Abstract
Tuberculosis remains a major global health problem that kills up to 2 million people annually. Central to the success of Mycobacterium tuberculosis (Mtb) as a pathogen is its ability to evade host immunity and to establish a chronic infection. Although its primary intracellular niche is within macrophages, the underlying molecular mechanisms are poorly understood. Here we show that Rv2224c, a cell envelope-associated predicted protease, is critical for Mtb virulence. Disruption of Rv2224c led to prolonged survival of infected mice and highly reduced lung pathology. Absence of Rv2224c enhanced host innate immune responses, compromised the intracellular survival of Mtb in macrophages, and increased its susceptibility to lysozyme. We provide insights into the molecular basis for Rv2224c function by showing that Rv2224c activity promotes processing and extracellular release of the Mtb protein, GroEL2. Inhibition of Rv2224c and its targets offers opportunities for therapeutic interventions and immune-modulatory strategies.
Keywords: macrophages, pathogen, intracellular, cell envelope
Mycobacterium tuberculosis (Mtb) is a highly successful human pathogen that elicits strong innate and adaptive immune responses. Tuberculosis (TB) is characterized by an inflammatory response that leads to containment, but not eradication, of bacteria within granulomas in the lung (1). The Th1-type response elicited by Mtb contributes to the extensive immunopathology, tissue necrosis, and lung damage associated with TB (2). Macrophages are central immune cells linking innate and adaptive immunity by promoting lymphocyte activation and recruitment (1). They also serve as the primary niche in vivo, because Mtb actively replicates within macrophages. Mtb inhibits fusion of phagosomes with lysosomes and interferes with antigen presentation, IFN-γ-mediated signaling pathways, and transcriptional responses (3). Thus, the ability of Mtb to subvert macrophage microbicidal functions likely contributes to the hallmark delayed T cell response in TB, but the bacterial factors that mediate these processes are poorly understood (2, 3). In recent genome-wide studies, we comprehensively identified the bacterial genes necessary for Mtb survival in vivo and in macrophages (4, 5). Our data predicted that one of these genes, Rv2224c, was required for both in vivo growth in mice and intracellular replication in macrophages (4). In this study, we show that Rv2224c is indeed critical for Mtb virulence in vivo, promotes survival in macrophages, and modulates innate immune control of infection.
Results
Rv2224c Is Associated with the Mycobacterial Cell Envelope.
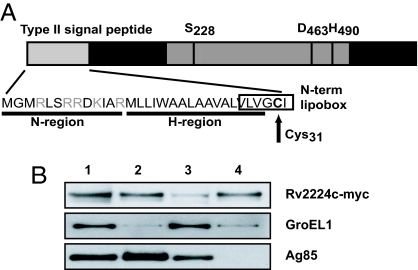
Rv2224c is predicted to encode a lipoprotein based on an N-terminal type II signal peptide and a cysteine lipoprotein anchor residue (Fig. 1A) (6). Rv2224c contains a GXSXG consensus sequence surrounding a putative serine active site (S228), which is characteristic of α/β hydrolase-fold family members, namely proteases, esterases, and lipases. Rv2224c is annotated as a tripeptidyl peptidase in the MEROPS peptidase database, and its sequence shows high sequence similarity to the serine proteases SlpD and SlpE from Streptomyces lividans, which are mycelium-associated lipoproteins involved in cell growth (7). Refolded recombinant Rv2224c also has been reported to display esterase activity in vitro (8). However, the physiological substrate(s) of Rv2224c remains to be established. Lipoproteins of Mtb typically associate with its complex lipid-rich cell envelope (6, 9). We therefore tested whether Rv2224c was present in mycobacterial cell wall and cell membrane fractions. We expressed myc epitope-tagged Rv2224c in Mycobacterium bovis bacillus Calmette–Guérin; separated protein extracts into whole-cell, cell wall, cytoplasmic, and cell membrane fractions; and found that Rv2224c is clearly enriched in cell wall and membrane fractions (Fig. 1B). However, Rv2224c was not detected in culture supernatants and is thus not likely to be secreted extracellularly (data not shown). As controls for fractionation, we show that cytoplasmic heat-shock protein, GroEL1, is largely absent from cell envelope fractions and that Ag85, a known cell wall-associated and extracellular protein, is present in cell wall fractions (Fig. 1B) (9).
Fig. 1.
Rv2224c is a cell envelope-associated predicted hydrolase. (A) Rv2224c is predicted to be a lipoprotein with an N-terminal type II signal peptide. The N region has lysine and/or arginine, and the H region has hydrophobic residues. Cysteine at amino acid residue 31 within the conserved “lipobox” sequence is the putative lipoprotein attachment site, where diglyceride units are added by thioester linkage, followed by cleavage at a site immediately preceding the lipidated cysteine, which becomes the N terminus of the mature lipoprotein (6). Rv2224c contains the α/β hydrolase fold sequence and a catalytic triad typically present in serine proteases, esterases, and lipases, consisting of the catalytic nucleophile serine active site (S228), which associates with the proton carrier histidine (H490), and a charge relaying aspartic acid (D463). (B) Cell fractionation of M. bovis bacillus Calmette–Guérin-expressing myc-tagged Rv2224c and Western blots probed to detect the myc epitope, GroEL1, or Ag85 proteins. Lane 1, whole-cell extract; lane 2, cell wall; lane 3, cytoplasm; lane 4, cell membrane.
