Abstract
For 50 years since their discovery, the malaria parasite liver stages (LS) have been difficult to analyze, impeding their utilization as a critical target for antiinfection vaccines and drugs. We have undertaken a comprehensive transcriptome analysis in combination with a proteomic survey of LS. Green fluorescent protein-tagged Plasmodium yoelii (PyGFP) was used to efficiently isolate LS-infected hepatocytes from the rodent host. Genome-wide LS gene expression was profiled and compared with other parasite life cycle stages. The analysis revealed ≈2,000 genes active during LS development, and proteomic analysis identified 816 proteins. A subset of proteins appeared to be expressed in LS only. The data revealed exported parasite proteins and LS metabolic pathways including expression of FASII pathway enzymes. The FASII inhibitor hexachlorophene and the antibiotics, tetracycline and rifampicin, that target the apicoplast inhibited LS development, identifying FASII and other pathways localized in the apicoplast as potential drug targets to prevent malaria infection.
Keywords: drug targeting, fatty acid synthesis, Plasmodium
Malaria, caused by intracellular Plasmodium parasites, remains a devastating disease. More than 500 million clinical cases occur each year (1), and millions of people die in developing countries. Efforts to control malaria have been foiled by the complex biology and ecology of parasites and their Anopheline vectors, drug-resistant parasite strains (2), and sophisticated parasite immune evasion strategies during blood-stage replication (3). The initial asymptomatic LS of Plasmodium are considered the most promising targets for vaccines and also good targets for prophylactic drugs, because parasite numbers are limited, and successful intervention at this stage prevents infection (4). However, despite decades of efforts, a fully protective recombinant malaria vaccine has remained elusive. The most advanced malaria vaccine, RTS/S, based on the circumsporozoite protein (CSP), conferred only limited protection against malaria infection (5). Strikingly, however, complete and long-lasting protection against human experimental malaria infection is achievable with live attenuated parasites, which can invade hepatocytes but cannot complete LS development (6). The antigens targeted by protective immune responses remain largely unidentified.
Because of their apparent distinct metabolism, LS are not susceptible to blood-stage antimalaria drugs such as chloroquine. One licensed drug, primaquine, targets LS but is contraindicated in pregnant women, a population with high malaria-related mortality, as well as individuals with glucose-6-phosphate dehydrogenase deficiency (7). Furthermore, Plasmodium vivax malaria parasites can lie dormant in the liver and relapse to blood infection after months or even years and cannot be targeted by any licensed drug except primaquine (7).
The sequencing of the human Plasmodium falciparum parasite genome (8) and rodent malaria model genomes (9, 10), in conjunction with subsequent comprehensive gene and protein expression surveys (10–13), gave great hope that effective rational strategies for vaccine and drug target identification were achievable. However, LS remained excluded from these studies because of the insurmountable obstacles that prevented access to this rare parasite form (14). Consequently, very little is known of the gene and protein expression during LS development. An EST library derived from laser-captured LS suggested expression of ≈600 genes (15). However, laser-capture microdissection (LCM) will be difficult to apply to early LS and will not produce adequate amounts of material for proteomic analysis. Additionally, an EST library from axenically transformed sporozoites has been published (16).
To overcome the obstacles for analyzing gene expression and protein composition of LS in their host cell environment, we have recently generated a Plasmodium yoelii rodent malaria parasite line that expresses GFP (PyGFP) during LS development, allowing efficient isolation of LS-infected hepatocytes from mouse infections using fluorescence activated cell sorting (17). Here, we used purified LS-infected hepatocytes to conduct a direct comprehensive transcriptome and proteome analysis of the asymptomatic initial malaria parasite infection in the liver.
Results and Discussion
Comprehensive LS Gene Expression Analysis.
