12). However, it remains unclear how the Formin proteins are activated in vivo, and one molecule shown to bind the Formin proteins is Profilin1, which functions in actin polymerization (12, 14).
Here we report on the mechanisms of activation of Daam1. We show that Daam1 exists in an autoinhibited state via intramolecular interactions between its amino-terminal GBD and carboxyl-terminal DAD. Daam1 is not significantly activated by binding of Rho but rather by binding of Dvl, which interacts with the DAD and relieves autoinhibition of Daam1. In the Xenopus embryo we show that coexpression of Dvl with Daam1 leads to activation of Daam1 as monitored by hyperactivation of the noncanonical Wnt pathway. Deletion or mutation of conserved amino acid residues of the DAD further converts Daam1 into an activated protein that can rescue defects in gastrulation induced by noncanonical Wnt signaling components. Our studies together uncover the role of Dvl as an important factor that mediates Daam1's activation.
Results
Daam1 Is Autoinhibited by Interactions Between the GBD and DAD.
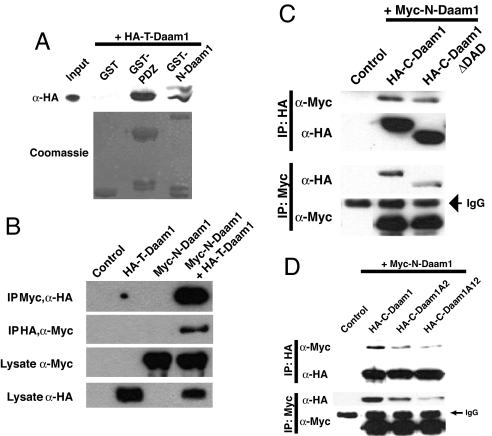
We had previously shown that the full-length Daam1 protein appeared to be autoinhibited because it was incapable of inducing Rho activation and cytoskeletal changes whereas a fragment of Daam1 termed C-Daam, harboring the FH1, FH2, and DAD, can induce Rho activation and cytoskeletal changes (7) [supporting information (SI) Fig. 6A]. We explored whether Daam1 was autoinhibited by interactions between its GBD and DAD similar to other Formin proteins (12). To test for interactions we performed GST pull-down assays. We generated GST fusion proteins that encompass the PDZ domain of Dvl (GST-PDZ) (7) and another containing the amino-terminal domain of Daam1 encompassing the GBD (GST-N-Daam) and used in vitro translated HA-T-Daam, which contains the DAD of Daam1. These pull-down assays demonstrated a direct and specific interaction between DAD and the PDZ domain of Dvl or the GBD domain of Daam1, because the DAD fragment did not bind to GST protein alone (Fig. 1A).
Fig. 1.
Autoinhibition of Daam1 is mediated by interactions between its amino-terminal and carboxyl-terminal regions. (A) GST pull-down assays reveal that T-Daam binds to the PDZ domain of Dvl and to N-Daam. Precipitated T-Daam was detected with Western blotting, and input of GST proteins was visualized by using Coomassie staining. (B–D) Coimmunoprecipitation assays. Plasmids encoding tagged Daam1 fragments were cotransfected into HEK293T cells, and cell lysates were immunoprecipitated (IP) with indicated Abs. Precipitates were then immunoblotted with indicated Abs. (B) N-Daam interacts with T-Daam. (C) N-Daam interacts with C-Daam, but this interaction is reduced with C-DaamΔDAD, which lacks the DAD. (D) Mutations within the DAD in the context of C-Daam reduces interaction with N-Daam.
To further delineate interactions between the GBD and DAD of Daam1, we examined interactions by coimmunoprecipitation using epitope-tagged proteins expressed in mammalian HEK393T cells (SI Fig. 6A). We find that N-Daam can bind to T-Daam and C-Daam, both of which contain the DAD, but its binding to C-DaamΔDAD, which lacks the DAD, was reduced (Fig. 1 B and C). This suggests that the DAD can bind to the GBD.
The DAD of Daam1 is positioned at amino acids 1030–1040 and contains two leucine residues (leucine-1036 and leucine-1040) known to be critical for maintaining intramolecular interaction between the GBD and DAD of mDia1 (13, 15, 16). We therefore mutated these two leucine residues to alanine (SI Fig. 6A) and tested for their ability to bind to the GBD of Daam1. These studies revealed that mutation in leucine-1040 (C-Daam A2) impaired binding between the DAD and GBD whereas mutation of both residues (C-Daam A12) significantly reduced interaction (Fig. 1D).
Because studies indicate the Formins can oligomerize mediated through an FH1 and FH2 domain-containing fragment (16, 17), we tested whether C-Daam, which contains the FH1 and FH2 domains, can also oligomerize. Coimmunoprecipitation experiments revealed that a Myc-tagged C-Daam could efficiently precipitate a HA-tagged C-Daam and that this was independent of the DAD (SI Fig. 6B). This shows that Daam1 can indeed oligomerize, similar to other Formins.