Rv2224c Is Required for Optimal Mtb Growth in Vivo.
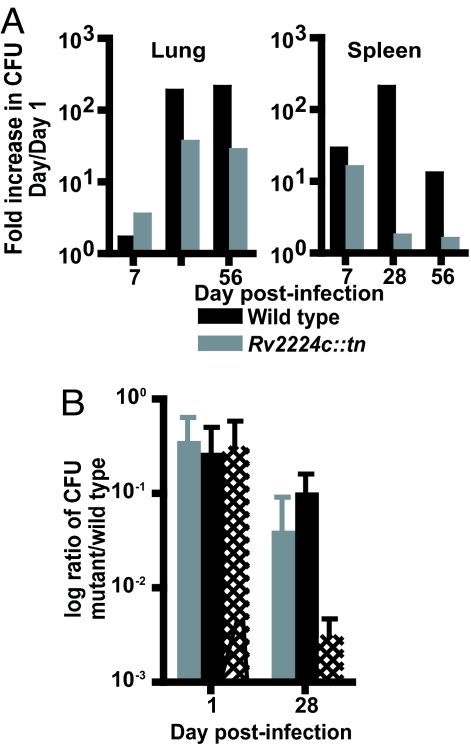
Many bacterial cell envelope- and cell surface-exposed proteins have access to host cellular compartments and modulate host responses (10). Thus, we sought to study the function of Rv2224c during infection in vivo. We isolated a transposon mutant disrupted in Rv2224c (Rv2224c::tn) and monitored its ability to compete for growth with wild-type Mtb in mice. We used mixed infections in a competition setting, because this allowed us to directly compare the fitness of the mutant strain relative to wild type within the same in vivo environment. We infected C57BL/6 mice with a 1:1 ratio of Rv2224c::tn mutant to wild type and monitored cfus of each strain from lungs and spleens as described in Materials and Methods (Fig. 2A). Decreases in mutant cfus were apparent by 28 days and continued to 56 days after infection, indicating that Rv2224c is required for optimal growth of Mtb in vivo. Both wild-type and mutant strains grew equally well in vitro in 7H9 medium over an 8-day growth period (data not shown).
Fig. 2.
Rv2224c is required for optimal Mtb growth in vivo. (A) Mixed infection (1:1, wild type:Rv2224c::tn), by using 5 × 105 cfus per strain, of C57BL/6 mice (four per time point) by i.v. infection. cfus were determined at 1, 7, 28, and 56 days after infection as described in Materials and Methods. Data are represented as fold increase in mean cfus relative to mean cfus on day 1. (B) Mixed infections to assess in vivo competition between wild type versus mutant Rv2224c::tn (wild type:Rv2224c::tn, gray bars), wild type versus mutant Rv2224c::tn complemented with intact Rv2224c (wild type:Rv2224c::tn + 2224c, black bars), or wild type versus mutant Rv2224c::tn-expressing Rv2224c containing the S228A mutation (wild type:Rv2224c::tn + S228A, hatched bars). cfus of each strain were determined at 1 and 28 days after infection from lungs. Five mice were used per strain for each time point. Data are represented as log ratios of mean cfus of mutant per mean cfus of wild type.
We next asked whether the mutant's growth defect could be complemented by expressing intact Rv2224c (Rv2224c::tn + 2224c). Mice were infected with wild-type Mtb mixed with either the Rv2224c::tn strain or Rv2224c::tn + 2224c (Fig. 2B). Lungs were harvested at 28 days after infection and plated for cfus. Data are represented as ratios of cfus of mutant/wild type. Full-length Rv2224c protein led to partial complementation of the growth defect after infection (Fig. 2B, black bars). We also constructed a mutant Rv2224c::tn strain expressing both Rv2224c and Rv2223c, which is present within a putative operon with Rv2224c and shares 49% identity. However, this strain grew very slowly in vitro relative to wild type and thus could not be used in complementation studies in vivo. We then tested whether expressing Rv2224c harboring a mutated serine active site (S228A) could complement the growth defect. Competing wild type with Rv2224c::tn + S228A resulted in a dramatically more severe defect in growth than competition with Rv2224c::tn alone (Fig. 2B, hatched bars), suggesting that Rv2224c enzymatic activity is required. Moreover, the active site mutation may disrupt the catalytic triad of Ser228–Asp463–His490 shared by many α/β hydrolases and generate a dominant-negative protein that either inhibits the activity of Rv2223c or some other related hydrolase, leading to a more severe phenotype.
Disruption of Rv2224c Prolongs Survival and Reduces Immunopathology of Infected Mice.
