Abstract
Changes in DNA methylation patterns are an important characteristic of human cancer. Tumors have reduced levels of genomic DNA methylation and contain hypermethylated CpG islands, but the full extent and sequence context of DNA hypomethylation and hypermethylation is unknown. Here, we used methylated CpG island recovery assay-assisted high-resolution genomic tiling and CpG island arrays to analyze methylation patterns in lung squamous cell carcinomas and matched normal lung tissue. Normal tissues from different individuals showed overall very similar DNA methylation patterns. Each tumor contained several hundred hypermethylated CpG islands. We identified and confirmed 11 CpG islands that were methylated in 80–100% of the SCC tumors, and many hold promise as effective biomarkers for early detection of lung cancer. In addition, we find that extensive DNA hypomethylation in tumors occurs specifically at repetitive sequences, including short and long interspersed nuclear elements and LTR elements, segmental duplications, and subtelomeric regions, but single-copy sequences rarely become demethylated. The results are consistent with a specific defect in methylation of repetitive DNA sequences in human cancer.
Keywords: DNA methylation, tiling arrays, CpG islands
Changes in DNA methylation patterns are frequent events in human tumors (1). DNA hypomethylation in cancer tissue was first observed more than two decades ago (2–6) and may be mechanistically linked to tumorigenesis (7). In the 1990s, researchers reported hypermethylation of CpG islands of several known and putative tumor suppressor genes and other genes involved in important genome defense pathways, such as DNA repair (1, 8–12). Today, there are many reports that have documented methylation of CpG islands associated with a large number of different genes, including almost every type of human cancer. In lung cancer, several CpG islands known to be methylated include those associated with CDKN2A, RASSF1A, RARbeta, MGMT, GSTP1, CDH13, APC, DAPK, TIMP3, and several others (13–17). The methylation frequency (i.e., the percentage of tumors analyzed that carry methylated alleles) ranges from <10% to ≈80%, but these numbers differ widely depending on the tumor histology, the study population, and/or the methodology used to assess methylation. Detection of methylated CpG islands in easily accessible biological materials such as serum or sputum has the potential to be useful for the early diagnosis of lung cancer and other malignancies (18–20).
Repetitive DNA elements, such as short and long interspersed nuclear elements (SINEs and LINEs, respectively) and simple repeat sequences, are often found hypomethylated in tumors (21–26). Although it seems clear that methylation-induced silencing of tumor suppressor genes can be an important event in tumorigenesis, the magnitude, exact sequence specificity, and biological significance of tumor-associated DNA hypomethylation is much less understood (21, 26). In particular, the extent and sequence context of single-copy gene and general genome hypomethylation is not known, and it is commonly assumed that all genomic sequences are hypomethylated in tumors.
Current research approaches are geared toward the characterization of the full complement of DNA methylation changes in cancer. Several techniques, including differentiation of methylated and unmethylated sequences by use of restriction enzymes or by precipitation with an anti-5-methylcytosine antibody have been introduced (27). We recently developed a methylation detection method, the methylated-CpG island recovery assay (MIRA), that does not depend on the use of sodium bisulfite but has similar sensitivity and specificity as bisulfite-based approaches (28). The MIRA method is based on the high affinity of the MBD2b/MBD3L1 protein complex for methylated CpG dinucleotides. Methylated double-stranded DNA sequences are enriched and are used to make probes for use with microarrays. For efficient pull down of methylated DNA by this method, two or more methylated CpG sites in a fragment of 50 or fewer base pairs are required (29). In the present study, we have used the MIRA method in combination with CpG island and genomic tiling arrays to characterize at high resolution the DNA methylation changes that occur in the genome of lung squamous cell carcinomas (SCCs). Tumor-specific CpG island DNA methylation markers are identified as well as a specific defect in methylation of repetitive DNA elements.
Results
Chromosomal DNA Methylation Patterns.