These studies together reveal that Daam1, similar to other Formin proteins, was an autoinhibited protein and that this autoinhibition was mediated via interactions between its GBD and DAD.
Dvl Binds to the DAD of Daam1.
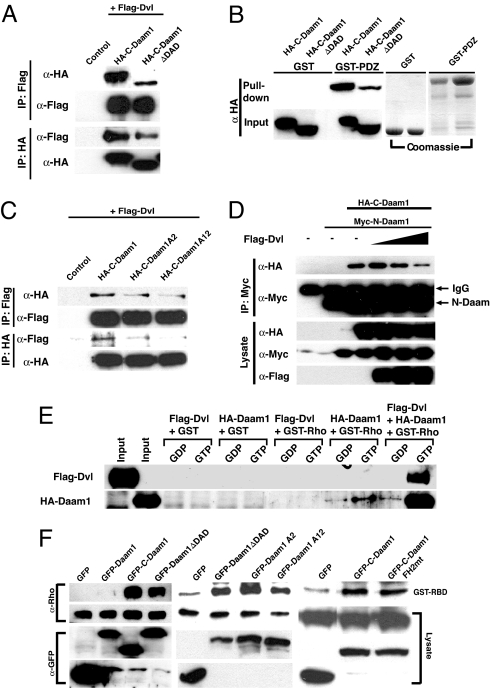
As the T-Daam fragment of Daam1 that contains the DAD binds to Dvl (Fig. 1A) (7), we tested whether Dvl binding to Daam1 depended on the DAD. We performed coimmunoprecipitation experiments using epitope-tagged proteins expressed in mammalian HEK293T cells. These studies revealed that Dvl also binds to C-Daam and T-Daam (7), but its binding to C-DaamΔDAD, which lacks the DAD, was strongly impaired (Fig. 2A). We further confirmed these results using GST pull-down assays and found that deletion of the DAD significantly impaired binding to the PDZ domain of Dvl (Fig. 2B).
Fig. 2.
Dishevelled binds to the DAD of Daam1 and activates Daam1. (A) Coimmunoprecipitation assays reveal that Dvl binds to C-Daam but that its binding to C-DaamΔDAD is impaired. Plasmids encoding tagged Daam1 fragments were cotransfected into HEK293T cells, and cell lysates were immunoprecipitated (IP) with and immunoblotted with indicated Abs. (B) GST pull-down assays reveal that the PDZ domain of Dvl binds to C-Daam but that its binding to C-DaamΔDAD is reduced. (C) Single mutations within the DAD reduce whereas double mutations strongly impair interactions between Dvl and Daam1. (D) Dvl disrupts interactions between N-Daam and C-Daam. Increasing doses of Dvl were cotransfected with N-Daam and C-Daam into HEK293T cells, cell lysates were immunoprecipitated (IP) with indicated Abs, and precipitates were then immunoblotted with indicated Abs. (E) GST pull-down assays show that Daam1 binds to Rho-GTP with a higher preference over Rho-GDP but that the binding of Daam1 to Rho-GTP is amplified in the presence of Dvl. (F) Expression of Daam1 does not induce Rho activation in lysates from HEK293T cells, but C-Daam does. Removal of the DAD (DaamΔDAD) or mutations within the DAD of Daam1 induces Rho activation to levels similar to DaamΔDAD, and mutation within the FH2 domain of Daam1 that abolished the ability of Daam1 to induce stress fibers does not impair Rho activation.
We further examined interactions between Dvl and the DAD of Daam1 using the C-Daam A2 and C-Daam A12 constructs (Fig. 1A). In coimmunoprecipitation experiments or in GST pull-down assays we found that the mutation in leucine-1040 impaired the binding between Dvl and the DAD whereas mutation of both residues significantly disrupted this interaction (Fig. 2C and data not shown).
Together, these studies reveal that the DAD was central for the interaction between Dvl and Daam1, a complex that is induced by Wnt stimulation (7).
Daam1 Autoinhibition Is Relieved by Its Interaction with Dvl.
Our results thus far suggest that the GBD of Daam1 interacts with the DAD, suggesting that, in the absence of Dvl, Daam1 exists in a closed autoinhibited conformation. Because Dvl binds to the DAD (Fig. 2 A and B) but not to the GBD (7), we suspected that Dvl binding might release Daam1 from its autoinhibition. We tested this hypothesis using GST pull-down assays and used GTP- or GDP- loaded GST-Rho fusion proteins and extracts from HEK293T cells overexpressing Daam1 or Dvl. Daam1 is observed to bind to both GTP- and GDP-loaded Rho with a higher preference toward GTP-Rho whereas Dvl does not bind to either forms of Rho (Fig. 2E). However, when Dvl was incubated with Daam1 and then binding was performed, a dramatic increase of binding of Daam1 to Rho-GTP was observed, and Dvl was now observed in a complex containing Rho-GTP and Daam1 (Fig. 2E).