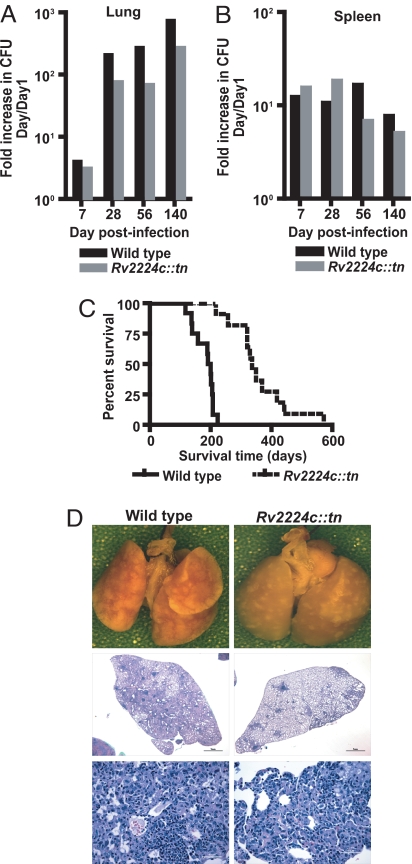
To study how disruption of Rv2224c impacts disease progression and survival of the host, we performed single infections with either wild-type or mutant strains. These experiments assess the ability of each individual strain to grow in vivo and its direct impact on the host. The Rv2224c::tn mutant showed higher bacterial burdens in spleens and lungs when infected singly relative to when mixed with wild type, suggesting that Rv2224::tn may be more susceptible to the immune response evoked by wild-type Mtb (Fig. 3 A and B). Despite modest reductions in bacterial burdens, however, mice infected with the mutant survived significantly longer than wild-type-infected mice (median survival time of 336 versus 195 days; P < 0.0001) with reduced lung pathology (Fig. 3C). Wild-type-infected lungs showed large granulomatous nodules and widespread tissue damage, with 80–90% of the tissue containing cellular infiltrates (Fig. 3D Middle) of mostly foamy macrophages and lymphocytes within the granulomatous lesions (Fig. 3D Bottom). In contrast, lungs of mutant-infected mice contained small, discrete foci of macrophage- and lymphocyte-rich lesions and retained ≈80% of alveolar space (Fig. 3D Middle). Overall, although the differences in bacterial burden between wild-type and mutant strains were relatively mild, the significant extension of survival and the less detrimental immunopathology suggest differential handling of the two strains by the host. Thus, absence of Rv2224c attenuates the virulence of Mtb in vivo.
Fig. 3.
Disruption of Rv2224c prolongs survival and reduces lung immunopathology of infected mice. (A and B) Single infections of C57BL/6 mice with wild type or Rv2224c::tn and Mtb cfus in lungs (A) and spleens (B) over time. Four mice were used per strain for each time point. cfu data are represented as fold increase in cfus relative to cfus on day 1. (C) Survival of C57BL/6 mice infected with wild type or Rv2224c::tn. Twelve mice were infected in each group. Median survival time was 195 days (wild type) and 336 days (Rv2224c::tn) (P < 0.0001 by log-rank test). (D) (Top) Gross pathology of lungs at 20 weeks after infection. (Middle and Bottom) H&E-stained lung tissue sections from wild-type- or mutant-infected mice at 20 weeks. (Scale bars, Middle, 1 mm; Bottom, 50 μm.)
Rv2224c Impacts Host Innate Immune Responses.
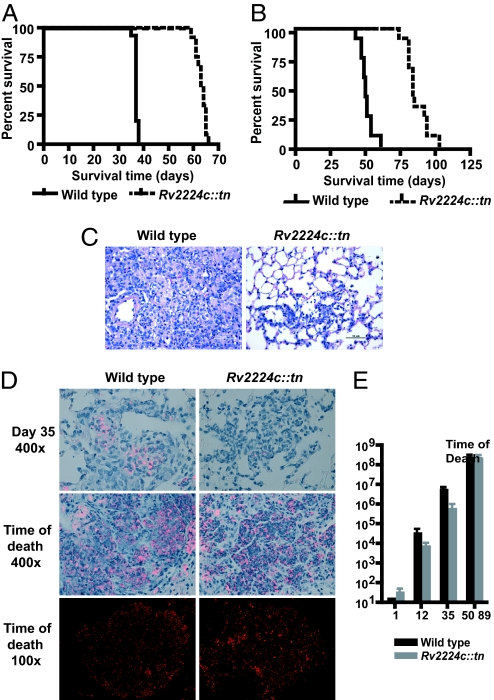
To evaluate the contribution of innate versus adaptive immunity to this outcome, we infected RAG−/− mice, which lack T and B cells, by aerosol and i.v. routes (Fig. 4 A and B). Immunodeficient RAG−/− mice were highly susceptible to Mtb, with as little as ≈10 cfus, and rapidly succumbed to infection, which is consistent with the critical role of T and B cells in controlling TB (11). Interestingly, infection with Rv2224c::tn by either route significantly prolonged survival of RAG−/− mice (Fig. 4 A and B). The lungs of RAG−/− mice infected with wild type were infiltrated with numerous macrophages and neutrophils; by 35 days after aerosol infection, these lesions occupied >90% of the alveolar space (Fig. 4C). Much smaller lesions of similar composition were observed in Rv2224c::tn-infected lungs and resulted in milder pathology (Fig. 4C). Notably, fewer bacteria were present within macrophages in lung sections, with a 10-fold decrease in viable bacteria 12 and 35 days after infection in infected lungs (Fig. 4 D and E). These results suggest that lung macrophages control the growth of Rv2224c::tn in the innate phase and limit intracellular growth. Interestingly, at the time of death, the numbers of bacteria recovered from the lungs of wild-type or mutant-infected mice were indistinguishable, as was the pathology (Fig. 4 D and E). Thus, mutant strains are not inherently defective in their ability to replicate but fail to do so in the face of early innate immune responses. However, they actively grow once innate immunity no longer controls growth. Taken together, these data provide evidence for enhanced early control of the Rv2224c::tn mutant burden by innate immune mechanisms.
