Abstract
Toll-like receptors (TLRs) initiate immune responses by recognizing pathogen-associated molecules, but the molecular basis for recognition is poorly understood. In particular, it is unclear how receptor-ligand interactions lead to the initiation of downstream signaling. Here, we describe the mechanism by which TLR3 recognizes its ligand, double-stranded RNA (dsRNA), and forms an active signaling complex. We show that dsRNA binds saturably, specifically, and reversibly to a defined ligand-binding site (or sites) on the TLR3 ectodomain (TLR3ecd). Binding affinities increase with both buffer acidity and ligand size. Purified TLR3ecd protein is exclusively monomeric in solution, but through a highly cooperative process, it forms dimers when bound to dsRNA, and multiple TLR3ecd dimers bind to long dsRNA strands. The smallest dsRNA oligonucleotides that form stable complexes with TLR3ecd (40–50 bp) each bind one TLR3ecd dimer, and these are also the smallest oligonucleotides that efficiently activate TLR3 in cells. We conclude that TLR3 assembles on dsRNA as stable dimers and that the minimal signaling unit is one TLR3 dimer.
Keywords: dsRNA, innate immunity, Toll-like receptors
The recognition of pathogen-derived substances by Toll-like receptors (TLRs) is crucial for the initiation of innate immune responses against bacterial, viral, and parasitic pathogens (1). However, the molecular mechanism by which recognition occurs and leads to signal induction is poorly understood. TLRs recognize pathogens via ectodomains (ecds) comprising 18–25 tandem copies of the leucine rich repeat motif, which adopt a configuration that resembles a long solenoid bent into the shape of a horseshoe (2). Although direct binding of pathogen-derived ligands to some TLRecds has been reported (3–10), it is not known how binding occurs, or indeed whether TLRs contain specific saturable binding sites. This is an important question, because a detailed knowledge of TLR-ligand interactions could greatly aid in the development of anti-inflammatory drugs (TLR antagonists) and adjuvants for vaccines (TLR agonists).
To date, the known TLRecd structures are those of TLR3ecd (9, 11) and the recently reported complexes of TLR4ecd-MD-2 (12) and a heterodimer of TLR1ecd-TLR2ecd bound to a triacylated lipopeptide (13). TLR3ecd recognizes double-stranded RNA (dsRNA), a molecular signature of viral pathogens (14, 15), and is usually located in the lumens of endosomes, although in some cells it is expressed on the plasma membrane (16–20). TLR3 responds to dsRNA by recruiting its sole cytosolic adaptor molecule, Toll/IL-1 receptor (TIR) domain-containing adaptor inducing interferon-β (TRIF), which initiates downstream signaling (21, 22). Subsequently, the transcription factors IFN regulatory factor (IRF)-3 and NF-κB are activated and induce the secretion of IFN-β and other inflammatory cytokines to trigger antiviral responses in the infected host (23). No crystal structure of TLR3ecd complexed with dsRNA is currently available, but preliminary studies indicate that dsRNA binds directly to the TLR3ecd and that binding requires acidic conditions (9, 10, 24).
We have previously shown that His 539 and Asn 541 on the glycan-free lateral face of TLR3 are essential for ligand binding and receptor function (25), which suggests that that a dsRNA molecule might bridge multiple TLR3ecds by binding to their lateral faces. Here, we test this hypothesis by determining how TLR3 binds dsRNA to form complexes and induce signaling in cells.
Results
Binding of dsRNA to TLR3 Is Saturable and Specific.
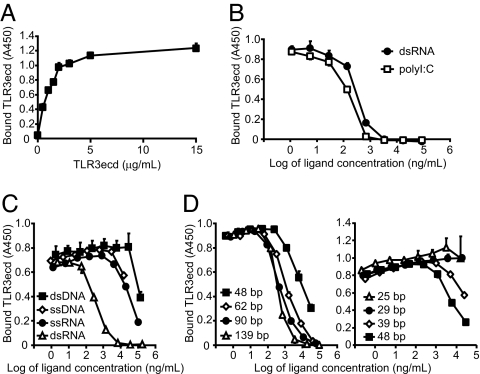
We first asked whether dsRNA binds to a defined site on TLR3ecd. As shown in Fig. 1 A and B, the binding of TLR3ecd to immobilized dsRNA reached a plateau at high TLR3ecd concentrations and was inhibited by free, soluble dsRNA or the synthetic dsRNA analogue, polyinosinic acid:polycytidylic acid (polyI:C). These results indicate that solution-phase dsRNA and polyI:C compete with immobilized dsRNA for a defined ligand-binding site (or sites) on TLR3ecd. Moreover, the observation that polyI:C and dsRNA inhibit TLR3ecd binding at similar concentrations indicates that TLR3 binding is independent of nucleotide sequence. Interestingly, soluble ssDNA, dsDNA, and ssRNA also inhibited TLR3ecd binding to some extent, even though these ligands do not activate TLR3 in cells [see below and Alexopoulou et al. (14)]. However, 100- to 1,000-fold higher concentrations of these oligonucleotides were required to inhibit as effectively as dsRNA (Fig. 1C). Therefore, dsRNA binds to a defined site on TLR3 by a specific and saturable mechanism.