Because the DAD of Daam1 is responsible for binding to Dvl, we further tested whether Dvl can interfere with the binding between N-Daam and C-Daam. Cotransfection of N-Daam and C-Daam along with increasing doses of Dvl in mammalian HEK293T cells resulted in a sharp diminution of interaction observed between N-Daam and C-Daam using coimmunoprecipitation assays (Fig. 2D).
These studies show that Dvl can inhibit GBD/DAD binding, providing a mechanism to relieve the autoinhibition of Daam1.
Removal of the DAD Converts Daam1 into an Active State.
If the DAD is central for maintaining Daam1 in an autoinhibited state, we reasoned that removal of this domain should convert Daam1 into an “activated” protein. We have shown that C-Daam behaves as an activated protein that can induce Rho activation (7), and we tested whether Daam1, Daam1 lacking DAD (DaamΔDAD), or Daam1 harboring DAD point mutations (Daam A2 and Daam A12) can activate Rho. To measure Rho activation, we used the Rho activation assay (18) and extracts of HEK293T mammalian cells transfected with Daam1, C-Daam, DaamΔDAD, or Daam1 harboring mutations within the DAD. Similar to previous studies, C-Daam expression induced Rho activation but expression of Daam1 did not (Fig. 2F) (7). However, expression of DaamΔDAD or Daam1 harboring DAD mutations induced Rho activation to levels similar to that of C-Daam (Fig. 2F).
To determine whether Rho activation mediated by activated forms of Daam1 depended on its ability to polymerize actin, we used a mutant construct that harbors an isoleucine-to-alanine mutation within the FH2 domain (isoleucine-698). This mutation abolished actin polymerization mediated by the FH2 domain within the context of mDia1 (19, 20). Interestingly, mutation within the FH2 domain did not impair Rho activation (Fig. 2F) whereas its ability to induce stress fibers was abolished [Fig. 3A and SI Fig. 7 A and B].
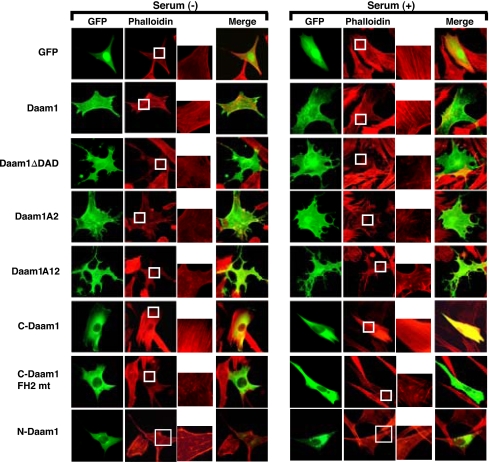
Fig. 3.
Removal or mutations of the DAD activates Daam1. Expression of GFP-Daam1 does not induce stress fiber formation in serum-starved NIH 3T3 cells or disrupt stress fibers found in NIH 3T3 cells cultured in the presence of serum. GFP-DaamΔDAD, GFP-Daam A2, and GFP-Daam A12 constructs induce morphological changes characterized by numerous protrusions and collapsed stress fibers in NIH 3T3 cells. GFP-C-Daam induces the formation of stress fibers in NIH 3T3 cells, and a mutation within the FH2 domain of C-Daam abolishes the ability of C-Daam to induce stress fibers.
We further explored whether removal of or mutations within the DAD allows Daam1 to induce cytoskeletal changes and stress fiber formation. We had shown that C-Daam but not Daam1 could induce stress fibers when expressed in NIH 3T3 cells (7). We therefore expressed Daam1, C-Daam, DaamΔDAD, Daam A2, and Daam A12 in NIH 3T3 cells and assayed for the ability of these transfected cDNAs to induce stress fiber formation in cells cultured in the absence of serum or their ability to collapse stress fibers in cells cultured in the presence of serum. From these studies we observed that Daam1 did not induce stress fibers but C-Daam did and that neither Daam1 nor C-Daam induced the formation of cell protrusions (Fig. 3A and SI Fig. 7 A and B). However, we observed that expression of DaamΔDAD, Daam A2, and Daam A12 did not induce stress fibers in these cells but rather strongly collapsed the existing stress fibers and induced numerous long membranous protrusions and branched structures (Fig. 3A and SI Fig. 7 A and B). Expression of N-Daam1, which harbors the GBD, did not induce such protrusions but strongly collapsed existing stress fibers (Fig. 3A and SI Fig. 7 A and B). To determine whether this induction of membranous protrusions and branches structures may be due to Rac activation, we used a Rac activation assay (18). We found that, similar to reported studies, Daam1, C-Daam1, or N-Daam1 did not induce Rac activation in HEK293T cells, but, surprisingly, expression of Daam1ΔDad was able to induce Rac activation similar to Dvl expression (SI Fig. 7E).