Fig. 4.
Rv2224c modulates innate immune responses. (A and B) Survival of RAG−/− mice infected by i.v. route at 2 × 106 cfus per strain (median survival: wild type, 37 days; Rv2224c::tn, 64 days; P < 0.0001 by log-rank test) (A) and aerosol route (median survival: wild type, 50 days; Rv2224c::tn, 84 days; P < 0.0001 by log-rank test) (B). Twelve mice per strain were infected in both A and B. (C) H&E-stained lung tissue sections from wild-type- or mutant-infected mice at 35 days after infection. (D) Auramine/rhodamine staining of lung tissue sections from aerosol-infected RAG−/− mice on day 35 (Top) and time of death (Middle and Bottom). (E) cfus from lungs of aerosol-infected RAG−/− mice. Mean time of death for each strain is indicated. Four mice were used per group per time point. Error bars represent standards deviation from the mean.
Rv2224c Modulates Host Macrophage Responses.
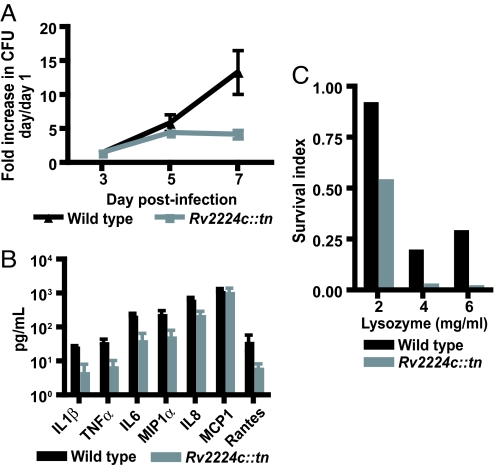
Because our data pointed to a role for Rv2224c in promoting survival within macrophages, we directly assessed the growth of Rv2224c::tn in primary macrophages. Intracellular growth of wild-type Mtb increased ≈10-fold after 7 days, but the Rv2224c::tn mutant failed to continue growing (Fig. 5A). Both wild-type and mutant strains grew equally well during the early stages (Fig. 5A) and in vitro in culture medium (data not shown). Thus, Mtb requires Rv2224c for sustained intracellular growth in macrophages. To assess whether the macrophage environment is altered upon infection with the mutant, we measured the levels of cytokines and chemokines secreted by infected macrophages. Two days after infection, when the bacterial burden of wild-type and of mutant strains were indistinguishable (data not shown), we found that key proinflammatory cytokines TNFα, IL1β, and IL6 were decreased in the absence of Rv2224c, as were chemokines IL8, MIP1α, and RANTES, which contribute to neutrophil and lymphocyte recruitment (Fig. 5B). This finding suggests that a dampened inflammatory milieu may be generated by Rv2224c::tn-infected macrophages in vivo.
Fig. 5.
Rv2224c is required for survival in macrophages. (A) Macrophages were infected with wild type or Rv2224c::tn at an moi of 1 in triplicate and lysed at 1, 3, 5, and 7 days after infection, and the intracellular bacteria were plated for cfus. Data represent four independent experiments and are shown as fold increases in cfus, normalized to cfu on day 1 for each strain. (B) Secretion of cytokines and chemokines by macrophages 48 h after infection with wild type or Rv2224c::tn. Data are representative of three independent experiments. Error bars represent standard deviation from the mean. (C) Survival index of wild-type Mtb or Rv2224c::tn in the presence of lysozyme (ratio of cfu of each strain in the presence/absence of lysozyme). Data are representative of four independent experiments, each in triplicate, per concentration of lysozyme.
We next wished to explore the basis for the apparent increased susceptibility of Rv2224c::tn to the macrophage environment. One possibility is increased sensitivity to antimicrobial agents produced by macrophages that limit mutant growth. The antimicrobial agent lysozyme is secreted abundantly by macrophages, neutrophils, and lung epithelial cells (12). Most Gram-positive bacteria are highly susceptible to lysozyme, which damages their cell walls by hydrolyzing glycosidic bonds in peptidoglycan or by displacing cell wall autolytic enzymes that normally mediate remodeling during cell division (12, 13). Mtb has an impermeable cell envelope and is extremely resistant to lysozyme. Wild-type Mtb was resistant to lysozyme at up to 2 mg/ml and showed decreased viability at 4 and 6 mg/ml (Fig. 5C). In contrast, the Rv2224::tn mutant was markedly more susceptible to lysozyme at all three concentrations (Fig. 5C). This finding suggests that the mutant cell envelope is more accessible to lysozyme in macrophages, leading to decreased bacterial viability.
Rv2224c Regulates the Processing of Mtb Stress-Induced Secreted Protein GroEL2.
