Abstract
Synaptosomal-associated protein of 25 kDa (SNAP-25) is a SNARE protein that regulates neurotransmission by the formation of a complex with syntaxin 1 and synaptobrevin/VAMP2. SNAP-25 also reduces neuronal calcium responses to stimuli, but neither the functional relevance nor the molecular mechanisms of this modulation have been clarified. In this study, we demonstrate that hippocampal slices from Snap25+/− mice display a significantly larger facilitation and that higher calcium peaks are reached after depolarization by Snap25−/− and Snap25+/− cultured neurons compared with wild type. We also show that SNAP-25b modulates calcium dynamics by inhibiting voltage-gated calcium channels (VGCCs) and that PKC phosphorylation of SNAP-25 at ser187 is essential for this process, as indicated by the use of phosphomimetic (S187E) or nonphosphorylated (S187A) mutants. Neuronal activity is the trigger that induces the transient phosphorylation of SNAP-25 at ser187. Indeed, enhancement of network activity increases the levels of phosphorylated SNAP-25, whereas network inhibition reduces the extent of protein phosphorylation. A transient peak of SNAP-25 phosphorylation also is detectable in rat hippocampus in vivo after i.p. injection with kainate to induce seizures. These findings demonstrate that differences in the expression levels of SNAP-25 impact on calcium dynamics and neuronal plasticity, and that SNAP-25 phosphorylation, by promoting inhibition of VGCCs, may mediate a negative feedback modulation of neuronal activity during intense activation.
Synaptosomal-associated protein of 25 KDa (SNAP-25) belongs to the SNARE superfamily of membrane proteins that participate in the regulation of neuronal exocytosis. SNAP-25 is present in two isoforms, a and b, resulting from alternative splicing of the exon 5 of the gene, which is differentially expressed during development. SNAP-25a is expressed at the embryonic stage, and SNAP-25b becomes the major isoform postnatally (1–3). SNAP-25 is anchored to the cytosolic face of membranes by palmitoyl side chains located in the central region of the molecule and contributes two α-helices to the exocytotic fusion complex, together with syntaxin-1 and synaptobrevin/VAMP2 (4–5). SNAP-25 also interacts with the synaptic vesicle protein, synaptotagmin I (6), a major calcium sensor that regulates neurotransmitter release (7, 8). Interaction of synaptotagmin with SNAP-25 is essential for the calcium-dependent triggering of membrane fusion (9) and for the control of fusion pore during the final steps of exocytosis (10). Furthermore, the C terminus of SNAP-25 is a target of G protein β- and γ-subunits that mediate presynaptic inhibition (11). Therefore, SNAP-25 represents a multifunctional protein involved in the control of secretion by multiple interactions. Besides its well characterized role in regulating exocytosis, there is increasing evidence that SNAP-25 modulates various ion channels (12, 13). In particular, SNAP-25 physically interacts with different types of voltage-gated calcium channels (VGCCs), including N type (14), P/Q type (15, 16), and L type (17), through a channel region known as the synaptic protein interaction (synprint) site. This interaction alters channel function by changing the N-type steady-state voltage dependence of inactivation in Xenopus oocytes (18) by inhibiting L-type currents in HIT-T15 cells (19) and by negatively shifting the steady-state voltage dependence of inactivation of P/Q-type channels in HEK293 cells (20).
We previously reported that differential SNAP-25 expression in hippocampal neurons is reflected by altered calcium responsiveness, and that overexpression of SNAP-25 significantly reduces calcium responses to depolarization (21). However, the molecular mechanism by which SNAP-25 modulates calcium dynamics and the precise region of the protein involved in such effect is still undefined, although in neuronal cells it comprises the residues from amino acids 180–197 (21). We now show that SNAP-25 phosphorylation on ser187 is responsible for its negative effect on calcium dynamics, and that such negative modulation occurs through the inhibition of VGCCs. Because ser187 phosphorylation is transiently induced by neuronal activity, these data reveal that SNAP-25 provides a negative feedback mechanism for controlling neuronal excitability.
Results
Calcium Responsiveness to Depolarization in Neurons from SNAP-25-Transgenic Animals.