Because we have uncovered that C-Daam1 was able to induce Rho activation, which was independent of its ability to induce stress fibers, we examined whether the ability of Daam1 to induce stress fibers depended on Rho activation. We transfected NIH 3T3 cells with C-Daam1 and used a cell-permeable Rho inhibitor. These experiments revealed that, in the absence of Rho inhibitor, C-Daam1 was able to induce stress fibers but that, in the presence of the Rho inhibitor, this ability was suppressed (Fig. 3B and SI Fig. 7 C and D). Thus, Daam1's ability to induce stress fibers depended on Rho activation, and this suggests that there are two pathways downstream of Daam1 that regulate actin polymerization. One is Rho activation-dependent, which is necessary but not sufficient, and the other is Rho activation-independent.
These studies together demonstrate that the DAD was central for maintaining Daam1 in an autoinhibited state and that removal of the DAD can convert Daam1 into an activated protein that induces Rho activation and mediate cytoskeletal changes.
Daam1 Synergizes with Dvl to Regulate Xenopus Gastrulation.
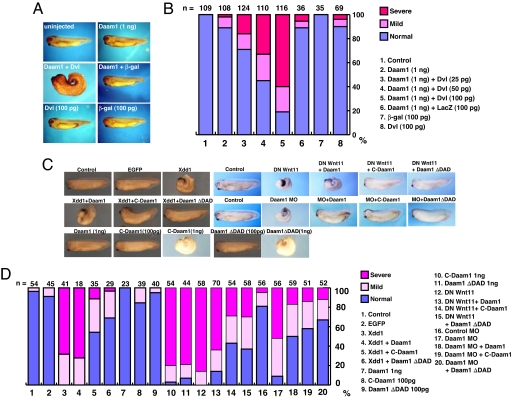
Because our above biochemical data demonstrate that Dvl can relieve the autoinhibition of Daam1, we explored this finding in the Xenopus embryo. Dorsal expression of 1 ng of Daam1 alone has little effect on development but when coexpressed with increasing doses (25–100 pg) of XDsh (Xenopus homologue of Dvl) leads to a significant increase in the numbers of embryos with gastrulation defects (Fig. 4 A and B). This effect was specific because expression of the highest doses of Dsh (100 pg) alone has little effect on development, and coexpression of 1 ng of Daam1 with 100 pg of β-Gal RNA did not result in such effects (Fig. 4 A and B). We interpret these findings to mean that coexpression of suboptimal doses of Dvl with Daam1 leads to the activation of Daam1 and that active Daam1 hyperactivates noncanonical Wnt signaling resulting in gastrulation defects.
Fig. 4.
Dishevelled activates Daam1, leading to gastrulation defects, and activated Daam1 rescues convergent extension defects. (A) Expression of subthreshold doses of Dsh (100 pg of RNA) or Daam1 (1 ng of RNA) does not interfere with gastrulation, but coinjection of these subthreshold doses of Dsh and Daam1 induces gastrulation defects. Embryos were injected into both dorsal blastomeres, and embryos were scored at stage 35. Embryos with an open blastopore, exposed endodermal tissue, and significantly reduced anterior–posterior (AP) axis were scored as severe embryos (Middle Left), and embryos with a small open blastopore or delayed blastopore closure and a slightly shortened AP axis or bent body axis were scored as mild. (B) Quantitation of the phenotypes of injected embryos in A. (C) Gastrulation in Xenopus embryos is inhibited by expression of dominant negative Dsh (Xdd1) (1 ng of RNA), dominant negative XWnt11 (DN-Wnt11) (2 ng of RNA), or Daam1 MO (100 ng), but this inhibition is rescued by C-Daam (100 pg of DNA) or DaamΔDAD (100 pg of DNA) coinjection but not Daam1 (1 ng of RNA) coinjection. (D) Quantitation of the results of phenotypic analysis in C. In B and D, the number of embryos scored (n) is shown at the top of each bar.