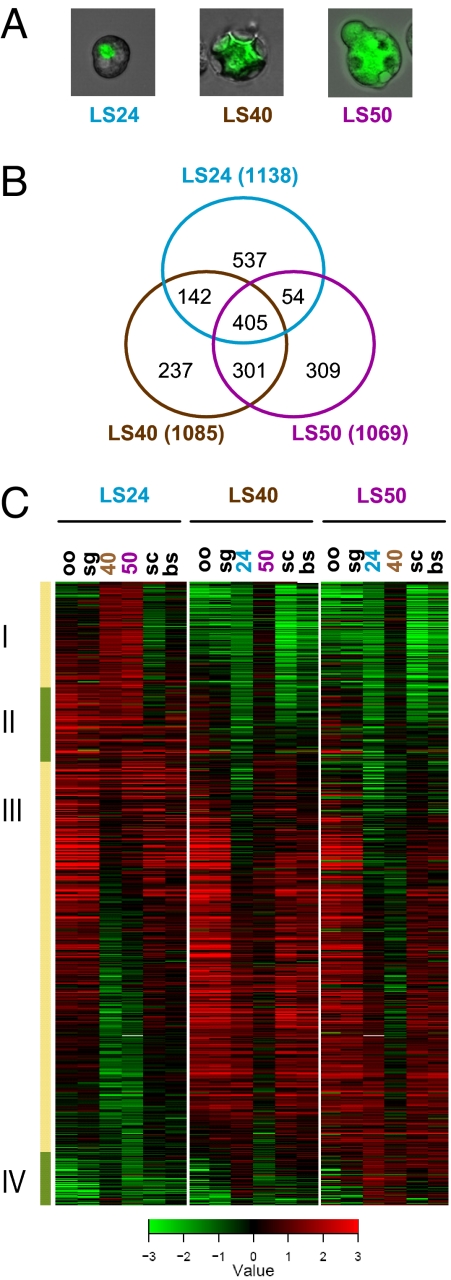
We used an oligonucleotide (65-mer) microarray, designed based on the annotated ORFs of the rodent malaria parasite P. yoelii (9), to analyze gene expression at three time points of LS development in vivo. LS-infected hepatocytes were obtained directly from infected mice at 24 h postinfection (pi), when the majority of LS parasites are at a small early schizont stage and have undergone two to four rounds of nuclear division (LS24); at 40 h pi, when the majority of LS parasites are at large late schizont stage and have undergone up to 13 rounds of nuclear division (LS40); and at 50 h pi, when the majority of LS parasites have commenced merozoite formation (LS50) [Fig. 1A; supporting information (SI) Fig. 5]. The gene expression of LS was compared with two mosquito stages (midgut-oocyst sporozoites and salivary gland sporozoites) and intraerythrocytic stages (mixed blood stages and blood-stage schizonts). Overall, the gene expression profiles of the three LS developmental time points were more similar to each other than to any other life cycle stage. Gene expression among LS40 and LS50 was more similar to each other than to LS24 (data not shown). The mammalian LS and intraerythrocytic stages were more similar to each other than to the mosquito-stage sporozoites, possibly reflecting differences in host environment and also in invasive vs. replicative lifestyles (data not shown). To identify genes active in LS, we considered genes that showed ≥2-fold expression level changes and a false-discovery rate threshold of 0.01 during any of the three time points of LS development when compared with the other stages assayed. Each of the three LS up-regulated >1,000 genes (Fig. 1B); ≈400 of these genes were up-regulated at all three time points, whereas ≈500 genes showed up-regulation in two of the three time points. In total, our analysis identified 1,985 P. yoelii genes in the LS active transcriptome, 66% of which have orthologs in P. falciparum (Table 1; SI Table 3). Almost half of the LS-expressed genes encoded hypothetical proteins. Many of the hypothetical genes have orthologs present in other Plasmodium species (hypothetical conserved; Table 1). LS up-regulated genes in each of the developmental stages examined were clustered by using a hierarchical algorithm (Fig. 1C). Organized in this manner, a developmental series of gene expression patterns that varied mainly in the timing and the duration for which the gene was active were discerned. Within the continuous pattern of expression, four main clusters of gene expression were recognized (Fig. 1C; SI Table 3). The first cluster (I) included LS genes mainly up-regulated in LS24 compared with LS40 and LS50, but that were also expressed in the mosquito and erythrocytic stages. Cluster II encompassed genes that were mainly up-regulated at LS24. Cluster III showed most of the genes up-regulated in all three LS time points queried. The last group (IV) showed genes mainly up-regulated in LS40 and LS50.
Fig. 1.
Comprehensive transcriptional profile of LS active genes. (A) Closeup images of LS-infected hepatocytes for each time point are shown. (B) Venn diagram of the overlap of LS active genes detected from LS24, LS40, and LS50. (C) Gene expression profiles of LS active genes show four clusters of peak expression. The heat map shows the expression ratios of each LS sample compared with all of the other life stages: oo, oocyst sporozoite; sg, salivary gland sporozoite; 24, LS24; 40, LS40; 50, LS50; bs, mixed blood stages; and sc, blood-stage schizonts. Green indicates down-regulation, whereas red indicates up-regulation of the gene in the LS sample compared with the other stages.
Table 1.
Summary of annotations for genes and proteins in LS
| Category | P. yoelii genes probed in microarray (% of total) | LS transcriptome (% of total) | LS proteome(% of total) |
|---|---|---|---|
| Plasmodium ortholog | 4,339 (68%) | 1,436 (72%) | 672 (94%) |
| P. falciparum ortholog | 3,792 (59%) | 1,305 (66%) | 654 (92%) |
| Hypothetical | 3,386 (53%) | 945 (48%) | 164 (23%) |
| Conserved hypothetical | 2,159 (34%) | 611 (31%) | 147 (21%) |
| Signal peptide | 839 (13%) | 312 (16%) | 93 (13%) |
| Transmembrane domain | 1,670 (26%) | 468 (24%) | 76 (11%) |
| Total number | 6,428 | 1,985 | 712 |
There are 104 additional proteins, which were identified by searching against the P. berghei sequence database, but their P. yoelii orthologs were not mapped in the study.
The LS Proteome.