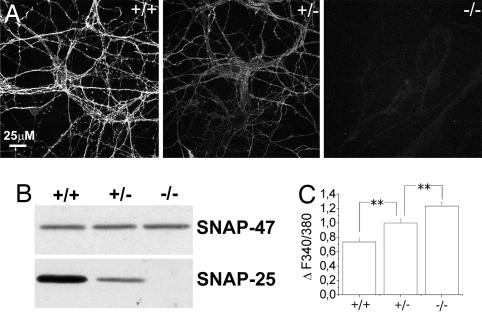
We previously found that the overexpression of SNAP-25b lowers neuronal calcium responsiveness to depolarization, and that SNAP-25 silencing in glutamatergic cells significantly increases it (21). To confirm that SNAP-25 modulates calcium responsiveness, primary hippocampal cultures from Snap25−/−, Snap25+/− mutants, and wild-type (WT) embryonic day 18 fetuses were loaded with the calcium-sensitive dye, Fura-2, and imaged by single-cell recording. Immunocytochemistry and Western blotting revealed that cultures from Snap25+/− mutants display levels of SNAP-25 intermediate between Snap25−/− and WT neurons (Fig. 1 A and B) (22). Notably, neuronal responsiveness to 30 mM KCl was inversely proportional to the amount of the expressed SNARE, with the highest and lowest calcium responsiveness being detectable in Snap25−/− and WT neurons, respectively (Fig. 1C), which is in line with a concentration-dependent modulation of calcium dynamics. Glutamatergic neurons, identified by morphological criteria or retrospective labeling, were selected for analysis.
Fig. 1.
Calcium responses recorded from hippocampal neurons established from SNAP-25 null and heterozygous mice. (A) Analysis of SNAP-25 immunoreactivity in hippocampal cultures established from WT (+/+), heterozygous (+/−), and KO (−/−) mice. (B) Western blot analysis of SNAP-25 expression in homogenates from (+/+), (+/−), and (−/−) mouse hippocampi. SNAP-47 was used as a loading control. (C) Quantitative analysis of peak calcium responses measured upon 30 mM KCl depolarization in 9–10 DIV hippocampal neurons from WT (+/+) (0.73 ± 0.05) (n = 46), heterozygous (+/−) (1 ± 0.05) (n = 58), and KO (−/−) (1.23 ± 0.05) (n = 71) mice. Values are normalized on calcium responses measured on (+/−) cultures. *, P < 0.05; **, P < 0.01.
Short-Term Plasticity in WT and Snap25+/− Hippocampal Slices.
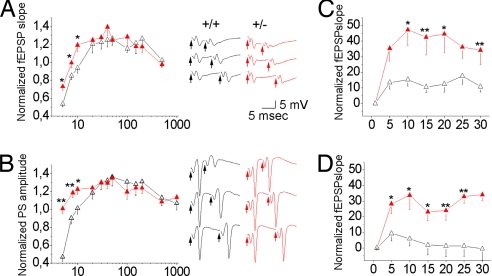
To determine the possible effect of reduced SNAP-25 expression on short-term synaptic plasticity, excitatory postsynaptic field potentials (fEPSPs) were recorded from CA1 pyramidal neurons in hippocampal slices from adult WT and SNAP-25+/− mice. Paired-pulse facilitation (PPF) was significantly increased in SNAP-25+/− slices relative to WT at brief interpulse intervals (IPIs), as evidenced by a significant difference between the fEPSP ratios in Snap25+/− and WT mice at 5, 7.5, and 10 ms (Fig. 2A). This increased short-term excitability also was reflected in the population spike (PS) amplitude, which was measured in the pyramidal layer (Fig. 2B). At longer IPIs, no significant difference in PP ratio of either fEPSP or PS between heterozygous and WT animals was observed, which is in agreement with previous results (23). Low-frequency stimulation protocol of 2 Hz for 15 s by using stimulus strengths capable of evoking maximal (Fig. 2C) and half-maximal (Fig. 2D) responses resulted in a transient frequency potentiation (P < 0.03 and 0.04; Mann–Whitney U test) of the responses to the first five stimuli in both SNAP-25+/− and WT slices. In the Snap25+/− mice, however, the potentiation was significantly larger and persisted throughout the 30-s stimulation, leading to a significant difference between the fEPSPs measured on WT and heterozygous mice until the end of stimulus protocol (P < 0.001) (Fig. 2 C and D).
Fig. 2.