Because deletion of the DAD converts Daam1 into an activated form, we tested the ability of DaamΔDAD to rescue defects in gastrulation induced by dominant negative Dishevelled (Xdd1), dominant negative Wnt11 (DN-Wnt11), or Daam1 MO. Dorsal expression of Xdd1, DN-Wnt-11, or Daam1 MO led to a significant number of embryos with gastrulation defects (Fig. 4 C and D). However, coexpression of Daam1 with Xdd1 or DN-Wnt11 did not rescue this phenotype whereas expression of C-Daam or DaamΔDAD strongly suppressed this phenotype (Fig. 4 C and D). Furthermore, coexpression of Daam1 with the Daam1 MO rescued the gastrulation defects induced by the Daam1 MO and coexpression of C-Daam or DaamΔDAD also suppressed the effects of the Daam1 MO (Fig. 4 C and D). Note that expression of Daam1, C-Daam, or DaamΔDAD at these doses did not induce any phenotypes (Fig. 4 C and D). We further tested that injections of the RNAs or MO used for our studies did not inhibit mesodermal and neural gene expression using in situ hybridization studies. We examined brachyury (Xbra), goosecoid (Gsc), Sox2, and Otx2 and found no changes in the levels of expression of these genes (SI Fig. 8A). Thus, at the phenotypic level, C-Daam and DaamΔDAD behave as activated forms of Daam1 and function epistatically downstream of Dsh to mediate cell movements independent of gene induction.
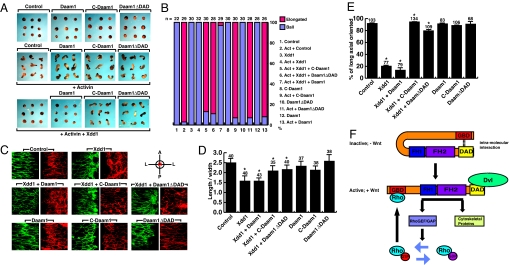
To test whether our DaamΔDAD construct can indeed function as activated Daam1 at the explant level, we coexpressed C-Daam, DaamΔDAD, or Daam1 constructs with dominant-negative Dishevelled (Xdd1) in activin-treated animal caps and scored for rescue of convergent extension movements. These studies revealed that both C-Daam and DaamΔDAD but not Daam1 were able to rescue the failure of convergent extension induced by Xdd1 in the animal caps (Fig. 5 A and B).
Fig. 5.
Activated Daam1 rescues defective cell behavior responsible for impaired convergent extension movements. (A) Extension of activin (Act)-treated animal caps is inhibited by expression of dominant negative Dsh (Xdd1) (1 ng of RNA), but this inhibition is rescued by C-Daam (100 pg of DNA) or DaamΔDAD (100 pg of DNA) coinjection but not Daam1 (1 ng of RNA) coinjection in convergent extension assays. (B) Quantitation of the extension of animal cap explants in A; the number of caps scored (n) is shown at the top of each bar. (C) Expression of dominant negative Dishevelled (Xdd1) (1 ng of RNA) impairs polarization, elongation, and mediolateral alignment of dorsal mesodermal cells undergoing convergent extension movements. These induced defects in cell behaviors are rescued by coexpression of Xdd1 with C-Daam (100 pg of DNA) or DaamΔDAD (100 pg of DNA) but not Daam1 (1 ng of RNA). Expression of C-Daam, DaamΔDAD, or Daam1 does not interfere with cell polarization, elongation, or mediolateral alignment at these injected doses. Orientation of the explants is shown in the upper right: A, anterior; P, posterior; L, lateral; M, midline. (D and E) Quantitation of cell polarization and mediolateral orientation of cells from the studies of C; the numbers of cells examined are shown at the top of each bar. *, statistically significant value (P < 0.005). (F) A model for how Daam1 is activated. Daam1 exists in the cytoplasm as an autoinhibited protein via intramolecular interactions between the GBD and the DAD. Wnt stimulation induces a complex formation between Dvl and Daam1, and Dvl binds to the DAD, disrupting interactions between the amino and carboxyl regions of Daam1. The binding of Dvl thus relieves Daam1's autoinhibition, and Daam1 with its downstream effectors triggers Rho activation and cytoskeletal changes to regulate gastrulation.
Last, we examined the polarization and cell shape changes of dorsal mesodermal cells undergoing convergent extension movements. During convergent extension movements, these cells adopt an elongated and polarized shape with a long axis oriented toward the midline. As shown (21–23), we observed that expression of dominant negative Dishevelled (Xdd1) inhibits both cell shape changes and mediolateral orientation of these cells as measured by a length-to-width ratio and long axial orientation (Fig. 5 C–E). However, coexpression of Xdd1 with C-Daam or DaamΔDAD but not Daam1 was able to rescue this inhibition of cell shape change and mediolateral orientation (Fig. 5 C–E). Thus, at the cellular level, C-Daam and DaamΔDAD behave as activated versions of Daam1 and rescue defects in cell behavior including polarization and mediolateral orientation responsible for normal convergent extension movements.
Discussion
The Formin proteins are critical modulators of the actin cytoskeleton and exist in an autoinhibited state mediated by interactions between their carboxyl-terminal DAD and a domain referred to as the diaphanous inhibitory domain (DID) harbored within the GBD (24–). Interaction studies have revealed that this autoinhibition is relieved by Rho-GTP binding, and crystal structure analyses have revealed that Rho-GTP binds within the GBD but not to the DID (19, 25). Thus, to rationalize how Rho-GTP binding relieves the interaction between the DAD and DID, it has been suggested that upon RhoA-GTP binding a conformational change occurs within the GBD interrupting the binding of the DAD to the DID (19, 27). Interestingly, biochemical studies have also shown that incubation of GTP-bound Rho does not fully activate the Formin protein mDia1 in vitro (12, 16, 28, 29), suggesting that other mechanisms are required for the full activation of the Formin proteins.
