Abstract
The accessory subunit of the heterodimeric mtDNA polymerase (polγ) from Drosophila embryos is required to maintain the structural integrity or catalytic efficiency of the holoenzyme. cDNAs for the accessory subunit from Drosophila, man, mouse, and rat have been identified, and comparative sequence alignment reveals that the C-terminal region of about 120 aa is the most conserved. Furthermore, we demonstrate that the accessory subunit of animal polγ has both sequence and structural similarity with class IIa aminoacyl-tRNA synthetases. Based on sequence similarity and fold recognition followed by homology modeling, we have developed a model of the three-dimensional structure of the C-terminal region of the accessory subunit of polγ. The model reveals a rare five-stranded β-sheet surrounded by four α-helices with structural homology to the anticodon-binding domain of class IIa aminoacyl-tRNA synthetases. We postulate that the accessory subunit plays a role in the recognition of RNA primers in mtDNA replication, to recruit polγ to the template–primer junction. A similar role is served by the γ-complex in Escherichia coli DNA polymerase III, and indeed our accessory subunit model shows structural similarity with the N-terminal domain of the δ′ subunit of the γ-complex. Structural similarity is also found with E. coli thioredoxin, the accessory subunit and processivity factor in bacteriophage T7 DNA polymerase. Thus, we propose that the accessory subunit of polγ is involved both in primer recognition and in processive DNA strand elongation.
mtDNA polymerase (polγ) is the key enzyme involved in the replication of the mtDNA genome, and its catalytic features appear to be conserved from yeast to man. In studying Drosophila as an animal model of mitochondrial function, we have shown that Drosophila polγ is a heterodimer of catalytic and accessory subunits of 125 and 35 kDa, respectively (1, 2). The large catalytic subunit contains both 5′ → 3′ DNA polymerase and 3′ → 5′ exonuclease activities (3). Typical of replicative DNA polymerases, the two-subunit Drosophila polγ is both highly processive and highly accurate in nucleotide polymerization (4–6). With the high processivity determined in in vitro biochemical studies, Drosophila polγ could replicate the entire mtDNA molecule on binding of the initiating primer on each DNA strand. Thus, the recruitment of polγ to the initiating primer and maintenance of enzyme processivity may be critical for efficient and faithful mtDNA replication.
To elucidate structure–function relationships in Drosophila polγ, we have cloned cDNAs for its subunits (3, 7). The catalytic subunit shares a high degree of sequence similarity with other DNA polymerase catalytic cores, both from the polγ family (3, 8) and from other members of the family A class of DNA polymerases that includes the two-subunit bacteriophage T7 DNA polymerase (9). In contrast, we found that the cDNA for the accessory subunit encodes a novel protein (7). We have identified human, mouse, and rat homologs of the Drosophila cDNA, providing evidence for a common heterodimeric structure among animal polγs (7).
In parallel with biochemical studies of polγ and its isolated subunits, we have pursued molecular modeling of the accessory subunit. Here we report a striking structural similarity between the C-terminal region of the accessory subunit of Drosophila polγ and those of several class IIa aminoacyl (aa)-tRNA synthetases (RSs). That this domain has been shown to be involved in tRNA binding in the latter suggests the accessory subunit of polγ plays a role in recognition of the unusual RNA primers generated at the mtDNA replication origin (10) to guide the catalytic subunit to the primer terminus. In addition, the accessory subunit model shows structural homology with thioredoxin, the accessory subunit of T7 DNA polymerase, despite their lack of amino acid sequence homology. Thus, we predict a dual role for the accessory subunit as a processivity clamp and as a primer recognition factor/clamp loader in mtDNA replication.
MATERIALS AND METHODS
Sequence Analysis of the Accessory Subunit of Drosophila polγ.
The SwissProt protein sequence database was searched by using the blast algorithm (11) through the National Center for Biotechnology Information web server (http://www.ncbi.nlm.nih.gov/) for homologs to the sequence of the accessory subunit of polγ (7). Sequence alignments were obtained with the gap and bestfit programs of the University of Wisconsin Genetics Computer Group software package (GCG) (12) by using the Needleman and Wunsch algorithm (with gap weight = 5 and length weight = 4). Residues 254–361 of the accessory subunit of polγ (polγ-β) were aligned by using gap to residues 288–386 of the sequence from the crystal structure of the Thermus thermophilus prolyl tRNA synthetase (Pro-RS; generously provided by S. Cusack of European Molecular Biology Laboratory, Grenoble, France) (13), corresponding to an independent folding domain in Escherichia coli pro-RS identified by the blast search as being similar to the polγ-β sequence. Minor modifications of this alignment were made so that insertions and deletions fell into regions between secondary structures.