Fig. 1.
TLR3 binding to dsRNA is saturable, specific, and a function of dsRNA size. Capture (A) and competition (B–D) ELISAs were performed by using immobilized 540-bp dsRNA at pH 6.0. (A) TLR3ecd binds saturably to dsRNA. (B) Soluble polyI:C and dsRNA (139 bp) inhibit the binding of TLR3ecd (1 μg/ml) to immobilized dsRNA at comparable concentrations. (C) dsDNA (532 bp), ssDNA (90 bp), and ssRNA (633 bp) inhibit binding much less effectively than dsRNA (540 bp). (D) Inhibition of TLR3ecd binding to immobilized dsRNA depends upon dsRNA length. Experiments were performed in triplicate, and error bars indicate one standard deviation.
Binding Depends on dsRNA Length and pH.
Naturally occuring dsRNA molecules may range in size from several thousand basepairs, which is typical for viral genomes, to only 20–22 bp, which is the case for siRNAs produced by the digestion of viral dsRNA by Dicer (26). It was therefore of interest to determine the minimum ligand size required for TLR3 binding. Accordingly, we performed competition ELISAs, using soluble dsRNA oligonucleotides of differing lengths (Fig. 1D). Although the smallest dsRNA ligand that inhibited TLR3ecd binding to immobilized dsRNA was ≈40 bp, inhibition increased with soluble ligand size. All dsRNAs >62 bp in length inhibited binding to immobilized dsRNA at similar concentrations.
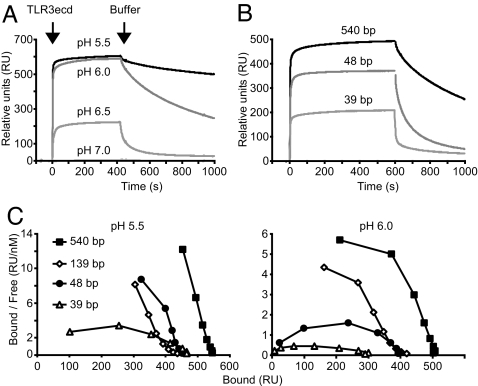
Because TLR3 is found in intracellular compartments in which the pH varies from ≈6.0 (early endosomes) to ≈5.5 or below (late endosomes) (27), we asked whether ligand binding is pH-dependent over this range. For these studies, TLR3ecd binding to immobilized dsRNA was quantified in real time by surface plasmon resonance (SPR). No binding to 540-bp dsRNA occurred at pH 7.0, but at pH 6.5 or below, the apparent affinity increased dramatically with buffer acidity, as indicated by both the larger amount of TLR3ecd bound at equilibrium and by slower dissociation (Fig. 2A). In addition, bound TLR3ecd immediately and completely dissociated from the dsRNA upon switching to buffer at pH 7.5 (data not shown), further indicating that the interaction of TLR3ecd with dsRNA is reversible. These data indicate that TLR3 binds dsRNA at pH values that correspond to those of acidic endosomes.
Fig. 2.
Binding affinity is a function of pH and dsRNA size. (A and B) TLR3ecd binding to immobilized dsRNA as determined by SPR. (A) Binding of TLR3ecd (40 μg/ml) to dsRNA (540 bp) increases with acidity. Arrows indicate the points at which TLR3ecd injection began and ended with the change back to buffer alone. (B) Affinity of TLR3ecd (40 μg/ml) binding at pH 6.0 increases with the length of the immobilized dsRNA. (C) The amount of TLR3ecd bound to dsRNA at equilibrium was determined by SPR, at several concentrations of soluble (free) TLR3ecd, and data were plotted for Scatchard analysis. Downward curvature suggests positive cooperativity.
To characterize the combined effects of pH and ligand size, dsRNA oligonucleotides of increasing length were immobilized, and TLR3ecd binding was analyzed by SPR. As competition ELISA data suggested (Fig. 1D), binding affinity increased with the length of immobilized dsRNA (Fig. 2B). We calculated an apparent equilibrium dissociation constant (Kd_apparent) for each dsRNA ligand and pH by fitting a set of SPR data to a single-step binding model (Table 1). High affinity binding (Kd_apparent < 100 nM) was observed at pH 5.5 for all oligonucleotides tested, but only the larger ligands bound with high affinity at pH 6.0. Changes in affinity between 39- and 48-bp ligands were especially pronounced, whereas additional increases in affinity with still larger ligands were more moderate. Finally, we looked for evidence of cooperativity in TLR3 binding by subjecting the SPR data to Scatchard analysis (Fig. 2C). Importantly, the prominent concave-down curvature of these plots provides evidence for strong positive cooperativity, indicating that the affinity increases with the number of TLR3ecds bound.
Table 1.