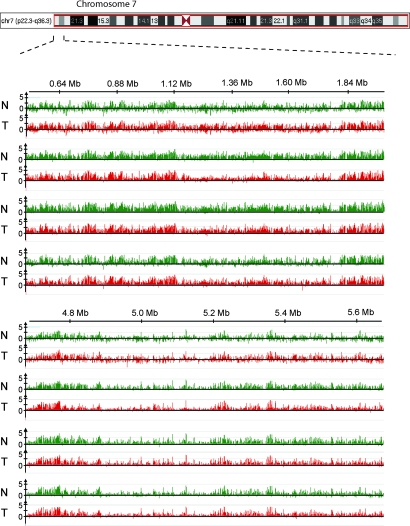
To analyze tumor-associated DNA methylation changes at the chromosomal level, we compared two stage I, one stage II, and one stage III lung SCCs with normal matched lung tissues. We used the MIRA-assisted microarray method (29) with genome tiling arrays for high-resolution DNA methylation analysis. MIRA-enriched and input fractions were cohybridized to NimbleGen arrays covering genomic regions at a resolution of 100 bp (Fig. 1). These MIRA microarrays provide complete and high-resolution mapping of DNA methylation patterns along chromosomes. We used arrays covering the entire short arm of chromosome 7 [57 megabases (Mb)], the entire short and part of the long arm of chromosome 8 (65 Mb), and regions of the long arm of chromosomes 6 (5 Mb) and 7 (12.7 Mb). DNA methylation profiles of normal lung and matched pairs of SCC samples were compared between different patients. We observed a striking conservation of overall chromosomal DNA methylation patterns [Fig. 1 and supporting information (SI) Fig. 5], in particular when comparing normal lung tissue from different individuals.
Fig. 1.
Conservation of global chromosomal DNA methylation patterns. MIRA-assisted microarrays were conducted for pairs of normal lung tissue (N, green) and corresponding SCCs (T, red). The profile of methylated DNA sequences is displayed as the ratio of MIRA-enriched DNA signal versus input DNA signal. The top two tumor samples are stage I SCCs, the third sample is stage II, and the bottom sample is a stage III tumor. Representative tiling array data are shown demonstrating the general conservation of chromosomal methylation profiles between different individuals and, at this level of resolution, also between normal and tumor tissue. A segment of the short arm of chromosome 7 is shown.
Hypermethylation in Tumors at CpG Islands.
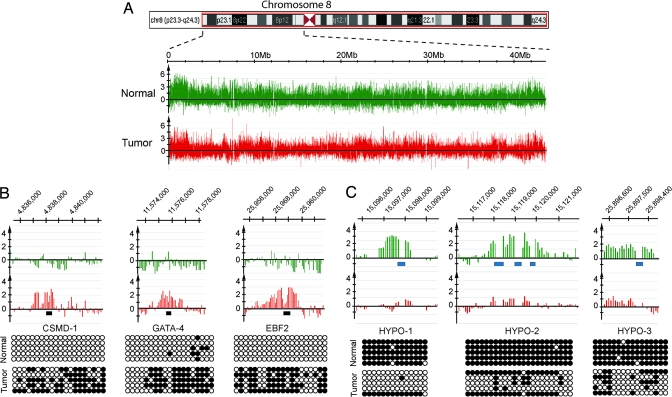
Regions near the centromeres and telomeres were more densely methylated than other loci in normal and tumor samples (Fig. 2A). Upon closer examination of the high-resolution methylation data, we detected 16 cancer-specifically hypermethylated regions on the short arm of chromosome 8 in one stage I SCC (tumor 2). All of them were CpG islands or CpG-rich regions, often overlapping or located in close proximity to promoter regions (SI Table 3). We analyzed the DNA methylation status of 11 of the 16 hypermethylated loci by bisulfite sequencing (examples are shown in Fig. 2B) and combined bisulfite restriction analysis (COBRA) assays (data not shown) and found this data to be entirely consistent with the array data. In stage II and stage III lung tumors, we detected a similar number of hypermethylated regions (SI Table 3). Hypermethylated targets from different patients were partially overlapping. Importantly, we found that, other than CpG islands, no other sequences were found to be hypermethylated in tumors relative to normal tissue.
Fig. 2.