Structural Modeling of the Accessory Subunit.
Initial prediction of the three-dimensional (3D) fold of the accessory subunit was obtained from the Protein Fold Recognition server (http://www.doe-mbi.ucla.edu/people/frsvr/preds.html) based on the algorithm of Fischer and Eisenberg (14). This approach uses both the secondary structure assigned to the sequence by phdsec (15) (http://www.embl-heidelberg.de/predictprotein/predictprotein.html) and the Gonnet amino acid substitution matrix to assess the similarity between the probe sequence (e.g., the accessory subunit) and sequences from the Protein Data Bank (PDB) (16, 17) of 3D protein structures. The use of both primary and predicted secondary structures in the search enhances its sensitivity in identifying the protein with the most similar 3D fold to that of the probe. When tested on a set of 68 proteins whose 3D structures were known but ignored, the fold recognition server can recognize successfully the 3D fold of a new sequence in 71% of the cases (14).
3D modeling was carried out by using Molecular Simulations’ (Waltham, MA) insightII and biopolymer software on a Silicon Graphics (Mountain View, CA) Indigo2 graphics workstation. By using the sequence alignment between the accessory subunit of Drosophila melanogaster polγ and the 2.4-Å crystal structure of residues 288–386 of T. thermophilus pro-RS, the backbone of pro-RS was used as a structural template, and side chains differing in polγ-β were substituted individually. For two of the three short loops between regular secondary structures that differ between polγ-β and pro-RS, the searchloop function in biopolymer was used to identify a loop of known structure with appropriate length, geometry, and packing to use as a structural template; for the third loop, between β-strands 4 and 5, the corresponding loop in the structure of T. thermophilus histidyl-RS (PDB entry 1adj) (18) was used as a template, followed by side-chain substitution where necessary. When side-chain substitutions resulted in steric collisions, they were resolved by minimal torsional-angle adjustments in insightII, followed by energy minimization on the loop regions only, by 100 steps of steepest descents minimization with the CVFF consistent valence force field by using discover software (Molecular Simulations, Waltham, MA). The stereochemistry of the final structural model of the C-terminal domain of polγ-β was validated by using procheck (19, 20), and the favorability of amino acid environments within this structure was assessed by using the 3D Profile method (21) via the verify3D server (http://www.doe-mbi.ucla.edu/Services/Verify3D.html). 3D structural comparisons between the accessory subunit model and the available crystallographic structures of thioredoxin and the δ′ subunit of DNA polymerase III were performed with the dali server (22) (http://www.embl-ebi.ac.uk/dali).
RESULTS AND DISCUSSION
Identification of Sequence Homologs of the Accessory Subunit of Polγ.
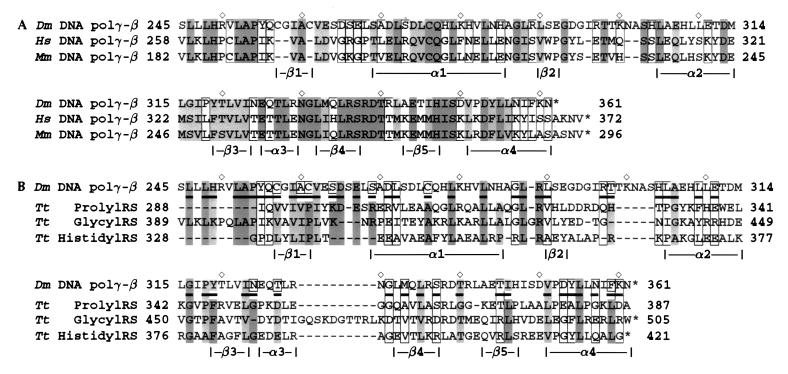
We have identified human (7) (GenBank accession no. U94703), mouse (GenBank accession no. AF006072), and rat (GenBank accession no. AA892950) homologs of the Drosophila cDNA of the accessory subunit of polγ. Comparative sequence alignment reveals that the C-terminal region of ≈120 aa is the most conserved (Fig. 1A).
Figure 1.