Short-term plasticity in WT and Snap25+/− hippocampal slices. (A and B) PPF profiles of fEPSP (A) and PS (B) recorded from the CA1 region of hippocampal slices. Note facilitation at short PPF intervals in heterozygous (n = 6) with respect to WT (n = 5) mice, which is reflected in both the fEPSP slope and PS amplitude. Representative fEPSP (upper traces) and PS (lower traces) evoked at 5, 7.5, and 10 ms IPI in WT (black lines) and heterozygous (red lines) mice are shown. Arrowheads indicate the stimulus pairs. (C and D) Frequency potentiation of fEPSP obtained by 2-Hz stimuli (delivered for 15 s) capable of evoking maximal (C) and half-maximal (D) responses. Note in both cases the significantly larger increase of the EPSP slope in heterozygous with respect to WT mice (each data point represents the mean of five responses normalized to the response to the first stimulus; data are from six slices from each mouse strain). All values are presented as mean ± SEM (Mann–Whitney U test: *, P < 0.05). Red triangles, heterozygous mice; black triangles, WT mice.
Negative Modulation of Calcium Dynamics Is Only Supported by SNAP-25b.
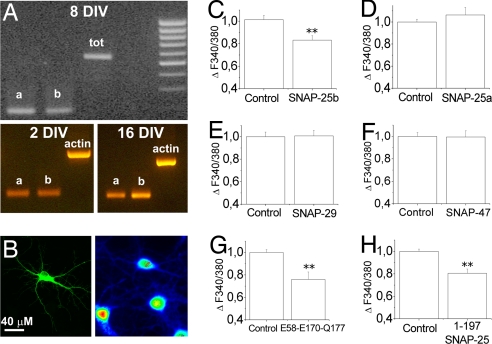
In hippocampal cultures from either mouse (Fig. 3A Upper) or rat (Fig. 3A Lower), mRNAs for both SNAP-25 isoforms are detectable, suggesting the presence of both protein isoforms in cultured neurons. To evaluate whether SNAP-25a, like SNAP-25b (21), is able to modulate neuronal calcium dynamics, hippocampal cultures were transfected with a bicistronic vector containing two cDNAs coding for SNAP-25a or SNAP-25b and for GFP, separated by the internal ribosome entry sites sequence. Analysis of calcium changes in glutamatergic neurons (Fig. 3B) showed that overexpression of SNAP-25b, but not SNAP-25a, significantly reduces calcium responses to 30 mM KCl (Fig. 3 C and D), thus indicating that the embryonic isoform is not able to modulate calcium dynamics. We then investigated whether the ability of SNAP-25b to modulate calcium responsiveness is shared by homologous proteins expressed in neurons, that is, SNAP-29 (24) and SNAP-47 (25). Cultures were transfected with cDNAs codifying for either SNAP-29 or SNAP-47, both fused to a GFP tag and imaged on depolarization. Fig. 3 E and F shows that, similarly to SNAP-23 (21), neither SNAP-29 nor SNAP-47 overexpression is able to reduce calcium responsiveness to depolarization, thus pointing to a specific role of SNAP-25b in this process. Finally, we investigated whether SNAP-25 mutations that reduce exocytosis also affect calcium dynamics. To address this issue, we measured depolarization-induced calcium responses in glutamatergic neurons transfected with a SNAP-25b mutant, D58A/E170A/Q177A, which is unable to support the fast Ca2+-dependent triggering of exocytosis (26). The quantitative analysis of calcium changes indicated that this mutant is still able to significantly reduce calcium responsiveness (Fig. 3G). Also, we confirmed that SNAP-25 1–197, the fragment generated from BoNT/A, which does not support exocytosis, is still able to negatively modulate calcium dynamics (Fig. 3H) (21). These data indicate independent roles of SNAP-25 in the control of exocytosis and the regulation of calcium dynamics.
Fig. 3.
Effect of different SNAP-25 isoforms and mutants on calcium responsiveness. (A) RT-PCR analysis for SNAP-25a and SNAP-25b performed on extracts from 8 DIV cultured mouse hippocampal neurons (Upper) or 2 (Lower Left) and 16 (Lower Right) DIV rat hippocampal neurons. (B–H) Cultured hippocampal neurons expressing different GFP-conjugated SNAP-25 isoforms or mutants were loaded with the calcium-sensitive dye FURA-2 and imaged by single-cell calcium imaging. (C–H) Quantitative analysis of intracellular calcium increases upon 30 mM KCl depolarization in neurons expressing SNAP-25B [Control (CTRL), 1.00 ± 0.03, n = 58; SNAP-25B-transfected, 0.81 ± 0.011, n = 16, P < 0.01] (C); SNAP-25A (CTRL, 1.00 ± 0.02, n = 62; SNAP-25A-transfected, 1.06 ± 0.06, n = 22) (D); SNAP-29 (CTRL, 1.00 ± 0.04, n = 28; SNAP-29-transfected, 1.00 ± 0.05, n = 8) (E); SNAP-47 (CTRL, 1.00 ± 0.03, n = 33; SNAP-47-transfected, 0.99 ± 0.05, n = 8) (F); the mutant E58-E170-Q177 (CTRL, 1 ± 0.02, n = 41; mutant E58-E170-Q177-transfected, 0.76 ± 0.06, n = 28; P < 0.01) (G); and the SNAP-25 fragment 1–197 (CTRL, 1 ± 0.02, n = 34; fragment 1–197-transfected, 0.8 ± 0.03, n = 18, P < 0.01) (H). An example of a SNAP-47-GFP-transfected neuron (Left) imaged at 380 nm (Right) is shown in B. *, P < 0.05; **, P < 0.01.