DNA methylation patterns along the short arm of chromosome 8. (A) Schematic display of a chromosome 8 region and the methylation signals (MIRA-enriched DNA versus input DNA) in normal lung tissue and a matching stage I SCC tumor (tumor 2). (B) Examples of hypermethylated CpG islands. The methylation array profiles are shown on the top, and the confirmation of tumor-associated methylation by bisulfite sequencing is shown near the bottom. Open circles, unmethylated CpG sites; closed circles, methylated CpG sites. The black bars indicate the regions analyzed by bisulfite sequencing. (C) Examples of hypomethylated SINE sequences in lung tumors. The array signals are shown on the top. Blue bars denote the location of SINE elements. Confirmation of the SINE methylation status by bisulfite sequencing is shown at the bottom.
Hypomethylation in Tumors at Repetitive DNA Sequences.
In addition to hypermethylation, the tiling arrays provided information on the extent and sequence specificity of DNA hypomethylation. SINEs and LINEs, together with human endogenous retroviruses (HERVs), make up >45% of the human genome (30). Transposable elements are highly methylated and mostly silenced in normal cells (25, 31). Although repetitive sequences are not directly represented as probes on the tiling arrays, we obtained information on the methylation status of SINE elements due to hybridization of flanking single copy DNA to adjacent probes after MseI digestion. In the MIRA technique, the highly methylated elements are captured by the MBD2b/MBD3L1 protein complex (29). After comparing the DNA methylation profiles of normal lung tissues and the matched SCC samples, we detected several thousand tumor-associated demethylation events of genomic regions carrying SINE elements (examples are shown Fig. 2C, Fig. 3, and SI Fig. 6). The methylation status of several arbitrarily chosen SINE elements was verified by bisulfite sequencing and COBRA assays. Primers for bisulfite sequencing were complementary to the flanking unique sequences, and the sequencing data reflects the methylation status of the repetitive element itself. The sequencing data confirmed the MIRA-assisted tiling array methylation profiles for SINE elements and their extensive hypomethylation in tumors. The cancer-specific hypomethylation of SINE elements was not well conserved between individual tumors; this reflects a degree of randomness for targeting individual SINE sequences for demethylation in cancer.
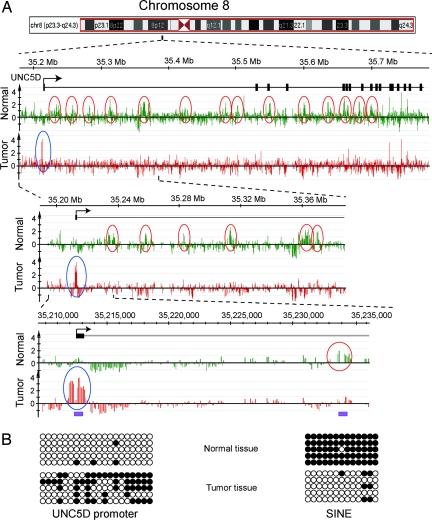
Fig. 3.
Promoter hypermethylation and intragenic SINE hypomethylation in the UNC5D gene. (A) This gene on chromosome 8 shows hypermethylation of the promoter-associated CpG island (blue circles) and hypomethylation of multiple intragenic SINE elements (red circles). (B) Bisulfite sequencing confirms the methylation status of the promoter and its proximal SINE element. The purple bars indicate the regions analyzed by bisulfite sequencing. Black boxes indicate exons, and the arrow shows the transcription start site.