Sequence comparisons of the accessory subunit of Drosophila polγ with mammalian homologs and with class IIa aaRSs. (A) Sequence alignment between the C-terminal regions of the accessory subunit of D. melanogaster polγ (Dm) (7) and its homologs from Homo sapiens (Hs) (7) and Mus musculus (Mm, GenBank accession no. AF006072). Residues are shaded based on degree of similarity, with dark shading indicating identical residues and light shading indicating conservative substitutions within the categories of the simplification matrix from the UW-GCG package (12): negatively-charged residues and derivatives (Asp, Asn, Glu, Gln); positively charged residues (His, Lys, Arg); small hydrophobic residues (Leu, Ile, Val, Met); large hydrophobic residues (Phe, Tyr, Trp); Cys; and other residues (Pro, Ala, Gly, Ser, Thr). Open boxes indicate loosely conserved residues, i.e., those conserved as hydrophobic or hydrophilic. ⋄ marks every 10th residue in the Drosophila polγ-β sequence. (B) Comparison of the Drosophila polγ-β sequence with the T. thermophilus (Tt) prolyl-RS (13), glycyl-RS (PDB ID 1ati) (24), and histidyl-RS sequences (PDB ID 1adj) (18). Sequences are shaded as in A, with underlined residues in polγ indicating conservation of hydrophobic/hydrophilic character between the accessory subunit and the RSs.
blast search in the SwissProt database suggested moderate sequence homology between a region of the accessory, or β, subunit of Drosophila polγ (polγ-β) and aaRSs from both prokaryotes and eukaryotes. The most similar was a probable glycyl-tRNA synthetase from Methanococcus jannaschii (SwissProt accession code Q57681), with a score of 84 and likelihood of 0.0018 over two aligned segments, reflecting 31% identity to residues 211–254 of polγ-β and 25% identity to residues 290–355. The top 10 matches, with up to 35% identity over ≈40 residues, were all members of the class II aaRS family (23). The blast sequence alignments (data not shown) between the accessory subunit of Drosophila polγ and these aaRSs were mostly limited to the C terminus (residues 290–360) of polγ-β (Fig. 1B).
Modeling the Accessory Subunit of Polγ Based on Structural Homology with tRNA Synthetases.
To obtain insights into the structure and function of the accessory subunit of polγ, 3D fold recognition (14) was used to identify the best structural match for polγ-β, based on the best alignment between its sequence and secondary structures with those of all the distinct structural folds in the PDB. The major benefit of the fold recognition server in the case of polγ-β was the identification of a structural homolog of the polγ-β protein that was not yet included in sequence databases, and therefore not identified by blast search. By using the sequence of the accessory subunit, the server indicated 3D structural similarity for polγ-β residues 24–359 with residues 73–504 of the glycyl-tRNA synthetase from T. thermophilus (PDB entry 1ati) (24). The Z score was 11.5, more than twice the threshold value (4.8 ± 1.0) for significant similarity.
aaRSs are enzymes that catalyze the attachment of each amino acid to its cognate tRNA (23, 25). The 20 aaRS enzymes can be divided into two different families of 10 members each based on their amino acid sequence and structural similarity, and crystallographic structures have been determined for members of both classes (refs. 13 and 23, and refs. therein). Besides Gly-RS, class II aaRSs include Ser-, Thr-, Pro-, His-, Asp-, Asn-, Lys-, Phe-, and Ala-RS. All are homodimers, with the catalytic domain built around a six-stranded antiparallel β-sheet. Most class II aaRSs have three conserved, class-defined motifs: motifs 2 and 3 are responsible for ATP binding and catalysis, whereas motif 1 is involved in dimerization essential for the enzymatic activity. Based on amino acid sequence comparison, Thr-, Pro-, His-, and most Gly-RS (including Gly-RS from T. thermophilus) have a homologous C-terminal domain attached to the catalytic core identifying them as members of subclass IIa. This domain interacts with the anticodon loop and stem of the tRNA substrate as elucidated by crystallography (13).
The sequence–structure alignments from fold recognition (data not shown) and sequence alignment (Fig. 1B) reveal that residues 254–360 of polγ-β map onto the anticodon-binding domain of class II aaRSs. The match of residues 24–253 of polγ-β to regions of the catalytic core of the synthetase proved more difficult to interpret because critical motifs 2 and 3 in the synthetase were not matched, and generally the alignment was substantially fragmented. Two β-strands in the catalytic core of Gly-RS are absent from polγ-β, which suggests that it would be difficult for the two to fold similarly in this region. In addition, the motif 1 structure in Gly-RS is missing in polγ-β. These results indicate that polγ-β lacks the central core structure for catalysis of tRNA aminoacylation.
Although these results suggest that the accessory subunit of polγ is not enzymatically homologous to the aaRS family, it is likely to share structural homology with the anticodon-binding domain of the synthetases, based on significant sequence identity in this region. Of the known structures of aaRSs, T. thermophilus Pro-RS has the highest sequence identity to polγ-β, with 28% identity (42.7% similarity) between the 81 C-terminal residues of Drosophila polγ-β and residues 302–381, which form the anticodon-binding domain in Pro-RS. The 3D structure of this C-terminal region of polγ-β is therefore expected to have a closely superimposible 3D structure with that of the anticodon-binding domain. This expectation is based on exceeding the degree of sequence identity required for structural similarity found from thousands of pairwise sequence and structural alignments between known protein structures (26) in the PDB. Once an appropriate structural template has been identified based on sequence identity, a homology model may be built by using this template for the main chain and by substituting side chains according to the sequence alignment.