SNAP-25 Reduces Calcium Responsiveness in Neurons by Inhibiting VGCCs.
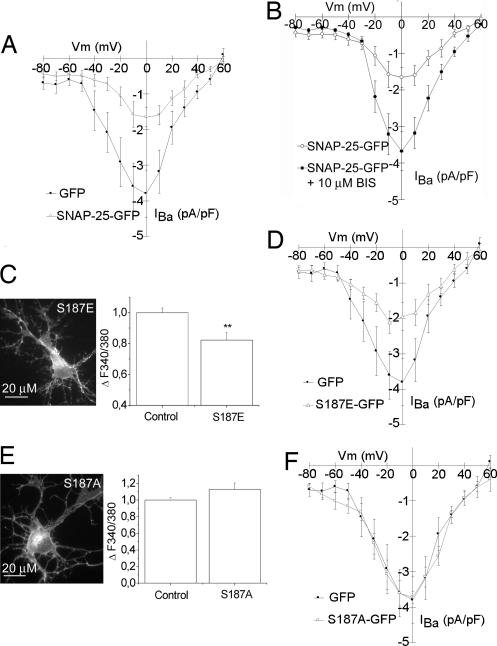
Biochemical and functional interactions between t-SNAREs and VGCCs have been previously described in various cell lines and overexpression systems (13). However, a direct inhibitory effect of SNAP-25 on calcium channels has not yet been observed in neurons. To directly prove that SNAP-25b modulates neuronal calcium dynamics by regulating VGCCs, peak calcium channel currents in neurons transfected with SNAP-25b-GFP or GFP alone were recorded by whole-cell patch clamp. Transfected glutamatergic neurons, identified by morphological and electrophysiological criteria, were selected for analysis. Fig. 4A shows that peak whole-cell Ba2+ currents were significantly reduced in SNAP-25b-GFP-transfected neurons, compared with GFP-transfected controls. No significant difference in peak Ba2+ current was observed between GFP-transfected and nontransfected controls (data not shown). These data demonstrate that SNAP-25b overexpression inhibits VGCCs.
Fig. 4.
Effect of SNAP-25 phosphomutants on VGCC currents in hippocampal neurons. (A and B) Mean I–V relationships of peak IBa recorded in SNAP-25-GFP- (n = 8) and GFP (n = 7)-expressing neurons (A) and in SNAP-25-GFP-expressing neurons in the presence (n = 10) or absence (n = 8) of 10 μM bisindolymaleimide I (B). (C–E) Quantitative analysis of intracellular calcium increases upon KCl depolarization in neurons expressing the phosphomimetic SNAP-25 mutant, S187E (CTRL, 1.00 ± 0.03, n = 66; S187E-transfected, 0.82 ± 0.04, n = 19, P < 0.01) (C) and the nonphosphorylated SNAP-25 S187A (CTRL, 1.00 ± 0.02, n = 54; S187A-transfected, 1.12 ± 0.07, n = 18) (E). Changes in intracellular calcium are normalized to nontransfected controls that were present in the same field of the GFP-expressing neurons. **, P < 0.05. (D and F) Mean IV relationship of peak IBa recorded in GFP-transfected control neurons (n = 7) and neurons expressing S187E-GFP (D) (n = 9) or S187A-GFP (F) (n = 8) SNAP-25 phosphorylation mutants. IBa current densities (pA/pF) are shown as mean ± SE of the number of recordings shown in parentheses.
Phosphorylation of SNAP-25 on Ser-187 Is Required for Negative Control of VGCCs.