To further dissect the biological function of Rv2224c, we sought to identify its physiological targets. Because Rv2224c is cell envelope-associated with predicted hydrolytic activity, we hypothesized a role in facilitating extracellular release of other Mtb cell envelope proteins that would be released into mycobacterial culture supernatant. We examined electrophoretically separated cell-free supernatants from wild-type or mutant short-term cultures and observed distinct banding patterns in the wild type that were missing in Rv2224c::tn samples (data not shown). We excised several bands and used mass spectrometric analysis to identify proteins that were absent from mutant supernatants. We focused on cell wall-associated heat-shock protein GroEL2 (Rv0440, hsp65), which was differentially present in supernatants from the two strains, and confirmed by Western blot analysis that the extracellular release of GroEL2 was aberrant in Rv2224c::tn (Fig. 6A). Whole-cell lysates from both wild-type and mutant bacteria expressed comparable amounts of GroEL2, but, in wild-type supernatants, a smaller form predominated (Fig. 6A). Mutant supernatants retained mostly full-length GroEL2 and appeared incapable of efficiently generating the smaller protein. This observation suggests that the smaller version is produced by cleavage of full-length GroEL2. The closely related GroEL1 is absent in supernatants from either strain, which is consistent with its known cytoplasmic localization and role in chaperone activities (14). In contrast, GroEL2 is present in cell wall fractions (Fig. 6B) and extracellularly in culture supernatants (Fig. 6A) (15, 16). GroEL2 is induced by heat, nutrient starvation, hypoxia, phagocytosis, and reactive oxygen radicals and is shown to modulate macrophage cytokine responses (14, 17–20).
Fig. 6.
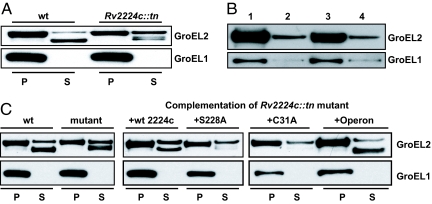
Rv2224c promotes processing of GroEL2. (A) Western blot showing GroEL2 in wild-type whole-cell pellets (P) and as a smaller form in the culture supernatant (S). In Rv2224c::tn pellets, the larger form of GroEL2 is present in both P and S, with defective processing of GroEL2 to the smaller form. GroEL1 is exclusively present in P and absent from S. (B) Localization of GroEL2 in cell fractions of M. bovis bacillus Calmette–Guérin. GroEL2 is present in cell wall, cell membrane, and cytoplasmic fractions. GroEL1 is largely cytoplasmic. Lane 1, whole-cell extract; lane 2, cell wall; lane 3, cytoplasm; lane 4, cell membrane. (C) Complementation of GroEL2 processing defect in Rv2224c::tn by expression in trans of wild-type Rv2224c, mutated versions (S228A and C31A), or both Rv2223c and Rv2224c (operon).
We sought to further delineate the role of Rv2224c in GroEL2 cleavage by structure-function analysis of the Rv2224c protein. We introduced wild-type and mutated versions of Rv2224c into Rv2224c::tn and tested their effects on GroEL2 processing in the corresponding supernatants (Fig. 6C). Expression of the intact Rv2224c protein complemented the defect, as seen by increased amounts of the processed form of GroEL2. Mutations in the putative serine active site of Rv2224c (S228A) failed to complement the defect, indicating that enzymatic activity is required for GroEL2 cleavage (Fig. 6C). Mutations in the cysteine anchor residue also failed to complement the defect, supporting the notion that positioning of Rv2224c in the cell wall is critical for its function. Finally, expression of the operon (Rv2224c-Rv2223c), which contains the closely related Rv2223c, fully restored GroEL2 processing to wild-type levels (Fig. 6C). Thus, the extracellular release of GroEL2 depends on Rv2224c function. Whether GroEL2 processing is a direct consequence of Rv2224c enzymatic activity or is an indirect consequence of regulation of intermediate proteins is unclear and awaits detailed biochemical characterization.
Discussion
Innate immune responses are critical for controlling infection and launching adaptive immunity. By avoiding macrophage microbicidal functions and by replicating intracellularly, Mtb interferes with the initiation of adaptive immunity (1). In this article, we demonstrate that Rv2224c is critical for Mtb disease progression and intracellular survival and modulates innate immune responses. Interestingly, aerosol infection of RAG−/− mice shows that, in the absence of Rv2224c, early control of Mtb replication is in fact possible (Fig. 4) and maps to growth limitation in macrophages (Fig. 5A). Although Rv2224c::tn mutant bacteria eventually overwhelm host innate responses, this early control leads to significant extension of survival. Thus, inhibiting Mtb factors such as Rv2224c could enhance innate immunity to TB. Macrophages infected with Rv2224c::tn also show dampened inflammatory milieu, and Rv2224c::tn was more susceptible to cell wall damage by lysozyme (Fig. 5 B and C). These results suggest that inhibition of Rv2224c could help drive the immune response away from its detrimental aspects and could simultaneously debilitate Mtb, resulting in more favorable outcomes for the host.
Competition experiments showed that Rv2224c is required for optimal growth in vivo (Fig. 2). Complementation with intact Rv2224c did not fully restore the growth defect (Fig. 2C), suggesting that disruption of Rv2224c may affect the function of the related Rv2223c. We can detect expression of Rv2223c mRNA in the Rv2224c::tn (data not shown), and therefore transcription of Rv2223c does not appear to be compromised. However, Rv2224c may be involved in posttranscriptional regulation of Rv2223c, and the two proteins may function in concert for optimal activity toward their targets. Unfortunately, complementation of the in vivo growth defect by expressing both Rv2224c and Rv2223c was not feasible because ectopic expression of the operon resulted in slow growth of the strain in vitro. Speculation that Rv2223c may in fact work in conjunction with Rv2224c is supported by the observation that expressing both genes results in better complementation of the GroEL2-processing defect (Fig. 6C). In addition, mutating the serine active site of Rv2224c resulted in a more severe growth defect in vivo, suggesting that disrupting Rv2224c activity may compromise Rv2223c function directly or indirectly by generating a dominant-negative protein that interferes with the function of Rv2223c or other related hydrolases.