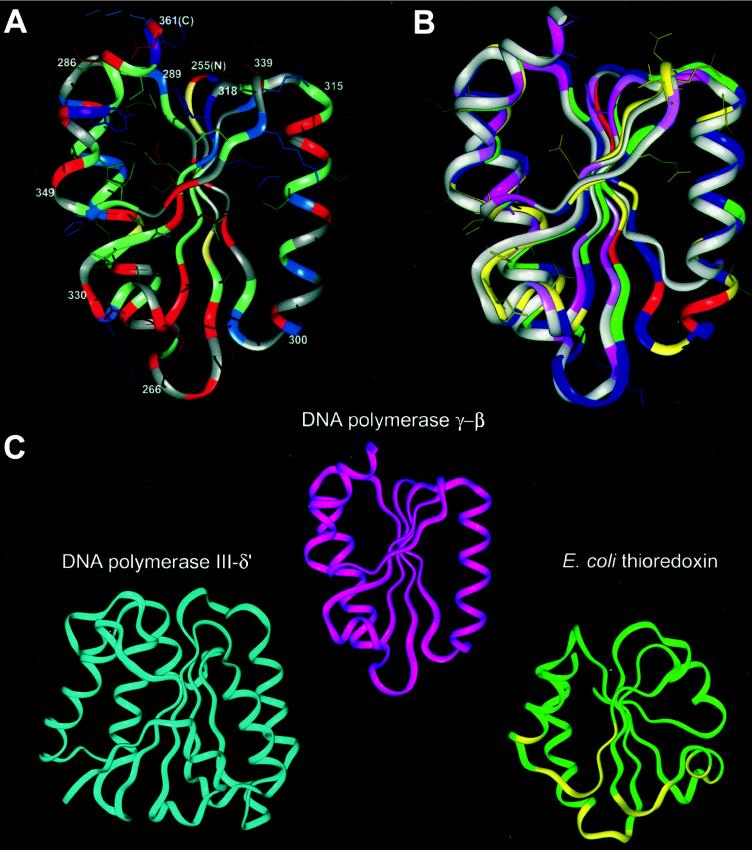
We used the anticodon-binding domain of T. thermophilus Pro-RS (coordinates provided by S. Cusack, European Molecular Biology Laboratory, Grenoble, France) as the structural template to build a homology model of the 3D structure of the C-terminal domain of the accessory subunit of polγ. Amino acid sequence comparison (Fig. 1A) indicated that the C-terminal domain of the accessory subunit of Drosophila polγ is also the most conserved region relative to its human and murine homologs, supporting its functional importance. Modeling was based on the bestfit sequence alignment between polγ-β and Pro-RS (Fig. 1B) and adjusted so that deletions or insertions fell in loop regions rather than in regular secondary structures. Residues 288–301 of Pro-RS were aligned manually to polγ-β based on the fold recognition alignment between Gly-RS and polγ-β. The aligned region shows 23–29% pairwise sequence identity between the aaRS sequences (Fig. 1B). Based on this alignment of Pro-RS to polγ-β, we used insightII to substitute polγ-β side chains on the Pro-RS main-chain structure, followed by side-chain and loop optimization (as described in Materials and Methods). The main-chain of the resulting model of the C-terminal region of polγ-β (residues 255–361; Fig. 2A) is the same as that of the anticodon binding domain of Pro-RS (0.21 Å rms deviation), except for three loop insertions: loop 1 between strand β1 and helix α1, loop 2 between β2 and α2, and loop 3 between β4 and β5 (Fig. 1B). This structural model was validated by using two techniques, procheck, which evaluates stereochemistry (favorable bond lengths, angles, and atomic packing), and verify3d, which assesses the favorability of residue environments within the folded structure. procheck found all residues to have favored or highly favored main-chain dihedral (Φ, Ψ) angles, and the model to have significantly better overall stereochemistry (G factor of 0.07) than typical crystal structures solved at the resolution of the Pro-RS structure (2.4 Å). Evaluation of residue environments with verify3d gave the accessory subunit model structure a score matching those of higher resolution (2.0 Å) crystal structures.
Figure 2.