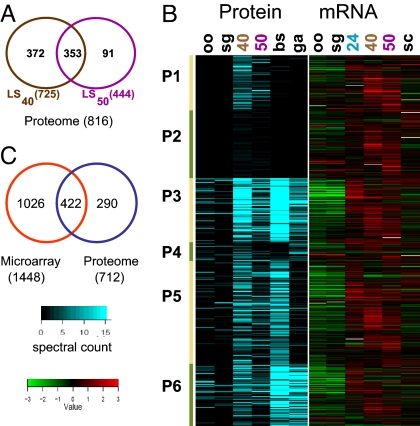
The availability of isolated LS-infected hepatocytes enabled proteomic analysis of the Plasmodium parasite in the liver host environment in vivo. A proteome survey of ≈40,000 LS40- and ≈40,000 LS50-infected hepatocytes was carried out by using a 1D LC/MS/MS shotgun proteomic approach. A majority of the proteins detected in LS50 were also detected in LS40, so henceforth we will refer to the combined detected proteins as the LS schizont proteome (Fig. 2A). We identified 712 LS schizont proteins by interrogating the P. yoelii annotated protein database and an additional 104 proteins from the interrogation of the annotated protein database of the sibling species Plasmodium berghei, indicating that these proteins are likely absent from the current P. yoelii database (Table 1 and SI Tables 4 and 5). More than 90% of the identified proteins in the LS schizont proteome had orthologs in P. falciparum, a much higher proportion than the 57% genome-wide orthology between P. yoelii and P. falciparum (10). Interestingly, the proportion of hypothetical proteins in the LS schizont proteome (23%) was less than half of the proportion observed for all annotated P. yoelii proteins (53%). This possibly reflects the lower abundance of hypothetical proteins in LS and the bias of tandem mass spectrometry in detecting abundant proteins with metabolic functions in the organism. Previously published comprehensive proteomic analyses of the mosquito and asexual erythrocytic and gametocytic stages of P. falciparum (12, 13) and P. berghei (10) revealed the presence of stage-specific as well as constitutively expressed proteins across the stages. The genome-wide amino acid sequence identity of the predicted proteomes (88%; ref. 10) between P. yoelii and P. berghei allowed us to compare the P. yoelii LS schizont proteome data with the P. berghei proteome data set (10) to gain a more extensive picture of the proteome of the parasite throughout its life cycle (SI Table 6). The LS schizont proteome showed 70% of the proteins detected had assigned P. berghei orthologs that were detected during the asexual intraerythrocytic stages of P. berghei (SI Table 6). This was even more pronounced for LS50, where close to 90% of the proteins were also detected in intraerythrocytic stages. The data indicate that, because the LS schizont ultimately produces the first generation of intraerythrocytic stages, it shifts protein composition toward a more erythrocytic stage-like profile. However, despite this similarity, we detected proteins that were more abundant and/or detected only in LS: Cluster P1 contained proteins with three times or higher spectral counts in LS compared with other stages, whereas cluster P2 contained proteins with one to two spectral counts in LS and not detected in any other stages (Fig. 2B). Within clusters P1 and P2, we found 174 proteins that appear unique to LS. The remaining clusters encompassed LS-detected proteins also detected in other life stages, particularly asexual erythrocytic stages (Cluster P3, P5, and P6 in Fig. 2B). Many of the proteins in these clusters have housekeeping functions falling into these main classes: ribosomal structure and translation components, chaperones, proteases, and metabolic enzymes. Cluster P4 included proteins mainly detected in both LS and gametocytes but not in the asexual erythrocytic stages (Fig. 2B).
Fig. 2.
Correlation between LS gene and protein expression. (A) Venn diagram of the overlap between LS40- and LS50-detected proteins. (B) LS-detected proteins are grouped into six clusters based on their detection patterns in other stages (10) as explained in the text: oo, P. berghei oocyst sporozoite; sg, P. berghei salivary gland sporozoite; 40, LS40; 50, LS50; bs, P. berghei asexual blood stages; and ga, P. berghei gametocytes. Estimation of protein abundance is based on the total spectral counts for each protein from the tandem mass spectrometry data and is indicated by the intensity of the blue shading. The corresponding expression profile for the genes encoding the LS detected proteins is shown (Right). The heat map for the gene expression profile is expressed as the ratio of gene expression for each stage by using the mixed blood-stage sample as reference: oo, oocyst sporozoite; sg, salivary gland sporozoite; 24, LS24; 40, LS40; 50, LS50; and sc, blood-stage schizonts. Green indicates down-regulation, whereas red indicates up-regulation of the gene in the LS sample compared with the other stages. (C) Venn diagram of the overlap between P. yoelii LS schizont (LS40 + LS50) transcriptome and proteome.
Integrative Analysis of LS.