Next, we surveyed all of the CpG islands on chromosome 8p in tumor SCC2 and its corresponding normal tissue. As expected, >98% (159/162) of the promoter-associated CpG islands were unmethylated in normal lung. In addition, there were 78 unmethylated intragenic and intergenic CpG islands. Further, we found 159 mostly short (<0.6 kb) methylated CpG islands in normal lung. Sixty-four of these methylated CpG islands were intragenic, and they generally did not become hypomethylated in the tumor. However, the majority of the methylated islands (a total of 95) were located between 0 and 2 Mb away from the chromosome end, overlapping the subtelomeric region, and these were not associated with a known gene. Almost all of the methylated subtelomeric CpG islands were composed of short direct or indirect repeat sequences. Fifty-four of the 95 subtelomeric methylated islands underwent demethylation in the tumor. Their demethylation is consistent with a specific defect of repetitive DNA methylation in cancer tissue. The repeat-rich subtelomeric region of chromosome 8, even outside of CpG islands, was substantially hypomethylated in the tumor (example shown in SI Fig. 7). Importantly, however, nonsubtelomeric single-sequence genes and intergenic regions were not demethylated in tumors. Within 157 Mb of DNA sequence analyzed, we could detect only one unique-sequence CpG-rich sequence that was cancer-specifically demethylated. This hypomethylated sequence is located at the 3′ end of an uncharacterized gene, C8orf72 (SI Fig. 8).
A particularly interesting example is the UNC5D gene, because cancer-specific hyper- and hypomethylation events occurred in the same gene. Its promoter was hypermethylated, whereas SINE sequences downstream in the intragenic region were all hypomethylated (Fig. 3). The UNC5D gene is frequently deleted in gastric cancer (32), suggesting a possible link between SINE-specific hypomethylation and chromosomal instability leading to loss of heterozygosity in this region. However, hypermethylation of a promoter was not always associated with demethylation of multiple elements in the gene body. The promoter-methylated ZNF703 and FGF17 genes, for example, are only 3.0 and 8.5 kb in length, respectively, and contain either no or just a single copy of intragenic SINE elements.
To get a more complete picture of the DNA methylation changes in other repetitive sequences, we next extended our analysis to LINE- and HERV-containing loci. LINEs, multiple-copy SINEs, and HERV-containing regions were not adequately covered by the microarray analysis because of their size and a lack of specific probes on the arrays. Therefore, we used a modified COBRA method (33) to explore methylation changes in LINE and HERV elements. Although this approach, unlike the MIRA-assisted tilling arrays, cannot provide information on a defined region, it can give an estimate for the global changes in methylation status of these elements. We analyzed 20 normal lung tissue and matching SCC samples (SI Fig. 9). We observed strong hypomethylation of LINEs in SCC samples. HERV promoter demethylation was not as pronounced as LINE demethylation but was still significant.
Another class of repeat sequences are segmental duplications that can be several kilobases in size. Chromosome 8p23 contains an area of a direct genomic duplication (30.5 kb direct repeat) that is also found on several other chromosomes. Although it is not possible to tell which particular chromosomal segment hybridizes to the probes on the array, it is clear that these duplicated sequences underwent extensive demethylation in the tumor sample (SI Fig. 10).
A Complete Set of Hypermethylated CpG Islands and Discovery of Highly Sensitive and Specific Lung Cancer DNA Methylation Biomarkers.
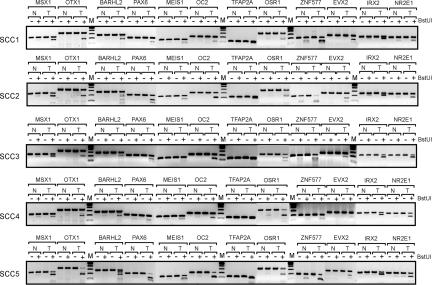
In addition to the chromosome tiling arrays, we used Agilent CpG island arrays, which cover most of the CpG islands in the human genome, to comprehensively analyze CpG island methylation. Five stage I SCCs were initially analyzed on these arrays. The number of methylated CpG islands ranged from 216 to 848 in the five individual tumors (Table 1). All methylated CpG islands are listed in SI Table 4. We identified 57 CpG islands that were methylated in five of five SCC tumors (SI Table 5). A large fraction of these methylated CpG islands were mapped to homeobox genes. The CpG island sequences and flanking 1-kb regions of the 15 most frequently methylated genes were analyzed for potential consensus DNA sequences, but we could not identify any significant consensus motifs. Because the most frequently methylated loci had excellent potential to be specific and sensitive methylation biomarkers, we analyzed 12 of these markers in a larger series of 20 SCCs (Fig. 4). The methylation frequency ranged from 14 of 20 (70%) to 20 of 20 (100%) of the tumors (Table 2). The OTX1- and NR2E1- associated CpG islands were methylated in all of the SCC tumors tested (100%). Several of these markers were highly specific for tumor-associated methylation, i.e., little or no methylation was observed in tumor-adjacent normal lung tissue. These markers included the CpG islands of the OTX1, BARHL2, MEIS1, OC2, PAX6, IRX2, TFAP2A, and EVX2 genes (Fig. 4). Most of these commonly methylated CpG islands were not methylated to a significant extent in noncancerous lung or leukocyte DNA (SI Fig. 11). The specificity and high methylation frequency of these genes make them excellent candidates for future diagnostic applications developed for early detection of lung cancer.