Structural modeling and comparison of the C-terminal domain of the accessory subunit of Drosophila polγ with other DNA polymerase accessory subunits. (A) Molecular model of the C-terminal domain of Drosophila polγ-β. The main-chain ribbon is shown along with protein side-chain atoms colored according to chemistry: negatively charged residues and derivatives (Asp, Asn, Glu, Gln), red; positively charged residues (His, Lys, Arg), blue; small hydrophobic residues (Leu, Ile, Val, Met), green; large hydrophobic residues (Phe, Tyr, Trp), purple; cysteine, yellow; and other residues (Pro, Ala, Gly, Ser, Thr), gray. The termini of the α-helices and β-strands are numbered according to the Drosophila polγ-β sequence in Fig. 1, with the N and C termini labeled as N and C. (B) Comparison of the structures of the modeled C-terminal domain of Drosophila polγ-β and the C-terminal domain of prolyl-RS, used as a template for the polγ-β model. The main-chain structure of the model, oriented as in A, is shown superimposed onto the anticodon-binding domain of T. thermophilus prolyl-RS. The accessory subunit is colored according to sequence conservation between the Drosophila and human sequences; residues conserved between the two sequences are shown in yellow along with their surface-accessible side chains, conservatively substituted residues are in green, nonconserved residues are in blue, and insertions in the fly sequence relative to the human sequence are in red. (There are no deletions in the fly sequence relative to human.) Conservation is defined as in Fig. 1. The prolyl-RS main-chain ribbon is colored by sequence conservation among the three related aaRSs shown in Fig. 1, with conserved residues in dark pink, similar residues in light pink, and nonconserved residues in grey. (C) Comparison of the structures of the accessory subunit of polγ, the δ′-subunit of E. coli DNA polymerase III, and E. coli thioredoxin. The N-terminal domain of DNA polymerase III δ′ (PDB ID 1a5t; ref. 36) is shown in blue, polγ-β in magenta, and thioredoxin (PDB ID 1t7p, chain B) (40) in green, with thioredoxin residues interacting with the catalytic subunit of T7 DNA polymerase shown in yellow. Optimally similar orientations were produced by using dali (22, 47).
The structural model of the C-terminal domain of the accessory subunit of Drosophila polγ reveals a rare α/β-fold comprising a five-stranded mixed β-sheet surrounded by four α-helices (Fig. 2A) found only in the anticodon-binding domains of class IIa aaRSs (13). The anticodon-binding pocket of the class II RSs, which faces the viewer in the lower half of Fig. 2B, is formed predominantly by the central β-sheet and by α-helices 2 and 3 (13); this β-sheet appears at center, with helix 2 appearing as the long helix at right and helix 3 as the short helix at lower left. Whereas residue conservation is often associated with side chains that bind substrates or ligands, and thus the anticodon-binding pocket might be expected to be conserved among aaRSs and between aaRSs and the accessory subunit, this is explicitly not the case. Different tRNA synthetases must recognize different anticodons, and the accessory subunit of polγ must recognize at least two distinct primers; thus, their ligand-binding residues are expected to vary to confer this specificity. Of the residues identical in the aligned region between polγ-β and Pro-RS (Fig. 1B), half are surface-exposed and half are somewhat or completely buried in the protein and do not colocalize to a specific region of the structure; for the additional residues that are chemically similar but not identical, ≈two thirds are exposed and the remainder are buried. Conserved residues at the surface of this domain in Pro-RS are not associated with an interface with the region N-terminal to this domain because the two domains are connected only by a loose tether in class IIa synthetases. Conserved surface-exposed residues of polγ-β (yellow side chains in Fig. 2B) may be involved in interactions with the N-terminal domain of the accessory subunit or with the catalytic subunit or nucleic acid substrates. An RNA-binding structural fold in the C terminus of the accessory subunit corresponding to that in class IIa aaRSs was unanticipated but is consistent with a function of the accessory subunit in interacting with RNA molecules containing tRNA-like structures.
The modeled region of the accessory subunit also includes two previously noted sequence motifs, a putative zinc finger and a putative leucine zipper (7). The segment containing the leucine zipper sequence in the model structure (residues 319–347) consists of one α-helical segment and two short β-strands, and thus it cannot function as a leucine zipper. The segment for the putative zinc-finger motif in polγ-β (residues 256–286) consists of a β-strand, a loop, and an α-helix, which is similar to the structure observed for some known zinc fingers (27). However, the side chains of Cys and His residues in the polγ-β structural model are not positioned such that they could bind zinc.
Biochemical Implications of the Structural Model.