The availability of LS microarray gene expression data allowed the examination of the temporal expression of genes encoding the proteins in the LS schizont proteome. Of the 712 P. yoelii proteins identified in the LS schizont proteome, 422 corresponded to genes that were transcriptionally up-regulated at the same time points indicating almost 60% concordance (Fig. 2C). For 401 of these LS schizont proteins, the genes encoding them were expressed in clusters III (genes up-regulated throughout LS) and IV (genes up-regulated at LS40 and LS50) (Fig. 1C). The 246 P. yoelii LS schizont proteins for which no active gene expression was discerned were encoded by genes with constitutive expression across all stages covered in this study (data not shown). These results suggest that transcription of LS genes is tightly coupled to translation similar to what has been observed for most other stages of Plasmodium (10, 11). A comparison between the LS transcriptome, LS proteome, and ESTs obtained through LCM of late LS reported by Sacci et al. (15) identified 136 genes present in all three data sets (SI Fig. 6). Overall, ≈50% of the 455 genes (which could be assigned to the LCM ESTs) are shared between the LCM and LS transcriptome, whereas ≈40% of the genes in the LCM data set encoded proteins also detected in the LS schizont proteome (SI Fig. 6).
We next analyzed the transcriptome and proteome data within the context of the annotated metabolic pathways and functional groupings available for P. falciparum (ref. 18; http://sites.huji.ac.il/malaria). We performed statistical analysis on the functional groupings and metabolic pathways overrepresented in the LS transcriptome and proteome. The results (Table 2) indicated that translation and its ancillary processes, such as ribosome structure, translational initiation, protein synthesis, and protein translocation, were overrepresented in LS. Genes encoding RNA polymerase components were enriched in the LS transcriptome but not in the LS schizont proteome, presumably because these proteins were less abundant and harder to detect by tandem mass spectrometry. Putative proteasomal and heat-shock proteins were well represented. The metabolic pathways enriched in LS include those involved in redox metabolism, the mitochondrial TCA cycle and electron transport system, and the fatty acid synthesis II (FASII) pathway (Table 2). These data, in combination with the functional profiles of the LS transcriptome and proteome (SI Fig. 7), reveal that LS are highly metabolically active. Our findings support the observation that malaria parasite LS undergo one of the fastest growth rates among eukaryotic cells, likely synthesizing large amounts of protein and fatty acids required to produce tens of thousands of exoerythrocytic merozoites. Our data also show that, similar to the gametocyte stages (and unlike the asexual erythrocytic stages), mitochondrial activity was up-regulated in LS (10).
Table 2.
Overrepresented functional groupings and pathways in LS (P < 0.05)
| Group description | Total* | No. LS transcriptome | P value | Total† | No. in LS proteome | P value |
|---|---|---|---|---|---|---|
| Translation | 5171 | 120 | 3.80E-11 | 192 | 114 | 2.20E-16 |
| Ribosome structure | 99 | 80 | 6.40E-13 | 111 | 69 | 3.20E-11 |
| Translation initiation | 24 | 15 | 0.092 | 25 | 16 | 0.0012 |
| Protein biosynthesis | 32 | 15 | 0.57 | 39 | 23 | 0.00058 |
| Posttranslational translocation in eukaryotes | 7 | 6 | 0.045 | 7 | 5 | 0.042 |
| RNA polymerase II transcription | 18 | 13 | 0.027 | 19 | 2 | 0.99 |
| Structure of RNA polymerases | 20 | 14 | 0.031 | 21 | 4 | 0.95 |
| Proteasome-mediated proteolysis | 48 | 34 | 0.00057 | 51 | 30 | 8.80E-05 |
| Chaperone-assisted protein folding | 26 | 20 | 0.0017 | 29 | 18 | 0.0032 |
| Redox metabolism | 21 | 14 | 0.054 | 23 | 15 | 0.0013 |
| Mitochondrial TCA cycle and electron transport | 32 | 21 | 0.025 | 33 | 17 | 0.019 |
| Fatty acid synthesis II | 20 | 17 | 0.00048 | 22 | 12 | 0.028 |
| Glycolysis | 19 | 11 | 0.23 | 20 | 14 | 0.00067 |
| Leucine, valine, and isoleucine metabolism | 7 | 3 | 0.72 | 7 | 5 | 0.042 |
| Protein traffic | 100 | 40 | 0.94 | 107 | 44 | 0.037 |
| Import and export through the nuclear pore | 14 | 5 | 0.87 | 14 | 10 | 0.0034 |
*Only genes represented on the microarray were considered.
†Only proteins belonging to orthologous groups that have a P. falciparum protein and at least one protein from P. yoelii or P. berghei were considered.
Potential LS Secretome.