Table 1.
Number of methylated CpG islands in stage I lung SCC
| Sample | Methylated CpG islands |
|---|---|
| SCC1 | 632 |
| SCC2 | 248 |
| SCC3 | 848 |
| SCC4 | 743 |
| SCC5 | 216 |
Fig. 4.
Frequently methylated CpG islands in lung SCCs. Verification of CpG island DNA methylation markers in normal lung tissue and matching stage I SCC samples was conducted by bisulfite-based COBRA assays. Methylation differences between SCCs (T) and matching normal tissues (N) were analyzed by COBRA assays of the indicated gene targets. −, control digestion with no BstUI; +, BstUI-digested samples. Digestion by BstUI indicates methylation of the sequence that was tested.
Table 2.
Frequency of methylation of 12 DNA methylation biomarkers in 20 lung SCCs
| SCC No. | Stage | MSX1 | OTX1 | BARHL2 | PAX6 | MEIS1 | OC2 | TFAP2A | OSR1 | ZNF577 | EVX2 | IRX2 | NR2E1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | I | + | + | + | + | + | + | + | + | + | + | + | + |
| 2 | I | + | + | + | + | + | + | + | + | + | + | + | + |
| 3 | I | + | + | + | + | + | + | + | + | + | + | + | + |
| 4 | I | + | + | + | + | + | + | + | + | + | + | + | + |
| 5 | I | + | + | + | + | + | + | + | + | + | + | + | + |
| 6 | I | + | + | + | + | + | + | + | + | + | + | + | + |
| 7 | I | + | + | + | + | + | + | + | + | + | + | + | + |
| 8 | I | + | + | − | − | + | − | + | + | + | + | + | + |
| 9 | I | + | + | + | + | + | + | + | + | + | + | + | + |
| 10 | I | + | + | + | + | + | − | + | + | + | + | + | + |
| 11 | I | + | + | + | + | + | + | + | + | + | + | + | + |
| 12 | II | + | + | + | + | − | + | + | + | + | + | + | + |
| 13 | II | + | + | + | − | + | + | + | + | − | − | + | + |
| 14 | II | − | + | − | − | − | − | + | + | + | − | − | + |
| 15 | II | + | + | + | + | + | − | + | + | + | + | + | + |
| 16 | II | + | + | + | + | − | − | − | + | + | − | + | + |
| 17 | III | + | + | + | + | + | + | + | + | + | + | + | + |
| 18 | III | + | + | − | + | + | + | + | + | − | − | + | + |
| 19 | III | + | + | + | + | + | + | + | + | + | + | + | + |
| 20 | III | + | + | + | + | + | − | + | + | + | + | + | + |
| Frequency | 19/20 | 20/20 | 17/20 | 17/20 | 17/20 | 14/20 | 19/20 | 20/20 | 18/20 | 16/20 | 19/20 | 20/20 |
+, methylated CpG island; −, unmethylated CpG island as determined by COBRA assay.