We have uncovered a role for Rv2224c in extracellular GroEL2 release from the cell wall of Mtb. GroEL2 is a highly expressed, immunodominant protein and induced by many physiological stresses related to intracellular growth, including phagocytosis, hypoxia, nutrient limitation, and reactive oxygen radicals (14, 21, 22). Recent purification of GroEL2 for crystallization studies revealed that it eluted as both a full-length and smaller N-terminally processed form, suggesting that physiological proteolysis might occur (23). Thus, the cleaved GroEL2 protein may constitute the functionally active form, whose release depends on Rv2224c activity. Whether or not GroEL2 is a direct substrate for Rv2224c remains unclear and awaits further biochemical characterization. Although it is unlikely to be the only target of Rv2224c, GroEL2 has been implicated in modulating proinflammatory cytokine responses in macrophages, which is consistent with our observations of dampened inflammatory milieu (Fig. 5C) (14, 19, 20). A detailed analysis of Rv2224c enzymatic activity toward GroEL2 and additional physiological substrates should help elucidate the precise biochemical function of Rv2224c.
Our results implicating Rv2224c in the processing of GroEL2 and the resistance to lysozyme suggest that other cell envelope components may be substrates for RV2224c activity. This idea also was advanced in studies from Flores et al. (24), who isolated a transposon mutant with insertions in Mycobacterium smegmatis expA, the homolog of Mtb Rv2224c, in a screen designed to study hypersusceptibility to β-lactam antibiotics, which act on bacterial cell wall peptidoglycan. Interestingly, M. smegmatis expA mutants showed increased susceptibility to lysozyme and displayed elongated cell morphology with swollen termini. The authors speculated that expA and expB (the Rv2223c homolog) might be directly involved in hydrolytic breakdown of cell wall components or indirectly involved by regulating proteins involved in autolysis or cell wall remodeling (24). Rv2224c is classified as an α/β hydrolase in a family with both esterase and amidase members (25). Its closest biochemically characterized homolog, the secreted SlpD triaminopeptidase from S. lividans, was identified by its activity against an amide (7). Protease activity is consistent with the requirement of Rv2224c for correct processing of GroEL2. However, several proteins related by homology to Rv2224c are carboxyesterases and lipases. Indeed, a recent study showed that refolded recombinant Rv2224c can hydrolyze ester compounds, which suggests that cell envelope lipids may be substrates (8). The chemical similarity of ester and amide hydrolysis is such that most proteases also catalyze the degradation of ester substrates, although the converse is not typically true (26). These possibilities will be clarified when physiological substrates of Rv2224c are established. Many Mtb proteins are exported to the cell envelope; some of these are secreted extracellularly by specialized secretion systems and in turn promote interactions with host cells (27, 28). Our studies suggest that cleavage of cell envelope proteins constitutes an additional mechanism by which Mtb can release critical extracellular factors. Enzymatic regulation of their release in vivo would facilitate rapid adaptation to changing immune environments.
In summary, we have shown that Rv2224c is a key determinant of disease outcome in vivo. Rv2224c modulates innate immune responses in macrophages and is required for extracellular release of the Mtb stress-induced protein, GroEL2. The cell surface accessibility of Rv2224c and its importance in host–pathogen interactions make it a target for therapeutics and immune-modulatory strategies.
Materials and Methods
Bacterial Strains and Media.
Mtb H37Rv strains and M. bovis bacillus Calmette–Guérin Pasteur were grown at 37°C in Middlebrook 7H9 broth or 7H10 agar as described in ref. 4. For the Rv2224c::tn mutant, 25 μg/ml kanamycin (Sigma–Aldrich) was added, and, for complemented strains, 50 μg/ml hygromycin (Roche) was added. The Mtb transposon library has been described in detail in ref. 29. The Rv2224::tn mutant was obtained by sequencing mutants from the library by using arbitrary primers as described in ref. 4. The mutant was maintained in the presence of 25 μg/ml kanamycin throughout.
Construction of Plasmids and Strains.
To construct myc-tagged Rv2224c driven by its own promoter, the gene was amplified by 5′-CCTCGGGCGATGGTCTAGATGCACCATGC-3′ and 5′-CGTTTCTTCGCCACTAGTGCACTTGGCG-3′ cloned into the XbaI and SpeI sites of pMV762 (hyg) with C-terminal c-myc tag. The C31S mutation was made with site-directed mutagenesis by using 5′-GTTCTTGTGGGCTCCATCCGCGTGGTC-3′ and S228A 5′-CTACCTGGGCTACGCGTACGGCACC-3′. The Rv2223c-2224c operon was cloned into the XbaI and SpeI sites of pMV762 (hyg) by using 5′-CCTCGGGCGATGGTCTAGATGCACCATGC-3′ and 5′-AAGGGGAGTGCGCGACTAGTCCAGGGCG-3′.
Preparation of Protein Extracts and Western Blotting.