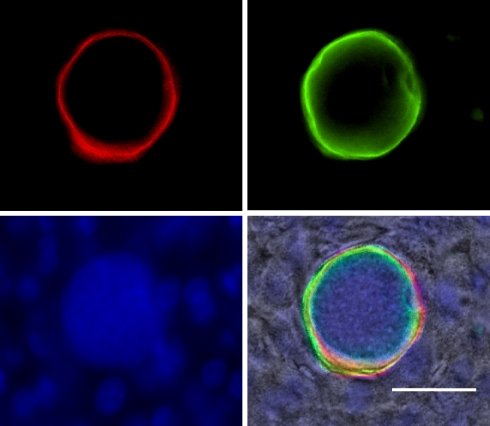
The Plasmodium LS is considered the most promising target for antiinfection vaccines. Radiation-attenuated (6, 19) or genetically attenuated (20–23) parasites arrest in LS development and are potent elicitors of immune responses that completely protect against infection. Protection is mainly mediated by CD8+ T cells (23, 24). For the most part, the protective antigens remain unknown but are likely expressed by LS, because treatment with primaquine, which disrupts LS schizogony, aborts the development of protection (25). These proteins might be accessible to the infected hepatocyte antigen presentation machinery and might be recognized by host effector immune responses. Among the 712 P. yoelii proteins detected in LS, 93 were annotated to contain a signal peptide (SP), indicating they may enter the LS secretory pathway (Table 1). Seventy-six LS proteins were predicted to be membrane-associated [with transmembrane domain (TM)] and were underrepresented in the LS proteome (11%), compared with their proportion in the genome (26%), possibly because membrane proteins are inherently more difficult to detect by tandem mass spectrometry. The only two previously identified LS parasitophorous vacuole (PV) membrane resident proteins Hep17 (PY04421; Exported protein 1; ref. 26) and UIS4 (21) were identified in the LS proteome. Hep17 is expressed in both LS and the erythrocytic stage, whereas UIS4 is expressed in salivary gland sporozoites and LS. Among the proteins identified at high abundance in LS was PY04499, a conserved hypothetical protein (PF14_0179) with a putative SP and TM. PY04499 appears active in LS only (data not shown). To localize the protein in LS, we performed immunofluorescence analysis on P. yoelii-infected liver sections using two independent peptide antisera against PY04499. PY04499 exhibited a circumferential staining in LS similar to Hep17, indicating it was also localized to the PV (Fig. 3 and data not shown). The anti-PY04499 antibody did not react with intraerythrocytic parasites or sporozoites in immunofluorescent studies (data not shown), indicating LS-specific expression. To date, the only LS-specific protein identified is LSA-1, a P. falciparum PV protein that is also a vaccine candidate with no rodent malaria ortholog (27). The combined LS transcriptome and proteome data will be valuable in the selection of antigens. A previous study testing 27 proteins present in the P. falciparum sporozoite proteome identified 16 of these proteins to be highly antigenic based on stimulation of immune cells obtained from volunteers immunized with radiation attenuated sporozoites (28). P. yoelii orthologs of two of these proteins (PY00455 and PY04544) were identified in the LS schizont proteome, whereas another two genes (PY03459 and PY00566) were present in the LS transcriptome.
Fig. 3.
PY04499 is an LS-specific protein localized to the parasitophorous vacuole (PV). Immunofluorescence assay was done by using tissue sections of 44-h pi infected mouse livers. The image shows a LS schizont stained with antisera against PY04499 (green) and a monoclonal antibody against Hep17 (red). The antisera against PY04499 were generated in rabbits by using the “KYLLLHHTNAFL” peptide. Hepatocyte and parasite nuclei were visualized with DAPI (blue). (Scale bar, 50 μm.)
LS Drug Targets.
Increased antimalarial drug resistance has made the search for new drugs a priority. Drugs targeting LS have the advantage that they provide true causal prophylaxis, preventing the erythrocytic stages and the clinical manifestations of malaria. Furthermore, LS drugs are critical to preventing relapsing infections of P. vivax hypnozoites. Primaquine remains the main licensed drug specifically used for LS (7). It also has activity against gametocytes but not against asexual erythrocytic stages (29). The mechanism of action of primaquine is not fully known but may involve the inhibition of mitochondrial processes by toxic metabolites (29). It is possible that the observed enrichment of proteins involved in the mitochondrial TCA and electron transport in the proteomes of LS (Table 2) and gametocytes (10) may account for the susceptibility of both stages to primaquine. In-depth analysis of the LS proteome revealed protein classes and metabolic pathways that are potential drug targets for malaria prophylaxis. For example, many of the annotated proteases were present in the LS transcriptome and proteome (SI Fig. 8 and SI Table 7). Proteases are attractive drug targets, because specific compound can be designed that fit tightly in their active site and act as strong competitive inhibitors (29). Among the putative proteases detected in LS were falcilysin, plasmepsins, the SERA cysteine proteases, and proteases in the proteasome proteolytic pathway (SI Fig. 8 and SI Table 7). Plasmepsin IV and falcilysin were previously implicated in hemoglobin degradation, and falcilysin more recently also in transit peptide degradation (30, 31). LS do not use hemoglobin, suggesting that these proteases have other functions in LS. The function of SERA proteases is unknown, but the demonstration that a P. berghei ortholog of SERA 8, ECP1, was critical for sporozoite egress from mosquito midgut oocysts (32) indicates a possible role of the SERA proteins in egress of exoerythrocytic merozoites (33).