The crucial region of SNAP-25 that controls neuronal calcium dynamics (residues 180–197) (21) contains a residue, ser187, which can undergo phosphorylation by PKC (27) in an activity-dependent manner (28). To evaluate whether SNAP-25 phosphorylation may be involved in the control of neuronal calcium dynamics, peak calcium currents were recorded in neurons transfected with SNAP-25b-GFP in the presence of PKC inhibitor Bisindolymaleimide I. Fig. 4B shows that peak whole-cell Ba2+ currents were significantly increased in neurons exposed to 10 μM PKC inhibitor. Because PKC phosphorylates (besides the 187 residue) the 138 residue of SNAP-25 (29), as well as additional synaptic substrates, including VGCCs (30–33), we generated two SNAP-25b mutants to evaluate the specific contribution of SNAP-25 phosphorylation at ser187: the mutant S187E, which mimics the constitutively phosphorylated state of SNAP-25, and the mutant S187A, which simulates the nonphosphorylated state of the protein. Neurons transfected with these GFP-tagged mutants were imaged for calcium responsiveness upon KCl exposure. Quantitative analysis of calcium changes in transfected neurons indicated that only the mutant S187E (Fig. 4C), but not the S187A (Fig. 4E), is effective in reducing calcium responses, thus indicating that phosphorylation of ser187 is critical for the calcium-regulating function of SNAP-25b. Accordingly, peak whole-cell currents were significantly reduced in S187E-transfected neurons, compared with GFP-transfected controls (Fig. 4D). S187A had no significant effect on neuronal calcium channel activity (Fig. 4F). These data demonstrate that phosphorylation of SNAP-25b on ser187 plays a crucial role in inhibiting VGCCs.
Neuronal Activity Induces Transient Phosphorylation of SNAP-25 on ser187 in Vitro.
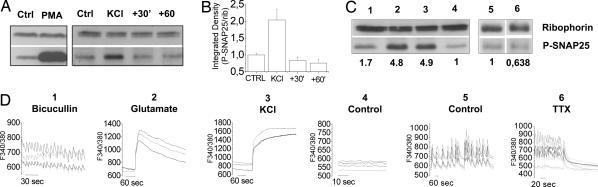
To address whether neuronal activity controls phosphorylation of SNAP-25, either in vitro or in vivo, we used an antibody specifically recognizing SNAP-25 phosphorylated at ser187 (34). PKC activation by 1 μM PMA resulted in an increase of the phosphorylation of SNAP-25 in hippocampal cultures (Fig. 5A), which is consistent with the evidence that PKC activation is responsible for SNAP-25 phosphorylation (27). Furthermore, stimulation of hippocampal cultures with 50 mM KCl for 5 min resulted in a significant increase in SNAP-25 phosphorylation (Fig. 5A) (28). Interestingly, SNAP-25 phosphorylation already returned to background levels 30 min after depolarization (Fig. 5 A and B). To provide direct evidence that SNAP-25 phosphorylation at ser187 is modulated by network activity, coverslips of hippocampal cultures were loaded with fura-2 and monitored under control conditions or upon pharmacological treatments. The recorded cultures were then analyzed by Western blotting to correlate levels of SNAP-25 phosphorylation with network activity in the same coverslip. Fig. 5 C and D shows that cultures not displaying significant spontaneous activity (4) are characterized by relatively modest levels of SNAP-25 phosphorylation, compared with sister cultures exposed to 100 μM bicuculline (1), 100 μM glutamate (2), or 50 mM KCl (3). Accordingly, when astrocyte-enriched cultures (35) displaying spontaneous activity (5) were compared with sister cultures electrically silenced with 1 μM TTX (6), a lower level of SNAP-25 phosphorylation could be detected.
Fig. 5.
SNAP-25 phosphorylation is modulated by network activity. (A) (Left) Western blot analysis of SNAP-25 phosphorylation in hippocampal cultures (12 DIV) in control conditions or after stimulation with 1 μM PMA for 30 min. (Right) Time course of SNAP-25 phosphorylation in cultures exposed to 30 mM KCl for 5 min, immediately solubilized (KCl), or washed and solubilized after 30 or 60 min. (B) Quantitative analysis of P-SNAP immunoreactivity normalized to ribophorin. (C and D) Calcium imaging (D) was coupled to Western blot analysis (C) to correlate calcium dynamics and SNAP-25 phosphorylation. Fura-2-loaded cultures were recorded by single-cell calcium imaging upon exposure to 40 μM bicucullin for 15 min (labeled no. 1), 100 μM glutamate for 5 min (labeled no. 2), 50 mM KCl for 5 min (labeled no. 3), and in control conditions (labeled no. 4). Astrocyte-enriched oscillating cultures were monitored under control conditions (labeled no. 5) or on incubation with 1 μM TTX for 30 min (labeled no. 6). (C) Afterward, recordings cultures were quickly solubilized and analyzed by Western blotting. Numbers above the blots indicate homogenates from the single coverslips recorded in D; numbers below the blots indicate quantitation of P-SNAP immunoreactivity, expressed as integrated density values normalized to controls.