The accessory subunit of polγ as a specialized primer recognition factor. mtDNA replication is thought to involve different priming mechanisms at the two origins. The leading DNA strand origin is located within the major noncoding region of the mtDNA genome in all animals examined to date (28). In man and mouse, transcripts initiated from the L strand promoter are processed for use as primers for leading DNA strand synthesis; RNase mitochondrial RNA processing cleaves the transcripts at one of several discrete sites, ≈100 nucleotides from the promoter (29, 30). The 3′ termini of these processed transcripts are then presumably elongated by polγ, but data regarding the mechanism of polγ-binding at the template–primer junction are lacking. Our structural model suggests that the accessory subunit may play a role in the recognition of these unusual RNA–DNA hybrids to recruit polγ to the template–primer junction. In particular, we propose that by virtue of its conserved C-terminal domain, homologous to the anticodon-binding domain in class IIa aaRSs, the accessory subunit recognizes secondary structural features in the processed RNA, thereby enhancing the template–primer binding affinity of polγ.
A similar mode of RNA–DNA hybrid recognition by polγ may occur at the lagging DNA strand origin. Priming of lagging DNA strand synthesis in mammals appears to be catalyzed by a primase (31) that recognizes a specific stem–loop structure formed in the displaced parental strand (32). Because the lagging DNA strand initiation site is nested within a cluster of five tRNA genes, the involvement of tRNA molecules cannot be excluded. Avian mtDNA genomes lack the noncoding sequence within the tRNA cluster that contains the stem–loop structure, suggesting a replication function for the tRNA genes (or a corresponding tRNA) (33). In Drosophila, the lagging DNA strand initiation site has been mapped within the noncoding A+T region, and in several species a stem–loop structure has been identified that may serve a replication function (34). Thus, although only in mammals has the stem–loop structure been documented to function in lagging DNA strand initiation, the situation may be similar in birds and in Drosophila. If so, the function of the accessory subunit of polγ in specialized primer recognition is likely very similar in both leading and lagging strand initiation in mtDNA replication and is generally conserved in animals.
The postulated role of the accessory subunit of polγ as a primer recognition factor parallels the function of the nonhomologous γ complex of E. coli DNA polymerase III and eukaryotic replication factor C. In contrast to the unidirectional and asymmetric mode of mtDNA replication, chromosomal replication in both prokaryotes and eukaryotes proceeds bidirectionally with symmetric synthesis of leading and lagging DNA strands. This involves multiple cycles of recognition of short oligonucleotide primers by the γ-complex and replication factor C, respectively. The γ-complex and replication factor C are multisubunit proteins that share both sequence and structural similarity (35, 36). Our structural model of the C terminus of the accessory subunit of polγ shows some similarity with the crystal structure determined for the δ′ subunit of the γ-complex, which forms a C shaped structure comprising three domains (36) (Fig. 2C Left). Although the N-terminal domain of δ′ is not highly similar to the C-terminal domain of polγ-β based on dali comparison (Z score of 1.0, below the significance threshold of 2.0), they are qualitatively similar, sharing an α/β-fold with a central β-sheet. This is consistent with our hypothesis of the accessory subunit as a primer-recognition factor and may also indicate a general structure–function theme in primer recognition factors in various replicative systems.
The accessory subunit of polγ as a processivity clamp.
The structural and functional features of Drosophila polγ are similar to those of the bacteriophage T7 DNA polymerase. The T7 DNA polymerase and polγ catalytic cores are members of the family A group of DNA polymerases (9). T7 DNA polymerase has a low intrinsic processivity, dissociating from the DNA after incorporation of only a few nucleotides (37). On infection, T7 DNA polymerase recruits a host-encoded protein, E. coli thioredoxin (12 kDa), to form a heterodimer that is capable of polymerizing thousands of nucleotides without dissociation (37). Likewise, we have found that the heterodimeric polγ is a highly processive enzyme (6). In the E. coli and eukaryotic replicative systems, DNA polymerase III and DNA polymerase δ use ring-shaped accessory subunit complexes, β (38) and proliferating-cell nuclear antigen (39), respectively, to convert them into highly processive enzymes. These “processivity clamps” are directed to the template-primer by the γ-complex and replication factor C, discussed above, which actually serve dual roles in primer recognition and clamp loading.
The recently determined crystal structure of a T7 DNA polymerase–thioredoxin–DNA complex revealed that thioredoxin is not a ring-shaped protein (40). Instead, thioredoxin binds to the tip of the thumb in the T7 DNA polymerase structure and from this position presumably clamps the template-primer. Corresponding biochemical studies indicate that this association results in an ≈80-fold increase in the affinity of T7 DNA polymerase for the primer terminus (37). Thus, the accessory subunit of T7 DNA polymerase apparently serves a dual function in primer binding and processivity enhancement. In fact, the mechanism of initiation at the T7 primary DNA replication origin shares similarity with that at the leading DNA strand origin in mitochondrial replication. In T7 replication, the T7 RNA polymerase initiates transcription at a site upstream of the origin and proceeds through it; T7 DNA polymerase then displaces the RNA polymerase and extends the 3′ end of the transcript (41). We find that although the accessory subunit of polγ and thioredoxin have low sequence similarity, our structural model of the C terminus of the accessory subunit is quite similar to the structure of thioredoxin (Fig. 2C Right), which consists of a central core of five β-strands surrounded by four helices. Although thioredoxin (PBD ID 1t7p) and the C-terminal domain of polγ-β do not have identical fold topologies, dali structural alignment indicates significant structural similarity between them (Z score of 2.7 over 71 residues).