Affinity of TLR3ecd binding to dsRNA
| dsRNA length, bp |
Kd_apparent, nM |
||
|---|---|---|---|
| pH 6.5 | pH 6.0 | pH 5.5 | |
| 39 | 2,250 ± 70 | 510 ± 23 | 72.6 ± 1.7 |
| 48 | 2,610 ± 80 | 156 ± 7 | 13.6 ± 0.3 |
| 139 | 487 ± 12 | 31.7 ± 0.6 | 9.19 ± 0.21 |
| 540 | 782 ± 15 | 28.2 ± 0.9 | 4.64 ± 0.11 |
Apparent equilibrium dissociation constants (Kd_apparent) were calculated for each dsRNA size and pH by fitting kinetic data from SPR experiments to a simple single-step binding model (A+B ↔ AB). See SI Table 2 for a similar analysis using a cooperative binding model. The standard error associated with each calculation is indicated.
Pairs of TLR3 Bind dsRNA to Form Stable Complexes.
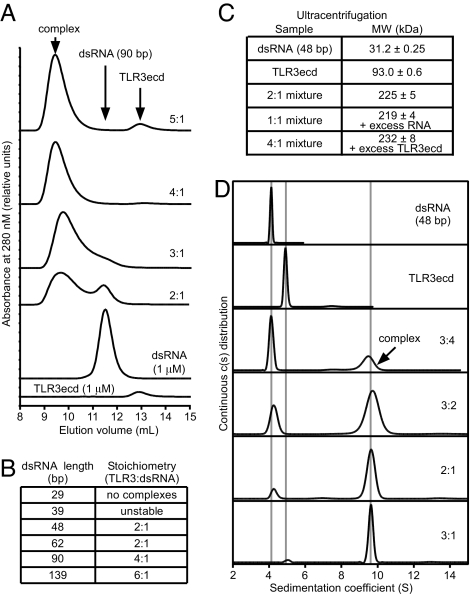
To identify the mechanism that generates cooperative binding, we next characterized the complexes formed by TLR3ecds binding to dsRNA. First, we used sedimentation equilibrium analysis to establish that ligand-free TLR3ecd is monomeric and monodisperse in solution [supporting information (SI) Appendix 1], which corroborates other reports (28, 29). The measured mass of TLR3ecd (93.0 ± 0.6 kDa), is consistent with the molecular mass previously determined by mass spectrometry (95.5 kDa) (9). To characterize receptor-ligand complexes, TLR3ecd and dsRNA were mixed and analyzed by gel filtration (Fig. 3A). By varying the molar ratio of TLR3ecd to dsRNA, we determined the maximum number of TLR3ecd molecules that can bind to a given dsRNA, which defines the stoichiometry of the complex. The smallest stable complex observed by gel filtration was a 2:1 TLR3ecd:dsRNA complex with 48-bp dsRNA. The stoichiometry increased with dsRNA length, but most strikingly, only even-numbered TLR3ecd:dsRNA stoichiometries were observed (Fig. 3B), which suggests that TLR3ecd binds to dsRNA as receptor pairs. We confirmed the composition of the 48-bp complex by sedimentation equilibrium analysis (Fig. 3C). A 2:1 mixture of TLR3ecd and dsRNA sedimented as a single 219-kDa species, which is consistent with a complex of two TLR3ecds and one 48-bp dsRNA. In addition, mixtures containing TLR3ecd:dsRNA ratios more or less than 2:1 were best fit assuming two species, one of ≈220–230 kDa and the other corresponding to the excess TLR3ecd or dsRNA, respectively. Finally, sedimentation velocity analysis confirmed that TLR3ecd and the 48-bp dsRNA form a single complex that saturates at a ratio of 2:1 (Fig. 3D). Importantly, there was very limited evidence for the formation of a 1:1 TLR3ecd:dsRNA complex. Taken together, our data indicate that monomeric TLR3ecd molecules bind to dsRNA and form stable pairs, that multiple pairs can bind to a single dsRNA, and that each TLR3ecd dimer requires ≈40–50 bp of ligand.
Fig. 3.
Composition of TLR3-dsRNA complexes. (A) The stoichiometry of complexes formed by TLR3ecd and 90-bp dsRNA was measured by titrating TLR3ecd against a fixed dsRNA concentration (1 μM) and analyzing the mixture by gel filtration (GF) at pH 6.0. For this ligand, the stoichiometry is 4:1 (TLR3ecd:dsRNA), because the only peak observed at this ratio corresponds to the complex, and, at higher and lower ratios, unbound TLR3ecd and dsRNA were apparent. (B) The stoichiometries of complexes formed by dsRNA of different lengths as determined by GF. Ligands smaller than 48 bp did not form stable complexes (SI Fig. 6). Sedimentation equilibrium (C) and sedimentation velocity analyses (D) of TLR3ecd and dsRNA (48 bp) mixtures at pH 5.5. At a 2:1 loading ratio, a single species is observed with the molecular mass expected for a complex that consists of two TLR3ecd and one dsRNA molecule (SI Appendix 1). Standard errors are indicated in C. Note that in A and D, peak heights reflect the fact that TLR3ecd's coefficient of absorption is less than that of dsRNA. Mixture compositions in A, C, and D are reported as the molar ratio of TLR3ecd:dsRNA. The predicted molecular mass of 48-bp dsRNA is 31.1 kDa.
Receptor Cross-Linking Activates TLR3.