Discussion
We have used the MIRA method in combination with genome tiling arrays for a comprehensive analysis of DNA methylation patterns in lung cancer genomes. The advantages of MIRA over antibody-based precipitation methods are its higher sensitivity (unpublished data) and high affinity to double-stranded methylated CpG targets. Its preference for binding to at least two methylated CpGs within ≈50 base pairs prevents overestimation of methylated DNA molecules. For the NimbleGen tiling arrays, hypermethylation of CpG islands could easily be detected and verified by bisulfite sequencing when 50–60% of the CpG in a particular region were methylated (Figs. 2 and 3). Given the intensity of the peaks in the tumors, it is likely that much lower methylation frequencies (20%) can readily be detected, but we have not found such areas. For hypomethylation on tiling arrays, we can easily pick up demethylation of 60–90% of a region's CpG sites (examples in Figs. 2, 3, and SI Fig. 8). The peak intensity differences suggest that we should be able to detect more moderate differences, probably ≥30% hypomethylation, but it is unlikely that the method would readily detect small amounts of hypomethylation, i.e., 5–25%, because such a region still is highly methylated and will be pulled down in the MIRA procedure. For the Agilent CpG island arrays, we were able to generally confirm hypermethylated CpG islands in tumors by bisulfite-based approaches when the fold-difference factor was >2 for multiple closely spaced probes. We also confirmed known SCC hypermethylated CpG islands such as CDKN2 and TCF21. Some methylated genes, however, may go undetected with this approach because the associated MseI fragments may be too large and may fail to amplify.
One of the most significant findings of this study is that cancer-specifically demethylated chromosomal regions were almost exclusively mapped to repetitive DNA-containing sequences (LINE, SINE, LTR elements, and segmental duplications) and subtelomeric regions. Subtelomeric DNA is composed of subtelomeric repeat sequences and segmental duplications and is densely methylated (34, 35). The terminal 2 Mb of chromosome 8p were substantially hypomethylated in the tumors. However, our analysis of chromosomes 7 and 8 suggests that demethylation of single copy, nonsubtelomeric DNA sequences is a very rare event. Where demethylation of single copy genes does occur, it may be mechanistically connected to demethylation of nearby repetitive DNA (although this formally remains to be tested). The limitations of the study are the number of samples and chromosomes analyzed. For example, there may be other single copy genes subject to hypomethylation in tumors, such as the MAGE genes (located on chromosomes 15 and X) (36) and the maspin gene (located on chromosome 18) (37, 38).
The specificity of hypomethylation for repetitive elements is consistent with a defect of a molecular mechanism that maintains methylation of repetitive DNA. The nature of this defect is currently unknown. One possibility is that repetitive DNA is actively demethylated in cancer cells, perhaps through reactivation of a DNA demethylase gene that normally is expressed only in early development. To date, the nature of the mammalian DNA demethylase has remained obscure (39). Alternatively, the maintenance process of methylation of repetitive DNA may be defective in cancer cells. The DNA methyltransferase DNMT1 is responsible for the reliable copying of existing DNA methylation patterns, but evidence for a role of altered DNMT1 function in cancer-associated DNA hypomethylation is not available. DNMT3A and DNMT3B are de novo DNA methyltransferases. DNMT3L, devoid of methyltransferase activity by itself, is capable of stimulating the activity of DNMT3A and DNMT3B (40). DNMT3L was shown to positively regulate DNA methylation at imprinted sequences and at repeat sequences in mouse germ cells (41). However, the role of these proteins in maintaining methylation of repeat sequences in somatic cells is not clear. Instead of invoking a defect in DNA methyltransferases themselves, another possibility is that their access to repetitive DNA in cancer cells may be impeded. Defects in two chromatin-associated DNA helicases, LSH and ATRX, have been associated with DNA hypomethylation in gene-targeted mice (42, 43). Although it was initially thought that LSH is required for total genomic DNA methylation (43), this defect may be most important for repetitive DNA (44). Deficiencies in either ATRX or LSH (also known as PASG, SMARCA6, or HELLS) in human tumors have been reported only rarely (45, 46). A double-stranded RNA-based mechanism that guides methylation of repetitive DNA through heterochromatin formation is also a formal possibility that warrants further investigation.