Herpes simplex virus type 1 DNA polymerase is also a two-subunit enzyme (42, 43). The catalytic subunit is a member of the family B class of DNA polymerases (9). Its accessory subunit, the processivity factor UL42, binds double-stranded DNA (44). Comparative sequence analysis reveals that residues 115–174 of UL42 bear 35% identity with the anticodon-binding domain (residues 329–383) of T. thermophilus ProRS (data not shown). Furthermore, UL42 residues 80–289 aligned with polγ-β residues 105–300, and of the 45 of these residues contained in the structural model, 31% are identical. Residues 129–163 and 202–337 in UL42 are required for stimulation of DNA polymerase activity (45), and residues within both regions are also required for DNA binding by UL42 (46). Taken together, these findings support our hypothesis of the role of the accessory subunit in polγ function.
In summary, the C-terminal domain of the accessory subunit of Drosophila polγ reveals a rare α/β-fold topology comprising a five-stranded mixed β-sheet surrounded by four α-helices, that is found only in the anticodon-binding domains of class IIa aaRSs (13). This structure may endow polγ-β with the capacity to bind RNA molecules with tRNA-like structures, as in aaRSs. This and the structural similarity with the δ′ subunit of E. coli DNA polymerase III, and particularly with thioredoxin, suggest that the accessory subunit may play a dual role in loading polγ onto the primer terminus and in enhancing its processivity. Considering that all of the proteins required for mtDNA replication are the products of nuclear genes, it is not surprising that the mitochondrion has evolved to require only a single polypeptide to perform the functions provided by both the multisubunit clamp loaders and processivity clamps in prokaryotic and nuclear replicative systems. Furthermore, this conservative mechanism for a cellular DNA polymerase can likely be generalized to animal polγs, because our sequence-based structural comparisons indicate that the proposed C-terminal structure is conserved in the accessory subunits from fly to man. Baculovirus overexpression shows that the accessory subunit is largely insoluble when expressed independently, whereas a purified form of the catalytic subunit alone exhibits a very low specific activity (48). Thus, the apparently critical functions of the accessory subunit may be assessed in mutant holoenzyme derivatives.
Acknowledgments
We thank Dr. Stephen Cusack (European Molecular Biology Laboratory, Grenoble, France) for generously providing structural coordinates for the anticodon binding domain of prolyl-RS and for comments on the manuscript. We also thank Dr. Michael Kron (Michigan State University) for sharing his expertise on aaRSs and for reviewing the manuscript. L.F. thanks Ya He for her enthusiastic support. This research was supported by National Institutes of Health Grant GM45295 to L.S.K. and an American Heart Association Established Investigatorship to L.A.K.
ABBREVIATIONS
- 3D
three dimensional
- PDB
Protein Data Bank
- RS
tRNA synthetase
- aaRS
aminoacyl-RS
- polγ
mtDNA polymerase
References
- 1.Wernette C M, Kaguni L S. J Biol Chem. 1986;261:14764–14770. [PubMed] [Google Scholar]
- 2.Olson M W, Wang Y, Elder R H, Kaguni L S. J Biol Chem. 1995;270:28932–28937. doi: 10.1074/jbc.270.48.28932. [DOI] [PubMed] [Google Scholar]
- 3.Lewis D L, Farr C L, Wang Y, Lagina A T, III, Kaguni L S. J Biol Chem. 1996;271:23389–23394. doi: 10.1074/jbc.271.38.23389. [DOI] [PubMed] [Google Scholar]
- 4.Wernette C M, Conway M C, Kaguni L S. Biochemistry. 1988;27:6046–6054. doi: 10.1021/bi00416a033. [DOI] [PubMed] [Google Scholar]
- 5.Kaguni L S, Wernette C M, Conway M C, Yang-Cashman P. In: Cancer Cells: Eukaryotic DNA Replication. Kelly T, Stillman B, editors. Vol. 6. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 425–432. [Google Scholar]
- 6.Williams A J, Wernette C M, Kaguni L S. J Biol Chem. 1993;268:24855–24862. [PubMed] [Google Scholar]
- 7.Wang Y, Farr C L, Kaguni L S. J Biol Chem. 1997;272:13640–13646. doi: 10.1074/jbc.272.21.13640. [DOI] [PubMed] [Google Scholar]
- 8.Lecrenier N, Van Der Bruggen P, Foury F. Gene. 1997;185:147–152. doi: 10.1016/s0378-1119(96)00663-4. [DOI] [PubMed] [Google Scholar]