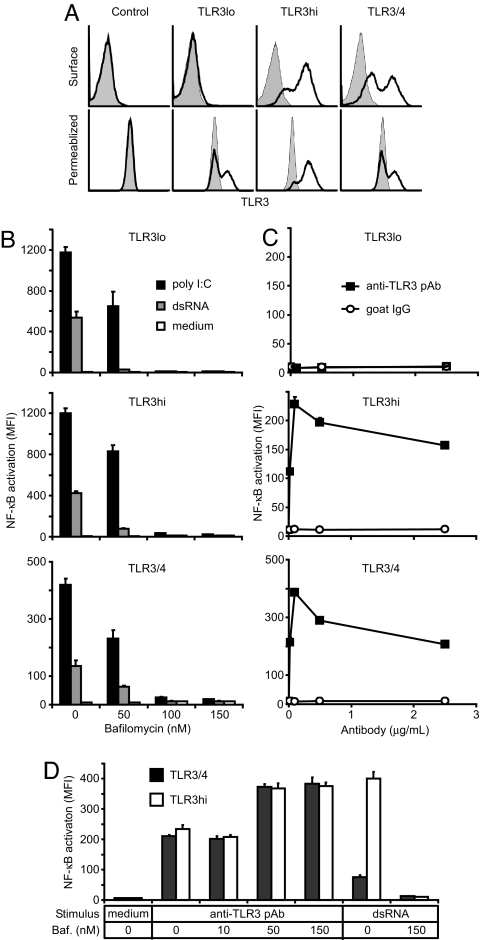
To test whether receptor cross-linking activates TLR3, we created a stable HEK293 reporter cell line that expresses TLR3 exclusively in intracellular compartments (TLR3lo), a line that also expresses TLR3 on the cell surface (TLR3hi), and a line expressing a chimera of TLR3ecd fused to TLR4 transmembrane and cytoplasmic domains (TLR3/4), some of which is on the cell surface (Fig. 4A). All three lines responded to dsRNA and polyI:C, but not to dsDNA, ssDNA, ssRNA, LPS (TLR4 ligand), R837 (TLR7 ligand) or CpG-D35 and CpG-2395 (two TLR9 ligands) (Fig. 4B and SI Fig. 7). Furthermore, the TLR3-deficient “control” cells, from which these lines were derived, did not respond to dsRNA (SI Fig. 7). Activation of the TLR3lo, TLR3hi, and TLR3/4 cells by dsRNA was inhibited by bafilomycin, a drug that specifically blocks endosome acidification, which indicates that ligand-mediated activation occurred entirely in acidic endosomes (Fig. 4B). Furthermore, even though TLR3/4 efficiently traffics to the cell surface and signals via the TLR4 intracellular domain, activation by dsRNA still requires that the TLR3ecd encounters its ligand in acidic endosomes. Polyclonal anti-TLR3 antibodies also stimulated cell lines that expressed TLR3 or TLR3/4 on the surface (Fig. 4C), presumably by cross-linking the receptors at the cell surface, and this stimulation was insensitive to bafilomycin treatment (Fig. 4D). Moreover, when the extracellular environment was adjusted to an acidic pH to promote dsRNA binding at the cell surface, the activation of surface TLR3-expressing lines (TLR3hi and TLR3/4) by polyI:C was not inhibited by bafilomycin (SI Fig. 8), indicating that ligand-mediated cross-linking at the cell surface can also activate TLR3 and TLR3/4. These results demonstrate that cross-linking per se activates TLR3, even in the absence of ligand, although cross-linking by dsRNA normally occurs only in acidic environments, such as endosomes, in agreement with the observed pH dependence of binding (Fig. 2A).
Fig. 4.
Receptor cross-linking activates TLR3. (A) Intracellular and surface expression of TLR3 in stable reporter cell lines, as assessed by FACS; shaded histograms are isotype controls. (B) TLR3-mediated activation of NF-κB in reporter cells by dsRNA (139 bp, 5 μg/ml) or polyI:C (1 μg/ml) and inhibition by bafilomycin. (C) TLR3 activation after cross-linking at the cell surface with anti-TLR3 polyclonal antibodies (pAb). (D) Cells preincubated in bafilomycin (baf) were activated by anti-TLR3 pAb (0.1 μg/ml) but not by dsRNA (as in B). Experiments were performed in triplicate, and error bars indicate one standard deviation.
TLR3 Dimer Formation Is the Minimal Signal for Activation.
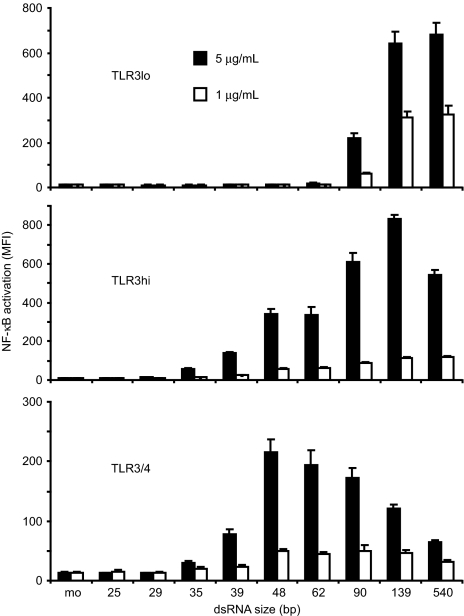
Finally, we treated the TLR3lo, TLR3hi, and TLR3/4 lines with dsRNA oligonucleotides of different lengths to determine the size of the smallest receptor-ligand complex that is capable of inducing signaling. As seen in Fig. 5, the TLR3lo cells required dsRNA of 90 bp or larger for activation, whereas TLR3hi and TLR3/4 cells responded robustly to 48-bp dsRNA and weakly to 39-bp dsRNA. Because the shortest dsRNA oligonucleotide that potently activated cells (48 bp) forms stable dimers, but not larger complexes, with TLR3ecd, we conclude that the minimal TLR3 signaling complex is a receptor dimer.