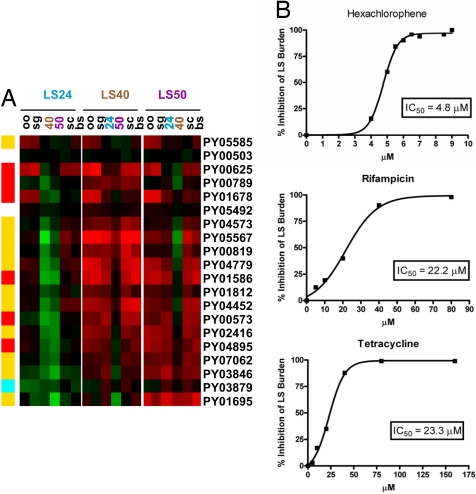
We also observed the overrepresentation of particular metabolic pathways in LS. The redox metabolism comprising the thioredoxin and glutathione systems was demonstrated to contribute to antioxidant defense and redox regulation in the erythrocytic stages (34). A putative thioredoxin reductase (PY02397) was detected in the LS transcriptome and proteome. The active site of the P. falciparum thioredoxin reductase harbors a solvent-exposed and highly reactive redox center compared with the mammalian counterpart, thus making it a good target for development of a selective inhibitor with antimalarial activity (35). One of the most conspicuous findings in the LS transcriptome and proteome was the specific overrepresentation of enzymes involved in fatty acid synthesis, including the FASII pathway (Table 2; Fig. 4A). The FASII pathway is found in plants, prokaryotes, and Archaea, where it is responsible for the synthesis of fatty acids up to C16 and C18 that are required for membrane biogenesis (36). In Plasmodium, the FASII pathway is localized in the apicoplast (36). The bacterial origin of this pathway and the absence of FASII in the human host make it an excellent target for antimalarial drugs (37). Biochemical assays and FASII inhibition studies have been done, indicating that the FASII pathway was active in P. falciparum erythrocytic stages in culture (37). Strikingly, however, many genes and proteins in this pathway were among those we detected in the LS-active transcriptome and proteome, but which were not detected in other stages of both P. berghei (ref. 10; Fig. 4A) and P. falciparum (12). These results suggest that the FASII pathway is extremely active in LS and might be targeted using available FASII inhibitors. We tested hexachlorophene, a FASII inhibitor that inhibits β-oxoacyl-ACP reductase (FabG), for its inhibitory potential against LS (38). Hexachlorophene inhibited LS development in vitro in a dose-dependent manner with an IC50 of 4.8 μM (Fig. 4B). Our data suggest the FASII pathway as a drug target for malaria prophylaxis. Further studies are necessary to determine whether the FASII pathway serves an essential function in LS and in erythrocytic stages. We also tested the ability of the antibacterials rifampicin and tetracycline for their ability to inhibit LS development, because both drugs have been shown to inhibit the growth of P. falciparum erythrocytic stages by interfering with mitochondrial and/or apicoplast function (39, 40), both of which may be important in LS. As shown in Fig. 4C, each drug inhibited LS development at similar IC50 concentrations, indicating that they may also be used for prophylaxis.
Fig. 4.
The fatty acid synthesis pathway in LS and drug inhibition studies. (A) Gene and protein expression involved in fatty acid synthesis, including the FASII pathway, is highly induced in LS. The heat map shows the expression ratios of each LS sample compared with all of the other life stages: oo, oocyst sporozoite; sg, salivary gland sporozoite; 24, LS24; 40, LS40; 50, LS50; bs, mixed blood stages; and sc, blood-stage schizonts. Green indicates down-regulation, whereas red indicates up-regulation of the gene in the LS sample compared with the other stages. The summary of gene and protein expression of proteins and enzymes in the fatty acid synthesis pathway is shown (Left): yellow indicates the gene is in the LS transcriptome, and the protein is detected in the LS proteome; red indicates gene is in the LS transcriptome; blue indicates protein is in the LS proteome; and white indicates no detection in either LS transcriptome or LS proteome. (B) Dose–response curves for the FASII inhibitor hexachlorophene and the antibacterials, rifampicin and tetracycline, for the inhibition of P. yoelii LS development in hepatoma cells measured at 40 h pi. Each drug inhibits LS growth efficiently.
For five decades since the discovery of malaria parasite LS, research into this most elusive stage of the complex Plasmodium life cycle has made little progress. This impeded elucidation of host–parasite interactions during the initial infection, the development of antiinfection vaccines, and the identification of novel drug targets. Our data represent previously undescribed in-depth analysis of the malaria parasite LS transcriptome and proteome. The data expose the antigenic and metabolic potential of LS and therefore provide a map that will guide and accelerate future recombinant vaccine development and drug development efforts. Our work fills the last “omics” gap in the parasite life cycle and may lead to interventions that alleviate the enormous suffering caused by malaria.
Materials and Methods
Detailed methods and procedures are provided in SI Text.
Parasite Isolation.
LS-infected hepatocytes were isolated from PyGFP-infected mice at 24, 40, and 50 h pi, as described (17). Sporozoites from P. yoelii (17XNL)-infected A. stephensi mosquitoes were isolated from midguts at day 10 and from salivary glands at day 15 after infectious blood meal. Parasites in the erythrocytic stages were harvested from PyGFP-infected mice (5–10% parasitemia). Purified blood-stage schizonts were prepared from P. yoelii-infected blood cultured as described (17).