Here we have focused on the Formin protein Daam1 (7). We show that Daam1 exists in an autoinhibited state. We further show that this autoinhibition is mediated by the carboxyl-terminal fragment of Daam1 harboring the DAD, which interacts with amino-terminal fragment of Daam1 encompassing the GBD and DID (Fig. 1 A–C). Removal of the DAD or mutations of critical hydrophobic leucine residues within the DAD impairs interaction between the DAD and GBD (Fig. 1 C and D). It is, however, important to note that deletion or mutation within the DAD does not completely abolish interactions, suggesting that other residue(s) outside of the DAD are required for this interaction.
In additional studies we observe that overexpression of full-length Daam1 does not induce any changes to the actin cytoskeleton in mammalian cells (Fig. 3 and SI Fig. 7 A and B), does not interfere with gastrulation, and does not rescue defects in convergent extension movements induced by dominant negative Dvl or cell behavior during Xenopus embryogenesis (Figs. 4 A–D and 5 A–E), consistent with being autoinhibited (7). These studies together reveal that the autoinhibition of Daam1 is mediated in a manner similar to other Formin proteins.
However, if Daam1 is autoinhibited similar to other Formin proteins and our studies show that Daam1 can activate Rho, how was it activated? Our studies demonstrated that Dvl and Daam1 formed a complex in response to Wnt stimulation, and it was reasonable to propose that Dvl binding may activate this protein (7). Here we show that Dvl binds specifically to the DAD of Daam1 (SI Fig. 6A and Fig. 2 A–C), and we find that incubation of Dvl with Daam1 allows Daam1 to significantly interact with Rho-GTP (Fig. 2E). Indeed, we found that Dvl can disrupt the DAD/DID interaction (Fig. 2D). We further substantiate this activation role of Daam1 by Dvl by showing that, in the embryo, expression of subthreshold doses of Daam1 or Dvl has no effect on development. But coexpression of subthreshold doses of Daam1 and increasing doses of Dvl dose-dependently inhibited gastrulation (Fig. 4 A and B). These findings further posit that Dvl and not Rho plays a central role in mediating the activation of Daam1.
Because a truncated construct of Daam1 lacking the GBD behaves as a constitutively active protein (7) and Dvl binds to the DAD to activate Daam1, we reasoned that removal of the DAD in the context of Daam1 will render the protein in an activated form. We show that removal of the DAD or mutations within the DAD allows Daam1 to now induce Rho activation similar to that of C-Daam (Fig. 2F) and rescue the inhibition of convergent extension defects or cell polarization and orientation induced by dominant-negative Dsh during gastrulation (Figs. 4 C and D and 5 A–E). Interestingly, expression of DaamΔDAD or Daam1 constructs harboring mutations within the DAD in contrast to C-Daam did not induce stress fiber formation in NIH 3T3 cells but rather induced the formation of numerous cellular protrusions and stress fiber collapse along with Rac activation (Fig. 3A and SI Fig. 7 A, B, and E). Because the DaamΔDAD construct also harbors the GBD, which by itself can induce the collapse of stress fibers and does not induce cellular protrusions (7), it is likely that a complex interplay among the GBD, FH1, and FH2 domains result in this phenotype.
It is important to note that that downstream of Daam1 two activities are apparent, Rho activation and cytoskeletal changes including stress fiber formation and cell shape changes. The FH2 domain within Daam1 is responsible for stress formation because mutation within the FH2 domain abolishes the ability of Daam1 to induce stress fiber formation (Fig. 3), similar to other Formin proteins (19, 20). However, mutation within the FH2 domain does not abolish Rho activation mediated by Daam1, but inhibition of Rho activity suppressed stress fiber formation, suggesting that these two activities are tightly integrated. Daam1 plays a central role in mediating noncanonical Wnt signaling to the actin cytoskeleton, and we propose that its effects on the actin cytoskeleton and cell shape changes in mammalian cells are translated into cell shape changes required for cell motility during gastrulation. Recent studies have uncovered differential localization of Daam1 during zebrafish gastrulation and implicated Daam1's function on cell shape changes for gastrulation (30).
Our studies together have shown that Dvl binds to the DAD of Daam1 and is a central factor for activating Daam1. This interaction disrupts the autoinhibition of Daam1 mediated by interactions between the GBD and DAD and relieves the autoinhibition of Daam1. Interestingly, it has been shown that the binding of Rho-GTP to other Formin proteins only partially activates these proteins, and whether other carboxyl-terminal binding proteins are responsible for their full activation remains to be uncovered (12, 16).