Fig. 5.
Activation of TLR3 depends on dsRNA size and receptor localization. Cells were stimulated with dsRNA ligands of varying length. mo, medium only. Experiments were performed in triplicate, and error bars indicate one standard deviation.
Discussion
In this study, we provide a detailed description of the dynamic interaction between a pathogen-derived substance (dsRNA) and a TLR (TLR3), which leads to the formation of complexes that initiate downstream signaling. We establish that the binding of dsRNA to TLR3 is saturable, specific, and reversible with an affinity that depends on both pH and dsRNA length (Figs. 1 and 2 and Table 1). Furthermore, we show that TLR3ecd monomers bind cooperatively to dsRNA to form stable dimeric complexes and that multiple dimers can bind to long dsRNA oligonucleotides (Figs. 2 and 3). Finally, we demonstrate that receptor cross-linking per se activates TLR3 (Fig. 4) and that the formation of a complex containing one dsRNA oligonucleotide and one TLR3 dimer is sufficient to initiate signaling (Fig. 5).
Previously, we proposed a model in which dsRNA phosphate groups bind to the protonated form of His-539 on the glycan-free surface of TLR3ecd, and in which the symmetry of the ligand allows two TLR3ecds to bind to opposite sides of a short stretch of dsRNA (25). This model explains the observed dependence of binding affinity on pH, since His residues are normally protonated only below pH 7 (pKa ≈ 6.0–6.5) (30), but it fails to predict the observed positive cooperativity of binding or explain the minimal dsRNA length required for dimer formation (40–50 bp or 100–130 Å).
The findings presented here suggest a revised model, in which two TLR3ecds still bind to opposite sides of a single dsRNA molecule but where the dimeric complex is stabilized by intermolecular interactions between previously unidentified homotypic interaction sites on the TLR3ecds. This homotypic interaction is of sufficiently low affinity that TLR3ecds do not efficiently dimerize without the additional stabilization conferred by dsRNA-binding. Similarly, dsRNA binding is weak without stabilization by homotypic interactions between the TLR3ecds. The homotypic interaction would also explain why binding is cooperative, because one TLR3ecd would bind weakly to dsRNA, but a pair of TLR3ecds would bind with high affinity. Our data also suggest that additional stabilization might be conferred by interactions between pairs of TLR3ecds on a long dsRNA molecule, because the binding affinity increased with dsRNA length beyond the 40–50 bp required for dimer formation (Table 1). One important consequence of cooperative binding is that mutations that directly disrupt dsRNA binding cannot easily be distinguished from those that disrupt the homotypic interactions required to form TLR3 dimers that stably bind dsRNA, and this may impact the interpretation of previous mutational studies (10, 25, 31). Presumably, the stable dimeric conformation places the outermost points at which each TLR3ecd in a pair contacts the dsRNA ≈100–130 Å apart along the oligonucleotide and juxtaposes the two TLR3 cytoplasmic domains such that they recruit TRIF to induce signaling. Because antibody-mediated cross-linking activated TLR3 even in the absence of ligand (Fig. 4B), receptor juxtaposition alone may suffice to recruit TRIF.
The smallest dsRNA ligands that activated TLR3 in cells were 39–48 bp long, but, interestingly, these ligands failed to activate the TLR3lo cells, in which TLR3 was exclusively intracellular. However, these cells responded to dsRNA ligands of 90 bp or longer. To explain this observation, we hypothesize that dsRNA encounters TLR3 exclusively in early endosomes in the TLR3lo line, whereas in the TLR3hi and TLR3/4 lines, some encounters occur in late endosomes. This could occur, for example, if some TLR3 and TLR3/4 at the plasma membrane traffics to late endosomes, as does dsRNA (20). The pH of early endosomes (≈6.0–6.5) (27) is conducive to TLR3 complex formation with dsRNA ligands ≥90 bp long (Fig. 3A), but not with a 48-bp ligand. However, the pH of late endosomes (≈5.5 or below) (27) is conducive to stable dimeric complex formation with 48-bp dsRNA (Fig. 6). According to this model, the activating signal in all cell lines is a TLR3 dimer, but, in early endosomes, a longer stretch of dsRNA is required to form these dimers.