RNA Preparation and Microarray Hybridization.
Total RNA from each sample was extracted and subjected to two rounds of linear amplification. For each parasite stage analyzed, two to five independent biological replicates were obtained. Cy3- and Cy5-labeled RNA were hybridized to P. yoelii microarray slides spotted with 65-mer oligonucleotide probes. Microarray hybridization and analysis of LS active genes are described in SI Text.
Protein Extraction and Proteomic Analysis.
Total protein was extracted from sorted LS-infected hepatocytes and separated on SDS/PAGE. Gel bands were cut, and in-gel trypsin digestion was performed. Mass spectra of the resulting peptides were obtained by nanoflow liquid chromatography tandem mass spectrometry (1D LC/MS/MS). The MS/MS data were searched against two sequence databases separately using the SEQUEST algorithm. One database contained annotated P. yoelii and Mus musculus protein sequences and the another annotated P. berghei and M. musculus sequences. Peptide and protein identification were done by using PeptideProphet and ProteinProphet, respectively (http://tools.proteomecenter.org/software.php).
LS Inhibition Studies.
LS inhibition studies by hexachlorophene and antibacterials were carried out in vitro by using HepG2:CD81 cells infected with PyGFP parasites. PyGFP LS infection and host cell viability were simultaneously determined by using flow cytometry analysis. IC50, the inhibitor concentration at which LS parasite growth was reduced by 50% compared with untreated LS infections, was estimated from four independent experiments.
Supplementary Material
ACKNOWLEDGMENTS.
We thank John Whistler for expert insectary technical assistance and Ashley Vaughan for editing the manuscript. This microarray and bioinformatics work is supported by the Bill and Melinda Gates Foundation through the Foundation at the National Institutes of Health Grand Challenges in Global Health Initiative and the Ellison Medical Foundation (S.H.I.K.). Design and construction of the P. yoelii microarray were supported by the National Institutes of Health (L.W.B.). Proteome analysis is supported by a Seattle Biomedical Research Institute Innovation grant.
Footnotes
Conflict of interest statement: S.H.I.K. is a coinventor on two patent applications relevant to this work (Genetically attenuated malaria vaccines, patent no. WO 2005/063991; and Plasmodium liver stage antigens, patent no. WO 2007/041216). The patents were filed for patent protection to promote the development and distribution of malaria vaccines to people in need worldwide, in accordance with a global access strategy.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus database (GEO), www.ncbi.nlm.nih.gov/geo (accession no. GSE8125).
This article contains supporting information online at www.pnas.org/cgi/content/full/0710780104/DC1.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenwood B. Malaria vaccines Evaluation and implementation. Acta Trop. 2005;95:298–304. doi: 10.1016/j.actatropica.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 3.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman SL, Doolan DL. Malaria vaccines targeting infected hepatocytes. Nat Med. 2000;6:1218–1219. doi: 10.1038/81315. [DOI] [PubMed] [Google Scholar]
- 5.Alonso PL, et al. Duration of protection with RTS,S/AS02A malaria vaccine in prevention of Plasmodium falciparum disease in Mozambican children: Single-blind extended follow-up of a randomised controlled trial. Lancet. 2005;366:2012–2018. doi: 10.1016/S0140-6736(05)67669-6. [DOI] [PubMed] [Google Scholar]
- 6.Luke TC, Hoffman SL. Rationale and plans for developing a nonreplicating, metabolically active, radiation-attenuated Plasmodium falciparum sporozoite vaccine. J Exp Biol. 2003;206:3803–3808. doi: 10.1242/jeb.00644. [DOI] [PubMed] [Google Scholar]
- 7.Baird JK, Hoffman SL. Primaquine therapy for malaria. Clin Infect Dis. 2004;39:1336–1345. doi: 10.1086/424663. [DOI] [PubMed] [Google Scholar]
- 8.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlton JM, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 10.Hall N, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 11.Bozdech Z, et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 13.Florens L, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 14.Blair PL, Carucci DJ. Functional proteome and expression analysis of sporozoites and hepatic stages of malaria development. Curr Top Microbiol Immunol. 2005;295:417–438. doi: 10.1007/3-540-29088-5_16. [DOI] [PubMed] [Google Scholar]
- 15.Sacci JB, Jr, et al. Transcriptional analysis of in vivo Plasmodium yoelii liver stage gene expression. Mol Biochem Parasitol. 2005;142:177–183. doi: 10.1016/j.molbiopara.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Brown S, Roos DS, Nussenzweig V, Bhanot P. Transcriptome of axenic liver stages of Plasmodium yoelii. Mol Biochem Parasitol. 2004;137:161–168. doi: 10.1016/j.molbiopara.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Tarun AS, et al. Quantitative isolation and in vivo imaging of malaria parasite liver stages. Int J Parasitol. 2006;36:1283–1293. doi: 10.1016/j.ijpara.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Ginsburg H. Progress in in silico functional genomics: the malaria metabolic pathways database. Trends Parasitol. 2006;22:238–240. doi: 10.1016/j.pt.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Nussenzweig RS, Vanderberg J, Most H, Orton C. Protective immunity produced by the injection of x-irradiated sporozoites of Plasmodium berghei. Nature. 1967;216:160–162. doi: 10.1038/216160a0. [DOI] [PubMed] [Google Scholar]