Each Mtb strain was grown to an OD600 of 0.8, pelleted, washed, and grown in Sautons' medium (30) plus 0.05% Tween 80 to an OD600 of 0.6, resuspended into Sautons' medium minus Tween 80, and grown for 24 h at 37°C. Supernatants were concentrated by using Centricon Plus-70 (Millipore). Each cell pellet was resuspended in 50 mM Tris, 10 mM NaCl (pH 8.0), and glass beads (Bio101) with bead beating for 45 s, was centrifuged at 16,000 × g for 15 min at 4°C, and 150 μl was removed for protein estimation. Remaining extracts were boiled with SDS protein-loading buffer for 20 min. Samples were separated by a 200-V SDS/PAGE by using NuPage 10% [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane or 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)-1,3-propanediol (Bistris) gels (Invitrogen) and transferred onto nitrocellulose at 30 V for 1 h. Antibodies used were mouse anti-GroEL2 (1:2,000 in 1% BSA; Imgenex), mouse anti-Ag85 (1:500 in 1% BSA; Abcam), GroEL1:rabbit anti-His (1:5,000 in 1% BSA; Novus Biologicals), rabbit anti-c-myc [1:10,000 in 3% 50 mM Tris·HCl (pH 7.4), 100 mM NaCl, and 5% nonfat dry milk (Blotto); Novus Biologicals], and HRP conjugated anti-mouse or anti-rabbit serum (1:10,000; Kirkegaard & Perry Laboratories) as a secondary antibody.
Fractionation of M. bovis Bacillus Calmette–Guérin.
Fractionation was performed essentially as described in ref. 31. Fractions were loaded onto gels normalized for cell numbers proportionally.
Macrophage Survival Assays and Cytokine Assays.
Macrophages were derived from bone marrow of C57BL/6 mice as described in ref. 4. Macrophages were plated onto 24-well plates (2 × 105 per well). Each strain was used for infection (in triplicate per time point) at an moi of 1 as described in ref. 4. Cell-free supernatants from macrophage monolayers 48 h after infection (moi of 5 in triplicate) were assayed by using a Luminex ELISA kit and were analyzed by using the Bioplex 200 Luminex system (Bio-Rad). Error bars represent standard deviation from the mean.
Lysozyme Susceptibility Assays.
These assays were developed in consultation with M. Pavelka. We incorporated hen egg lysozyme (MP Biomedicals) directly into 7H10 plates and plated serial dilutions of Mtb strains onto plates containing 0, 2, 4, or 6 mg/ml lysozyme. Plates were incubated for 4–5 weeks at 37°C and scored for cfus.
Infection of Mice with Mtb.
All animal experiments were approved by the Institutional Animal Care and Use Committee of Harvard University. For aerosol infections, RAG−/− mice (C67BL/6) were infected by using an aerosol apparatus manufactured by the College of Engineering Shops at the University of Wisconsin, Madison. The mice were then exposed for 40 min, resulting in 10 cfus per mouse lung (determined by plating lungs on day 1 after infection; four mice per group). For i.v. infections, mice were infected by tail-vein injection, with 5 × 105 cfus of each strain for mixed infections and 2 × 106 cfus for single infections (four mice per time point). Organs were homogenized, and serial dilutions were plated for cfus onto 7H10, 7H10 kanamycin (for the mutant Rv2224c::tn), or 7H10 kanamycin plus hygromycin (for Rv2224c::tn complemented).
Statistical Analysis.
GraphPad Prizm software, Version 4.0, was used for all analyses.
Histology and Staining of Tissue Sections.
Organs were fixed in 10% buffered formalin and embedded in paraffin, and 6-μm sections were stained with H&E. For detecting Mtb in tissue sections, slides were baked at 60°C and washed twice in xylenes; decreasing concentrations of ethanol were used to dehydrate tissue, and slide sections were incubated in auramine–rhodamine at 37°C for 15 min, washed in 1% HCl/70% EtOH, and counterstained by using Harris' modified haematoxylin. Excess stain was removed by sequential washing in acidic and basic solutions, rehydrated by reversing sequence in ethanol, and cleared in xylenes. Images were captured on a Nikon Eclipse 80i microscope by using a SPOT digital camera.
ACKNOWLEDGMENTS.