It is not yet clear whether other TLRs form signaling complexes similar to that of TLR3. The recently described structure of a TLR1-TLR2 heterodimer bound to tri-acylated lipopeptide involves both TLR-ligand contacts and interactions between the receptors (13), which is similar to our model of TLR3 binding to dsRNA. However, it remains to be seen whether the identified TLR1–TLR2 contacts mediate cooperative binding as we observed with TLR3. Another recent study reports that TLR9, which recognizes unmethylated bacterial CpG DNA and localizes intracellularly, exists as a preformed dimer that subsequently changes conformation upon ligand binding (32). Although TLR3 does not exist as a preformed dimer, our model neither requires nor excludes a conformational change in TLR3 upon ligand binding. Also, like TLR3, TLR9 is regulated by endosomal trafficking of ligands (33) and receptor localization (34). The homolog of vertebrate TLRs, Drosophila Toll, interacts with its ligand, Spätzle, by a mechanism that differs markedly from that of TLR3. Ligand-free Toll exists in equilibrium between monomers and inactive dimers, and Toll's N-terminal leucine-rich repeats prevent the formation of an active signaling complex until Spätzle binding alleviates this inhibition (35). Moreover, the binding of Spätzle exhibits negative cooperativity, which may provide a mechanism for tuning the amount of signaling through Toll over a large dynamic range of Spätzle concentrations. In contrast, the positive cooperativity required for TLR3 binding causes this receptor to function as a switch, preventing signaling in the absence of ligand while ensuring that low concentrations of dsRNA still generate robust signaling. Because TLR ligands vary widely in size and symmetry, the signaling complexes might reflect these differences, but it is possible that other TLRs are also activated by a similar mechanism of dimer formation.
Materials and Methods
Protein and Nucleic Acid Synthesis and Purification.
Recombinant human TLR3ecd protein, containing C-terminal FLAG and His6 affinity tags, was described in ref. 9. dsRNA oligonucleotides of defined lengths were synthesized enzymatically, using the T7 Ribomax kit (Promega) as described (SI Appendix 2) and purified by gel filtration on a Superdex 200 10/300 GL column (GE Healthcare), followed by ethanol-precipitation. All dsRNA sequences were based on an arbitrarily selected portion of the West Nile virus (WNV) envelope gene derived from pFastBac-CPrME (36). polyI:C was made by annealing a mixture of polyinosinic and polycytidylic acids (GE Healthcare) in PBS. dsRNA was end-labeled with biotin by treatment with Antarctic Phosphatase (NEB), incorporation of thiophosphate groups at the 5′ ends using adenosine 5′-(gamma-thio) triphosphate (Sigma) and polynucleotide kinase (NEB), and reaction with maleimide-PEO2-biotin (Pierce; 4 mg per 100 μg of dsRNA) per the manufacturers' instructions. Biotinylated ligands were repurified by gel filtration.
TLR3 Binding ELISAs.
Reacti-Bind Streptavidin HBC strip-wells (Pierce) were coated overnight at 4°C with biotinylated 540-bp dsRNA at 1 μg/ml in Pipes-buffered saline (PiBS) [20 mM Pipes (Sigma) and 150 mM NaCl] at pH 6.0. For all ELISAs, binding and wash steps occurred at pH 6.0 and room temperature. In each assay, wells were washed [three rinses with PiBS and 0.1% Tween 20 (Sigma)], incubated with TLR3ecd in PiBS for 2 h, washed, labeled 1 h with anti-FLAG M2-HRP mAb (Sigma) diluted 1:8,000 in PiBS and 0.1% BSA, and washed. Wells were developed with HRP Substrate Reagent (R&D Systems) and halted with 1M H2SO4, and absorbance at 450 nm was read. In competition ELISAs, soluble ligands were added to wells immediately before the TLR3ecd (1 μg/ml).
SPR.
SPR experiments were performed with a BIAcore 2000 instrument (BIAcore/GE Healthcare). Streptavidin-conjugated Sensor Chip SA integrated flow cells (BIAcore/GE Healthcare) were preconditioned with 1 M NaCl and 50 mM NaOH, and flow cells were coated one at a time with 10 nM dsRNA ligand in PiBS (pH 6.0) until a mass of 80–100 relative units was deposited. Excess streptavidin was blocked with 20 μM biotin (NEB), and one flow cell per chip was coated with biotin alone to serve as a blank. All binding experiments were run at 25°C in PiBS at the indicated pH, with a flow rate of 20 μl per minute. Flow cells were regenerated with PiBS at pH 7.5. To assess binding constants, serial dilutions of soluble TLR3ecd in pH-adjusted PiBS (from 4.8 μM to 37 nM), were injected over SPR chip surfaces coated with 39-, 48-, 139-, or 540-bp dsRNA. Background was subtracted from each run using the blank to correct for shifts in bulk refractive index. Apparent equilibrium dissociation constants (Kd_apparent) were calculated for each dsRNA size and pH by using BIAevalulation software, Version 4.1 (BIAcore/GE Healthcare) to fit kinetic data to a simple single-step binding model (A+B ↔ AB). Because long dsRNA molecules are polyvalent ligands and TLR3ecd bound to dsRNA cooperatively, binding was described in terms of “apparent affinity” to indicate that multiple molecular interactions are probably involved and that the single-step binding model provides only an effective value for measured binding constants.
Analytical Gel Filtration.