In summary, we propose a model of how Daam1 is activated and functions in noncanonical Wnt signaling during gastrulation. Daam1 exists in an autoinhibited state in the cellular cytoplasm via intramolecular interactions between the DAD and GBD, and with Wnt stimulation Dvl binds to the DAD of Daam1. The binding of Dvl to the DAD disrupts this intramolecular interaction and leads to the activation of Daam1. Activated Daam1 can then interact with downstream effector molecules including Profilin1 (14) and likely a RhoGEF and RhoGAP to modulate Rho activity. Together the complex interplay between the individual domains and effectors of Daam1 is required for Rho activation and modulation of the actin cytoskeleton for noncanonical Wnt signaling and cell motility during vertebrate gastrulation (Fig. 5D).
Experimental Procedures
Materials.
mAbs against RhoA (26C4), Myc (9E10), and GFP (B-2) and polyclonal Abs against Myc (N-262) were from Santa Cruz Biotechnology, and mAb against HA (Anti-HA High Affinity) was from Roche. mAb against Flag (M2) was from Sigma. Alexa Fluor anti-mouse and anti-rabbit Abs and Texas Red X-Phalloidin were from Molecular Probes.
Plasmids and Oligonucleotides.
The human Daam1 and mutant fragments of Daam1 were generated by restriction digestion or a PCR approach and subcloned in pCS2+MT or pcDNA-HA, or pCS2+GFP vector. Details of plasmids are available upon request. A Flag-tagged Dvl2 construct was used for our studies (7, 9).
Transfections.
All studies were done by using mammalian HEK293T cells or NIH 3T3 cells. Cells were transfected by using Polyfect reagent (Qiagen) with 1–2 μg of each indicated plasmid. Transfected DNA amounts were equalized via vectors without inserts.
GST Pull-Down Assays and Immunoprecipitation.
HA-tagged proteins used in GST pull-down assays were generated by TNT Quick-Coupled Transcription/Translation systems (Promega). GST pull-down assays and immunoprecipitation assays were performed as described (7, 9).
Immunocytochemistry.
Immunocytochemistry was carried out as described (7, 14, 31). Images were obtained by using an Olympus IX70 fluorescent microscope with a ×100 objective lens or a Zeiss Axiovert 100 confocal microscope. For quantification of effects on stress fibers, a base line of 10 stress fibers per cell was used; thus, any cell containing more than or fewer than 10 fibers was scored as an increase or decrease, respectively. For the Rho inhibitor assay, NIH 3T3 cells were trypsinized 8 h after transfection, cells were resuspended in DMEM containing 0.1% FBS, and the cell-permeable Rho inhibitor (Cytoskeleton) was added to a final concentration of 2 μg/ml. Cells were replated on coverslips and incubated for 6 h, and immunocytochemistry was performed.
Embryo Manipulations and Explant Assays.
Embryo manipulations and explant assays were performed as described (7, 9, 32). Embryo injections were done with in vitro transcribed RNAs or cDNAs. Convergent extension assays in explants were performed as described (9).
Embryo Dissection and in Vivo Imaging.
Microinjection and microdissection of Xenopus embryos were performed as described (33–36). Briefly, RNAs (0.5–2 ng) encoding GFP-CAAX and membrane-tethered Cherry were microinjected separately into dorsal blastomeres of four-cell stage embryos, alone or with RNAs encoding Xdd1 and Daam1 or with DNAs encoding C-Daam and DaamΔDAD. “Shaved” Keller explants were prepared at stage 12 and kept flat with mesoderm layer up by a coverslip. The explants were then examined with a confocal microscope at the neurula stage.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Sunita Kramer and the R.H. laboratory for critical comments. We thank Dr. Xi He (Children's Hospital, Boston) in whose laboratory these studies were initiated. We thank Drs. Jeffrey Miller, Chenbei Chang, Karen Symes, and Sergei Sokol for reagents, and we are grateful to Drs. William Wadsworth, Michael Matisse, and Noriko Kane-Goldsmith for use of their microscopes. This work was supported by grants from the Uehara Memorial Foundation and the New Jersey Commission for Cancer Research (to A.S.). R.H. is the recipient of an American Heart Association Scientist Development Grant, a Basil O'Connor Starter Scholar Award, National Science Foundation Grant 0544061, and National Institutes of Health Grant GM078172.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707277105/DC1.