The second important aspect of this paper is the comprehensive analysis of the CpG islands in human lung cancer by using microarrays. We were able to measure the methylation levels at >27,000 CpG islands and found that between 216 and 848 of these islands were methylated in our lung SCC samples. These numbers are compatible with earlier estimates derived from analysis of a subset of CpG islands methylated in cancer (47). We found that CpG islands with different CpG densities can become hypermethylated in tumors (SI Table 3). It is clear that not all of these methylated genes can be tumor suppressor genes. For example, consistent with earlier observations, a substantial subset of the methylated genes (20–40%, depending on the tumor) were homeobox genes (48). Homeobox gene-associated CpG islands were among the best stage I disease DNA methylation markers identified in this study. We found that the CpG islands of the OTX1, PAX6, IRX2, OC2, TFAP2A, and EVX2 genes are tumor-specifically methylated with very little methylation found in normal lung tissue or in blood DNA. Methylation of the OTX1, IRX2, OC2, and EVX2 genes has not yet been reported in human cancers. Also, importantly, the methylation frequency of these markers (80–100% of the tumors were methylated for 11 of 12 markers tested; 70% for OC2; Table 2) is much higher than methylation frequencies of other lung cancer DNA methylation markers reported previously. For example, we find that OTX1 was tumor-specifically methylated in 20 of 20 (100%) of the tumors. Such markers are excellent candidates for clinical or diagnostic applications aimed at either detection of early disease in body fluids such as blood or sputum or at disease management and follow-up by using molecular diagnostic testing.
Materials and Methods
Genomic DNA Preparation.
Lung SCC samples and matching normal tissues removed with surgery were obtained from the frozen tumor bank of the City of Hope National Medical Center. Genomic DNA was purified from tissues by standard procedures by using phenol chloroform extraction and ethanol precipitation.
MIRA-Assisted Tiling Array Analysis.
MIRA was performed as described in ref. 48. NimbleGen whole-chromosomal tiling arrays covering the entire short arms of human chromosome 7 (HG18Tiling_Set17) and 8 (HG18Tiling_Set19) were used in the DNA methylation profile analysis. MIRA-enriched DNA fractions were compared with input DNA. The labeling of dsDNA, microarray hybridization, and scanning were performed by the NimbleGen Service Group (Reykjavik, Iceland). Data were extracted from scanned images by using NimbleScan 2.3 extraction software (NimbleGen Systems). Human CpG island microarrays, which contain 237,000 oligonucleotide probes covering 27,800 CpG islands, were purchased from Agilent Technologies. Two micrograms each of the amplicons from MIRA-enriched tumor DNA and normal control samples were labeled with a BioPrime Array CGH Genomic Labeling kit (Invitrogen) with either Cy5-dCTP (tumor) or Cy3-dCTP (control) in 87.5-μl reactions (both Cy3- and Cy5-dCTP were obtained from GE Healthcare). The purified labeled samples were then mixed, and microarray hybridization was performed according to the Agilent ChIP-on-chip protocol (v.9.0). The hybridized arrays were scanned on an Axon 4000B microarray scanner, and the images were analyzed with Axon GenePix software v.5.1. Image and data analyses were performed as described in ref. 29. Individual CpG islands were considered methylation-positive when at least two adjacent probes allowing a one-probe gap within the CpG island scored a fold-difference factor of >2.0 when comparing tumor and normal tissue DNA.
DNA Methylation Analysis Using COBRA and Bisulfite Sequencing.
The COBRA assays were performed according to the method of Xiong and Laird (49) by using digestion with BstUI for analysis of single copy genes, HinfI for analysis of LINE elements, or TaqI for analysis of HERV sequences. DNA was treated and purified with the EpiTect bisulfite kit (Qiagen). The PCR primers used to amplify HERV sequences after bisulfite conversion were 5′-TTTATAGGTGTGTAGGGGTAATTTATTTT and 5′-AATAAAAAACATATTTACTTTTAATTTTAC. LINE elements were analyzed by using consensus primers as described by Yang et al. (33). Other PCR primers used for amplification of specific targets in bisulfite-treated DNA are available upon request. For sequence analysis, the PCR products obtained after bisulfite conversion were cloned into the pDrive PCR cloning vector (Qiagen), and five individual clones were sequenced.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by National Institutes of Health Grants CA104967 and CA128495 (to G.P.P.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE9622).