A Superdex 200 10/300 GL column (GE Healthcare) was run with an Akta liquid chromatography system (GE Healthcare). For analyzing complex formation, TLR3ecd and dsRNA were mixed in PiBS at the specified pH and concentration in a total volume of 100 μl per reaction, incubated for 1 h, and run on the column with the same PiBS at a flow rate of 0.7 ml/min. All steps were performed at room temperature.
Vector Construction and Production.
The TLR3/4 chimera was created by fusing the human TLR3ecd (pUNO hTLR3 amino acids 1–704; Invitrogen) to the human TLR4 transmembrane and TIR domains (pUNO hTLR4 amino acids 633–839; Invitrogen). The TLR3 and TLR3/4 expression cassettes were then cloned from pUNO into HIV CS CN lentiviral vectors (D. Schaffer, University of California, Berkeley, CA). The NF-κB-responsive reporter was constructed by replacing the luciferase gene in pNifty2-Luc (Invitrogen) with EGFP from pEGFP-N3 (BD Bioscience), and this construct was subcloned into pLenti6.2/V5-Dest (Invitrogen). Details are provided in SI Appendix 2. Lentiviral particles were produced as described in ref. 37.
Stable Cell Lines.
Culture medium was DMEM with 4 mM l-glutamine (Invitrogen), 10% qualified FBS (Invitrogen), 1% penicillin-streptomycin (Invitrogen), 0.1 mM MEM nonessential amino acids (Sigma), and 1 mM sodium pyruvate (Sigma). HEK293 cells were transduced with the Lenti6.2 reporter vector at a multiplicity of infection (MOI) of 0.1, then cultured in 10 μg/ml blasticidin (Invitrogen) for 2 weeks to produce “control” cells. Control cells were transduced with either the TLR3 or TLR3/4 vector at an MOI of 0.1 and cultured in 500 μg/ml G418 sulfate (Cellgro) and blasticidin for 2 weeks to produce TLR3lo or TLR3/4 cells, respectively. To force surface expression of TLR3, TLR3lo cells were repeatedly transduced with the TLR3 vector at an MOI of 5 at 3-day intervals a total of five times. The resulting cells were labeled with anti-hTLR3ecd mAb TLR3.7 (eBioscience) and an APC-conjugated goat-anti-mouse secondary (Invitrogen), and cells with high surface expression of TLR3 (TLR3hi) were isolated by FACS. Retention of surface TLR3 was confirmed 1 week after sorting. All lines were analyzed for TLR3 expression, using the antibodies described above: 1 μg of anti-TLR3 mAb or isotype control (eBioscience) plus 10 μg of human Ig (Sigma) and 0.5 μg of secondary per 106 cells. Cells were permeablized by using the Cytofix/Cytoperm kit (BD Biosciences PharMingen), and permeablization/wash buffer was supplemented with saponin (Sigma) to a final concentration of 0.5%. TLR3 expression was also confirmed by Western blot analysis (data not shown).
Stimulation Assays.
Cells (105) were plated in a 24-well plate in 0.25 ml of medium for 3 h, ligand or polyclonal anti-TLR3 or goat IgG control (R&D Systems) was added, and 24 h later NF-κB activation was quantified by measuring the mean GFP fluorescence intensity (MFI) by FACS. Where used, bafilomycin A1 (Sigma) was added to cell cultures 1 h before stimulation.
Supplementary Material
ACKNOWLEDGMENTS.
We thank T. Jake Liang (Liver Diseases Branch, National Institute of Diabetes, Digestive, and Kidney Diseases) for pFastBac-CprME, David V. Schaffer (Department of Chemical Engineering, University of California, Berkeley, CA) for HIV CS CN lentiviral vector, and Daniela Verthelyi (Laboratory of Immunology, Food and Drug Administration Center for Drug Evaluation and Research) for D35 and C2395 CpG oligonucleotides. Joseph Shiloach (Biotechnology Unit, National Institute of Diabetes, Digestive, and Kidney Diseases) performed large-scale insect cell fermentations, Lisa Boyd (Laboratory of Immunology, National Institute of Allergy and Infectious Diseases) assisted with SPR experiments, and Yan Wang (Experimental Immunology Branch, National Cancer Institute) performed Western blots. We thank Dr. Paul Roche (Experimental Immunology Branch, National Cancer Institute) and Alessandra Mazzoni (Laboratory of Experimental Immunology, National Cancer Institute) for thoughtfully reviewing the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health (National Cancer Institute and National Institute of Diabetes, Digestive, and Kidney Diseases) and by a National Institutes of Health/Food and Drug Administration Intramural Biodefense Award from National Institute of Allergy and Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710779105/DC1.