- 20.Mueller AK, Labaied M, Kappe SH, Matuschewski K. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature. 2005;433:164–167. doi: 10.1038/nature03188. [DOI] [PubMed] [Google Scholar]
- 21.Mueller AK, et al. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc Natl Acad Sci USA. 2005;102:3022–3027. doi: 10.1073/pnas.0408442102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labaied M, et al. Plasmodium yoelii sporozoites with simultaneous deletion of P52 and P36 are completely attenuated and confer sterile immunity against infection. Infect Immun. 2007;75:3758–3768. doi: 10.1128/IAI.00225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarun AS, et al. Protracted sterile protection with Plasmodium yoelii pre-erythrocytic GAP malaria vaccines is independent of significant liver stage persistence and is mediated by CD8+ T cells. J Infect Dis. 2007;196:608–616. doi: 10.1086/519742. [DOI] [PubMed] [Google Scholar]
- 24.Mueller AK, et al. Genetically Attenuated Plasmodium berghei Liver Stages Persist and Elicit Sterile Protection Primarily via CD8 T Cells. Am J Pathol. 2007;171:107–115. doi: 10.2353/ajpath.2007.060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheller LF, Azad AF. Maintenance of protective immunity against malaria by persistent hepatic parasites derived from irradiated sporozoites. Proc Natl Acad Sci USA. 1995;92:4066–4068. doi: 10.1073/pnas.92.9.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doolan DL, et al. Identification and characterization of the protective hepatocyte erythrocyte protein 17-kDa gene of Plasmodium yoelii, homolog of Plasmodium falciparum exported protein 1. J Biol Chem. 1996;271:17861–17868. doi: 10.1074/jbc.271.30.17861. [DOI] [PubMed] [Google Scholar]
- 27.Guerin-Marchand C, et al. A liver-stage-specific antigen of Plasmodium falciparum characterized by gene cloning. Nature. 1987;329:164–167. doi: 10.1038/329164a0. [DOI] [PubMed] [Google Scholar]
- 28.Doolan DL, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci USA. 2003;100:9952–9957. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiesner J, Ortmann R, Jomaa H, Schlitzer M. New antimalarial drugs. Angew Chem Int Ed Engl. 2003;42:5274–5293. doi: 10.1002/anie.200200569. [DOI] [PubMed] [Google Scholar]
- 30.Banerjee R, et al. Four plasmepsins are active in the Plasmodium falciparum food vacuole, including a protease with an active-site histidine. Proc Natl Acad Sci USA. 2002;99:990–995. doi: 10.1073/pnas.022630099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponpuak M, et al. A role for falcilysin in transit peptide degradation in the Plasmodium falciparum apicoplast. Mol Microbiol. 2007;63:314–334. doi: 10.1111/j.1365-2958.2006.05443.x. [DOI] [PubMed] [Google Scholar]
- 32.Aly AS, Matuschewski K. A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J Exp Med. 2005;202:225–230. doi: 10.1084/jem.20050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sturm A, Heussler V. Live and let die: manipulation of host hepatocytes by exoerythrocytic Plasmodium parasites. Med Microbiol Immunol. 2007;196:127–133. doi: 10.1007/s00430-007-0044-3. [DOI] [PubMed] [Google Scholar]
- 34.Becker K, et al. Oxidative stress in malaria parasite-infected erythrocytes: host-parasite interactions. Int J Parasitol. 2004;34:163–189. doi: 10.1016/j.ijpara.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Andricopulo AD, et al. Specific inhibitors of Plasmodium falciparum thioredoxin reductase as potential antimalarial agents. Bioorg Med Chem Lett. 2006;16:2283–2292. doi: 10.1016/j.bmcl.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 36.Ralph SA, et al. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 37.Surolia N, Surolia A. Triclosan offers protection against blood stages of malaria by inhibiting enoyl-ACP reductase of Plasmodium falciparum. Nat Med. 2001;7:167–173. doi: 10.1038/84612. [DOI] [PubMed] [Google Scholar]
- 38.Wickramasinghe SR, et al. Kinetic, inhibition and structural studies on 3-oxoacyl-ACP reductase from Plasmodium falciparum, a key enzyme in fatty acid biosynthesis. Biochem J. 2006;393:447–457. doi: 10.1042/BJ20050832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman CD, Su V, McFadden GI. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 2007;152:181–191. doi: 10.1016/j.molbiopara.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 40.Dahl EL, et al. Tetracyclines specifically target the apicoplast of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2006;50:3124–3131. doi: 10.1128/AAC.00394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.