We thank Dr. R. Bronson (Harvard Rodent Pathology Core) for his expertise and insights; Drs. G. Petsko, D. Ringe, J. Naffin, G. Brandt, and S. Sampson for helpful discussions; Dr. C. Sassetti for isolation of the Rv2224c:tn mutant; Drs. D. Sarracino, M. Chase, and S. Fortune for their input in mass spectrometry experiments; Ms. I. Breiterene for technical assistance; and Dr. Martin Pavelka for expert advice on lysozyme-susceptibility plate assays. This work was supported by National Institutes of Health grants (to B.R.B., L.H.G., and E.J.R.) and the New York Community Trust Heiser Program (J.R.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Bhatt K, Salgame P. Host innate immune response to Mycobacterium tuberculosis. J Clin Immunol. 2007;27:347–362. doi: 10.1007/s10875-007-9084-0. [DOI] [PubMed] [Google Scholar]
- 2.Flynn JL. Immunology of tuberculosis and implications in vaccine development. Tuberculosis (Edinburgh) 2004;84:93–101. doi: 10.1016/j.tube.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Houben EN, Nguyen L, Pieters J. Interaction of pathogenic mycobacteria with the host immune system. Curr Opin Microbiol. 2006;9:76–85. doi: 10.1016/j.mib.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci USA. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutcliffe IC, Harrington DJ. Lipoproteins of Mycobacterium tuberculosis: An abundant and functionally diverse class of cell envelope components. FEMS Microbiol Rev. 2004;28:645–659. doi: 10.1016/j.femsre.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Binnie C, et al. Isolation and characterization of two genes encoding proteases associated with the mycelium of Streptomyces lividans 66. J Bacteriol. 1995;177:6033–6040. doi: 10.1128/jb.177.21.6033-6040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lun S, Bishai WR. Characterization of a novel cell wall-anchored protein with carboxyesterase activity required for virulence in Mycobacterium tuberculosis. J Biol Chem. 2007;282:18348–18356. doi: 10.1074/jbc.M700035200. [DOI] [PubMed] [Google Scholar]
- 9.Lee RE, Brennan PJ, Besra GS. Mycobacterium tuberculosis cell envelope. Curr Top Microbiol Immunol. 1996;215:1–27. doi: 10.1007/978-3-642-80166-2_1. [DOI] [PubMed] [Google Scholar]
- 10.Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 12.Ganz T. Antimicrobial polypeptides in host defense of the respiratory tract. J Clin Invest. 2002;109:693–697. doi: 10.1172/JCI15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keep NH, Ward JM, Cohen-Gonsaud M, Henderson B. Wake up! Peptidoglycan lysis and bacterial non-growth states. Trends Microbiol. 2006;14:271–276. doi: 10.1016/j.tim.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Qamra R, Mande, Coates AR, Henderson B. The unusual chaperonins of Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 2005;85:385–394. doi: 10.1016/j.tube.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Marques MA, Chitale S, Brennan PJ, Pessolani MC. Mapping and identification of the major cell wall-associated components of Mycobacterium leprae. Infect Immun. 1998;66:2625–2631. doi: 10.1128/iai.66.6.2625-2631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenkrands I, et al. Mapping and identification of Mycobacterium tuberculosis proteins by two-dimensional gel electrophoresis, microsequencing and immunodetection. Electrophoresis. 2000;21:935–948. doi: 10.1002/(SICI)1522-2683(20000301)21:5<935::AID-ELPS935>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 17.Henderson B, Allan, Coates AR. Stress wars: The direct role of host and bacterial molecular chaperones in bacterial infection. Infect Immun. 2006;74:3693–3706. doi: 10.1128/IAI.01882-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart GR, et al. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002;148:3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 19.Lewthwaite JC, et al. Mycobacterium tuberculosis chaperonin 60.1 is a more potent cytokine stimulator than chaperonin 60.2 (Hsp 65) and contains a CD14-binding domain. Infect Immun. 2001;69:7349–7355. doi: 10.1128/IAI.69.12.7349-7355.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zugel U, Kaufmann SH. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin Microbiol Rev. 1999;12:19–39. doi: 10.1128/cmr.12.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinnick TM. The 65-kilodalton antigen of Mycobacterium tuberculosis. J Bacteriol. 1987;169:1080–1088. doi: 10.1128/jb.169.3.1080-1088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinke de Wit TF, et al. Mycobacteria contain two groEL genes: The second Mycobacterium leprae groEL gene is arranged in an operon with groES. Mol Microbiol. 1992;6:1995–2007. doi: 10.1111/j.1365-2958.1992.tb01372.x. [DOI] [PubMed] [Google Scholar]
- 23.Qamra R, Mande SC. Crystal structure of the 65-kilodalton heat shock protein, chaperonin 60.2, of Mycobacterium tuberculosis. J Bacteriol. 2004;186:8105–8113. doi: 10.1128/JB.186.23.8105-8113.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flores AR, Parsons LM, Pavelka MS., Jr Characterization of novel Mycobacterium tuberculosis and Mycobacterium smegmatis mutants hypersusceptible to β-lactam antibiotics. J Bacteriol. 2005;187:1892–1900. doi: 10.1128/JB.187.6.1892-1900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koschorreck M, Fischer M, Barth S, Pleiss J. How to find soluble proteins: A comprehensive analysis of alpha/beta hydrolases for recombinant expression in E. coli. BMC Genomics. 2005;6:49. doi: 10.1186/1471-2164-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujii R, Nakagawa Y, Hiratake J, Sogabe A, Sakata K. Directed evolution of Pseudomonas aeruginosa lipase for improved amide-hydrolyzing activity. Protein Eng Des Sel. 2005;18:93–101. doi: 10.1093/protein/gzi001. [DOI] [PubMed] [Google Scholar]
- 27.Gey Van Pittius NC, et al. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0044. RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallen MJ. The ESAT-6/WXG100 superfamily–and a new Gram-positive secretion system?. Trends Microbiol. 2002;10:209–212. doi: 10.1016/s0966-842x(02)02345-4. [DOI] [PubMed] [Google Scholar]
- 29.Sassetti C, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci USA. 2001;98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen BW. In: Mycobacteria Protocols. Parish T, Stoker NG, editors. Totowa, NJ: Humana; 1998. pp. 15–31. [Google Scholar]
- 31.Parish T, Wheeler PR. In: Mycobacteria Protocols. Parish T, Stoker NG, editors. Totowa, NJ: Humana; 1998. pp. 77–90. [Google Scholar]