References
- 1.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 2.Bell JK, et al. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–533. doi: 10.1016/s1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 3.Mizel SB, West AP, Hantgan RR. Identification of a sequence in human toll-like receptor 5 required for the binding of Gram-negative flagellin. J Biol Chem. 2003;278:23624–23629. doi: 10.1074/jbc.M303481200. [DOI] [PubMed] [Google Scholar]
- 4.Sato M, et al. Direct binding of Toll-like receptor 2 to zymosan, and zymosan-induced NF-kappa B activation and TNF-alpha secretion are down-regulated by lung collectin surfactant protein A. J Immunol. 2003;171:417–425. doi: 10.4049/jimmunol.171.1.417. [DOI] [PubMed] [Google Scholar]
- 5.Cornelie S, et al. Direct evidence that Toll-like receptor 9 (TLR9) functionally binds plasmid DNA by specific cytosine-phosphate-guanine motif recognition. J Biol Chem. 2004;279:15124–15129. doi: 10.1074/jbc.M313406200. [DOI] [PubMed] [Google Scholar]
- 6.Meng G, et al. Antagonistic antibody prevents toll-like receptor 2-driven lethal shock-like syndromes. J Clin Invest. 2004;113:1473–1481. doi: 10.1172/JCI20762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rutz M, et al. Toll-like receptor 9 binds single-stranded CpG-DNA in a sequence- and pH-dependent manner. Eur J Immunol. 2004;34:2541–2550. doi: 10.1002/eji.200425218. [DOI] [PubMed] [Google Scholar]
- 8.Vasselon T, Detmers PA, Charron D, Haziot A. TLR2 recognizes a bacterial lipopeptide through direct binding. J Immunol. 2004;173:7401–7405. doi: 10.4049/jimmunol.173.12.7401. [DOI] [PubMed] [Google Scholar]
- 9.Bell JK, et al. The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc Natl Acad Sci USA. 2005;102:10976–10980. doi: 10.1073/pnas.0505077102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranjith-Kumar CT, et al. Biochemical and functional analyses of the human Toll-like receptor 3 ectodomain. J Biol Chem. 2007;282:7668–7678. doi: 10.1074/jbc.M610946200. [DOI] [PubMed] [Google Scholar]
- 11.Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 12.Kim HM, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Jin MS, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA, activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 15.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto M, Kikkawa S, Kohase M, Miyake K, Seya T. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem Biophys Res Commun. 2002;293:1364–1369. doi: 10.1016/S0006-291X(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto M, et al. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J Immunol. 2003;171:3154–3162. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- 18.Kulka M, Alexopoulou L, Flavell RA, Metcalfe DD. Activation of mast cells by double-stranded RNA: evidence for activation through Toll-like receptor 3. J Allergy Clin Immunol. 2004;114:174–182. doi: 10.1016/j.jaci.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 19.Orinska Z, et al. TLR3-induced activation of mast cells modulates CD8+ T-cell recruitment. Blood. 2005;106:978–987. doi: 10.1182/blood-2004-07-2656. [DOI] [PubMed] [Google Scholar]
- 20.Johnsen IB, et al. Toll-like receptor 3 associates with c-Src tyrosine kinase on endosomes to initiate antiviral signaling. EMBO J. 2006;25:3335–3346. doi: 10.1038/sj.emboj.7601222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoebe K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 23.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 24.de Bouteiller O, et al. Recognition of double-stranded RNA by human toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J Biol Chem. 2005;280:38133–38145. doi: 10.1074/jbc.M507163200. [DOI] [PubMed] [Google Scholar]
- 25.Bell JK, Askins J, Hall PR, Davies DR, Segal DM. The dsRNA binding site of human Toll-like receptor 3. Proc Natl Acad Sci USA. 2006;103:8792–8797. doi: 10.1073/pnas.0603245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonard JN, Schaffer DV. Antiviral RNAi therapy: emerging approaches for hitting a moving target. Gene Ther. 2006;13:532–540. doi: 10.1038/sj.gt.3302645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cain CC, Sipe DM, Murphy RF. Regulation of endocytic pH by the Na+,K+-ATPase in living cells. Proc Natl Acad Sci USA. 1989;86:544–548. doi: 10.1073/pnas.86.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, et al. Structural and functional analyses of the human Toll-like receptor 3. Role of glycosylation. J Biol Chem. 2006;281:11144–11151. doi: 10.1074/jbc.M510442200. [DOI] [PubMed] [Google Scholar]
- 29.Takada E, et al. C-terminal LRRs of human Toll-like receptor 3 control receptor dimerization and signal transmission. Mol Immunol. 2007;44:3633–3640. doi: 10.1016/j.molimm.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 30.Edgcomb SP, Murphy KP. Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins. 2002;49:1–6. doi: 10.1002/prot.10177. [DOI] [PubMed] [Google Scholar]
- 31.Ranjith-Kumar CT, et al. Effects of single nucleotide polymorphisms on Toll-like receptor 3 activity and expression in cultured cells. J Biol Chem. 2007;282:17696–17705. doi: 10.1074/jbc.M700209200. [DOI] [PubMed] [Google Scholar]
- 32.Latz E, et al. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 33.Honda K, et al. Spatiotemporal regulation of MyD88-IRF-7 signaling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 34.Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7:49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- 35.Gay NJ, Gangloff M, Weber AN. Toll-like receptors as molecular switches. Nat Rev Immunol. 2006;6:693–698. doi: 10.1038/nri1916. [DOI] [PubMed] [Google Scholar]
- 36.Qiao M, et al. Induction of sterilizing immunity against West Nile Virus (WNV), by immunization with WNV-like particles produced in insect cells. J Infect Dis. 2004;190:2104–2108. doi: 10.1086/425933. [DOI] [PubMed] [Google Scholar]
- 37.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell. 2005;122:169–182. doi: 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.