This article contains supporting information online at www.pnas.org/cgi/content/full/0710735105/DC1.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lapeyre JN, Becker FF. 5-Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun. 1979;87:698–705. doi: 10.1016/0006-291x(79)92015-1. [DOI] [PubMed] [Google Scholar]
- 3.Romanov GA, Vanyushin BF. Methylation of reiterated sequences in mammalian DNAs. Effects of the tissue type, age, malignancy and hormonal induction. Biochim Biophys Acta. 1981;653:204–218. doi: 10.1016/0005-2787(81)90156-8. [DOI] [PubMed] [Google Scholar]
- 4.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 5.Gama-Sosa MA, et al. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983;740:212–219. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- 6.Riggs AD, Jones PA. 5-Methylcytosine, gene regulation, and cancer. Adv Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- 7.Gaudet F, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Zulueta M, et al. Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 9.Herman JG, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 10.Merlo A, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 11.Kane MF, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 12.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 13.Dammann R, et al. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 14.Zochbauer-Muller S, et al. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- 15.Yanagawa N, et al. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci. 2003;94:589–592. doi: 10.1111/j.1349-7006.2003.tb01487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Topaloglu O, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–2288. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- 17.Dammann R, et al. CpG island methylation and expression of tumour-associated genes in lung carcinoma. Eur J Cancer. 2005;41:1223–1236. doi: 10.1016/j.ejca.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 19.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 20.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 21.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 22.Weisenberger DJ, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cadieux B, Ching TT, Vandenberg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez J, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 25.Estecio MR, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS ONE. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 28.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 29.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 30.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 31.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 32.Koed K, et al. High-density single nucleotide polymorphism array defines novel stage and location-dependent allelic imbalances in human bladder tumors. Cancer Res. 2005;65:34–45. [PubMed] [Google Scholar]
- 33.Yang AS, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brock GJ, Charlton J, Bird A. Densely methylated sequences that are preferentially localized at telomere-proximal regions of human chromosomes. Gene. 1999;240:269–277. doi: 10.1016/s0378-1119(99)00442-4. [DOI] [PubMed] [Google Scholar]
- 35.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 36.De Smet C, Loriot A, Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5′ region of gene MAGE-A1 in tumor cells. Mol Cell Biol. 2004;24:4781–4790. doi: 10.1128/MCB.24.11.4781-4790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogasawara S, et al. Disruption of cell-type-specific methylation at the Maspin gene promoter is frequently involved in undifferentiated thyroid cancers. Oncogene. 2004;23:1117–1124. doi: 10.1038/sj.onc.1207211. [DOI] [PubMed] [Google Scholar]
- 38.Sato N, Fukushima N, Matsubayashi H, Goggins M. Identification of maspin and S100P as novel hypomethylation targets in pancreatic cancer using global gene expression profiling. Oncogene. 2004;23:1531–1538. doi: 10.1038/sj.onc.1207269. [DOI] [PubMed] [Google Scholar]
- 39.Chen ZX, Riggs AD. Maintenance and regulation of DNA methylation patterns in mammals. Biochem Cell Biol. 2005;83:438–448. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- 40.Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chedin F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J Cell Biochem. 2005;95:902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- 41.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 42.Gibbons RJ, et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 43.Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, et al. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res. 2004;32:5019–5028. doi: 10.1093/nar/gkh821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee DW, et al. Proliferation-associated SNF2-like gene (PASG): a SNF2 family member altered in leukemia. Cancer Res. 2000;60:3612–3622. [PubMed] [Google Scholar]
- 46.Yano M, et al. Tumor-specific exon creation of the HELLS/SMARCA6 gene in non-small cell lung cancer. Int J Cancer. 2004;112:8–13. doi: 10.1002/ijc.20407. [DOI] [PubMed] [Google Scholar]
- 47.Costello JF, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 48.Rauch T, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci USA. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.