References
- 1.Keller R. Science. 2002;298:1950–1954. doi: 10.1126/science.1079478. [DOI] [PubMed] [Google Scholar]
- 2.Harland R, Gerhart J. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- 3.Veeman MT, Axelrod JD, Moon RT. Dev Cell. 2003;5:367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 4.Wallingford JB, Fraser SE, Harland RM. Dev Cell. 2002;2:695–706. doi: 10.1016/s1534-5807(02)00197-1. [DOI] [PubMed] [Google Scholar]
- 5.Wallingford JB, Habas R. Development (Cambridge, UK) 2005;132:4421–4436. doi: 10.1242/dev.02068. [DOI] [PubMed] [Google Scholar]
- 6.Mlodzik M. Trends Genet. 2002;18:564–571. doi: 10.1016/s0168-9525(02)02770-1. [DOI] [PubMed] [Google Scholar]
- 7.Habas R, Kato Y, He X. Cell. 2001;107:843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 8.Marlow F, Topczewski J, Sepich D, Solnica-Krezel L. Curr Biol. 2002;12:876–884. doi: 10.1016/s0960-9822(02)00864-3. [DOI] [PubMed] [Google Scholar]
- 9.Habas R, Dawid IB, He X. Genes Dev. 2003;17:295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Yuan H, Xie W, Mao J, Caruso AM, McMahon A, Sussman DJ, Wu D. J Biol Chem. 1999;274:129–134. doi: 10.1074/jbc.274.1.129. [DOI] [PubMed] [Google Scholar]
- 11.Yamanaka H, Moriguchi T, Masuyama N, Kusakabe M, Hanafusa H, Takada R, Takada S, Nishida E. EMBO Rep. 2002;3:69–75. doi: 10.1093/embo-reports/kvf008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goode BL, Eck MJ. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 13.Higgs HN. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 14.Sato A, Khadka DK, Liu W, Bharti R, Runnels LW, Dawid IB, Habas R. Development (Cambridge, UK) 2006;133:4219–4231. doi: 10.1242/dev.02590. [DOI] [PubMed] [Google Scholar]
- 15.Alberts AS. J Biol Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- 16.Li F, Higgs HN. J Biol Chem. 2005;280:6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- 17.Copeland JW, Copeland SJ, Treisman R. J Biol Chem. 2004;279:50250–50256. doi: 10.1074/jbc.M404429200. [DOI] [PubMed] [Google Scholar]
- 18.Habas R, He X. Methods Enzymol. 2006;406:500–511. doi: 10.1016/S0076-6879(06)06038-1. [DOI] [PubMed] [Google Scholar]
- 19.Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Mol Cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 21.Park TJ, Gray RS, Sato A, Habas R, Wallingford JB. Curr Biol. 2005;15:1039–1044. doi: 10.1016/j.cub.2005.04.062. [DOI] [PubMed] [Google Scholar]
- 22.Wallingford JB, Harland RM. Development (Cambridge, UK) 2001;128:2581–2592. doi: 10.1242/dev.128.13.2581. [DOI] [PubMed] [Google Scholar]
- 23.Wallingford JB, Rowning BA, Vogeli KM, Rothbacher U, Fraser SE, Harland RM. Nature. 2000;405:81–85. doi: 10.1038/35011077. [DOI] [PubMed] [Google Scholar]
- 24.Faix J, Grosse R. Dev Cell. 2006;10:693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Nature. 2005;435:513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- 26.Wallar BJ, Stropich BN, Schoenherr JA, Holman HA, Kitchen SM, Alberts AS. J Biol Chem. 2006;281:4300–4307. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- 27.Nezami AG, Poy F, Eck MJ. Structure (London) 2006;14:257–263. doi: 10.1016/j.str.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Li F, Higgs HN. Curr Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- 29.Seth A, Otomo C, Rosen MK. J Cell Biol. 2006;174:701–713. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kida YS, Sato T, Miyasaka KY, Suto A, Ogura T. Proc Natl Acad Sci USA. 2007;104:6708–6713. doi: 10.1073/pnas.0608946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capelluto DG, Kutateladze TG, Habas R, Finkielstein CV, He X, Overduin M. Nature. 2002;419:726–729. doi: 10.1038/nature01056. [DOI] [PubMed] [Google Scholar]
- 32.Kato Y, Habas R, Katsuyama Y, Naar AM, He X. Nature. 2002;418:641–646. doi: 10.1038/nature00969. [DOI] [PubMed] [Google Scholar]
- 33.Ataliotis P, Symes K, Chou MM, Ho L, Mercola M. Development (Cambridge, UK) 1995;121:3099–3110. doi: 10.1242/dev.121.9.3099. [DOI] [PubMed] [Google Scholar]
- 34.Nie S, Chang C. Dev Biol. 2007;303:93–107. doi: 10.1016/j.ydbio.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tahinci E, Symes K. Dev Biol. 2003;259:318–335. doi: 10.1016/s0012-1606(03)00206-9. [DOI] [PubMed] [Google Scholar]
- 36.Wilson PA, Oster G, Keller R. Development (Cambridge, UK) 1989;105:155–166. doi: 10.1242/dev.105.1.155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.