- 9.Braithwaite D K, Ito J. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee D Y, Clayton D A. J Biol Chem. 1998;273:30614–30621. doi: 10.1074/jbc.273.46.30614. [DOI] [PubMed] [Google Scholar]
- 11.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 12.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cusack S, Yaremchuk A, Krikliviy I, Tukalo M. Structure (London) 1998;6:101–108. doi: 10.1016/s0969-2126(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 14.Fischer D, Eisenberg D. Protein Sci. 1996;5:947–955. doi: 10.1002/pro.5560050516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rost B, Sander C. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 16.Sussman J L, Lin D, Jiang J, Manning N O, Prilusky J, Ritter O, Abola E E. Acta Crystallogr D. 1998;54:1078–1084. doi: 10.1107/s0907444998009378. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein F C, Koetzle T F, Williams G J, Meyer E F, Jr, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. Eur J Biochem. 1977;80:319–324. doi: 10.1111/j.1432-1033.1977.tb11885.x. [DOI] [PubMed] [Google Scholar]
- 18.Aberg A, Yaremchuk A, Tukalo M, Rasmussen B, Cusack S. Biochemistry. 1997;36:3084–3094. doi: 10.1021/bi9618373. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 20.Morris A L, MacArthur M W, Hutchinson E G, Thornton J M. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 21.Luthy R, Bowie J U, Eisenberg D. Nature (London) 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 23.Cusack S. Nat Struct Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 24.Logan D T, Mazauric M H, Kern D, Moras D. EMBO J. 1995;14:4156–4167. doi: 10.1002/j.1460-2075.1995.tb00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnez J G, Moras D. Trends Biochem Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 26.Sander C, Schneider R. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 27.Pavletich N P, Pabo C O. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 28.Clayton D A. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 29.Chang D D, Clayton D A. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- 30.Lee D Y, Clayton D A. Genes Dev. 1997;11:582–592. doi: 10.1101/gad.11.5.582. [DOI] [PubMed] [Google Scholar]
- 31.Wong T W, Clayton D A. J Biol Chem. 1985;260:11530–11535. [PubMed] [Google Scholar]
- 32.Wong T W, Clayton D A. Cell. 1985;42:951–958. doi: 10.1016/0092-8674(85)90291-0. [DOI] [PubMed] [Google Scholar]
- 33.Desjardins P, Morais R. J Mol Biol. 1990;212:599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- 34.Clary D O, Wolstenholme D R. J Mol Evol. 1987;25:116–125. doi: 10.1007/BF02101753. [DOI] [PubMed] [Google Scholar]
- 35.O’Donnell M, Onrust R, Dean F B, Chen M, Hurwitz J. Nucleic Acids Res. 1993;21:1–3. doi: 10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guenther B, Onrust R, Sali A, O’Donnell M, Kuriyan J. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 37.Huber H E, Tabor S, Richardson C C. J Biol Chem. 1987;262:16224–16232. [PubMed] [Google Scholar]
- 38.Kong X P, Onrust R, O’Donnell M, Kuriyan J. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 39.Krishna T S, Fenyo D, Kong X P, Gary S, Chait B T, Burgers P, Kuriyan J. J Mol Biol. 1994;241:265–268. doi: 10.1006/jmbi.1994.1495. [DOI] [PubMed] [Google Scholar]
- 40.Doublie S, Tabor S, Long A M, Richardson C C, Ellenberger T. Nature(London) 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 41.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 42.Gottlieb J, Marcy A I, Coen D M, Challberg M D. J Virol. 1990;64:5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez T R, Lehman I R. J Biol Chem. 1990;265:11227–11232. [PubMed] [Google Scholar]
- 44.Gallo M L, Jackwood D H, Murphy M, Marsden H S, Parris D S. J Virol. 1988;62:2874–2883. doi: 10.1128/jvi.62.8.2874-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monahan S J, Barlam T F, Crumpacker C S, Parris D S. J Virol. 1993;67:5922–5931. doi: 10.1128/jvi.67.10.5922-5931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chow C S, Coen D M. J Virol. 1995;69:6965–6971. doi: 10.1128/jvi.69.11.6965-6971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holm L, Sander C. Nucleic Acids Res. 1994;22:3600–3609. [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, Y. & Kuhn, L. S. (1999) J. Biol. Chem. 274, in press.