Neuronal Activity Induces Transient Phosphorylation of SNAP-25 on ser187 in Vivo.
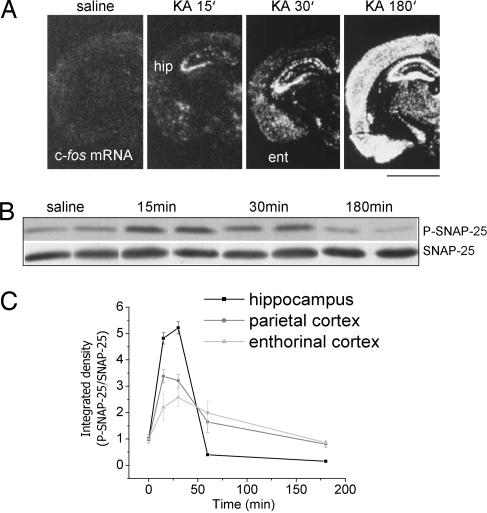
To assess whether SNAP-25 phosphorylation occurs in vivo in an activity-dependent manner, protein phosphorylation was monitored in different brain areas of mice in vivo after systemic administration of kainic acid (KA). A 30 mg/kg convulsant dose of KA i.p. rapidly (15 min) induced a marked activation of the hippocampus, as indicated by c-fos mRNA induction in this area (Fig. 6A). Shortly after (30 min), a c-fos signal also was detected in the entorhinal cortex (Fig. 6A). According to this profile of activation of limbic structures, at these time points, all mice showed behavioral signs of focal epileptic activity (stages 1–3). One hour after KA injection, the epileptic activity generalized in all treated mice, leading to a continuous status epilepticus that lasted for hours (stages 5–6). Accordingly, c-fos mRNA in situ hybridization at 180 min after KA revealed a widespread activation of many brain areas, including the hippocampus, cerebral cortex, and thalamus (Fig. 6A). Phosphorylation of SNAP-25 in the hippocampus, evaluated by immunoblotting, was rapidly (15–30 min) induced by KA administration and then returned to basal levels. The total amount of SNAP-25 did not change over time after treatment (Fig. 6B). Quantification of SNAP-25 phosphorylation in the hippocampus, enthorinal, and parietal cortex revealed that SNAP-25 is rapidly (15–30 min) phosphorylated in all three structures in response to systemic KA, the effect being stronger in the hippocampus (Fig. 6C). Levels of phospho-SNAP-25 markedly decreased in all three structures 60 min after KA and returned to basal levels (or even below in the case of the hippocampus) at 3 h, in line with previous reports (36). These data indicate that SNAP-25 phosphorylation is transiently enhanced by neuronal firing, suggesting that the protein can mediate a negative feedback modulation of neuronal activity during intense activation.
Fig. 6.
SNAP-25 phosphorylation after systemic KA treatment in mice. (A) Representative in situ hybridizations on coronal brain sections showing c-fos mRNA induction in the brain of mice treated with i.p. saline or KA (30 mg/kg). ent, entorhinal cortex; hip, hippocampus. (Scale bar: 2 mm.) (B) Representative immunoblots showing SNAP-25 phosphorylation in the hippocampus at different time points after 30 mg/kg KA i.p. Nonphosphorylated SNAP-25 was used as a control. (C) Quantification of SNAP-25 phosphorylation in the hippocampus, parietal cortex, and entorhinal cortex at different time points after KA treatment. Values are mean ± SE (n = 3–4 animals per time point).
Discussion
SNAP-25 is a multifunctional component of synapses that plays an essential role in neurotransmitter release and modulates calcium dynamics in response to depolarization (21). By using Snap25−/−, Snap25−/+, and Snap25 WT mice, we show that neuronal responsiveness to depolarization is inversely proportional to the amount of the expressed SNARE, in line with a concentration-dependent modulation of calcium dynamics. We also demonstrate a significant genotype effect on PPF, with increased facilitation for Snap25+/− mutants at short IPIs. PPF is accepted to be a measure of presynaptic facilitation probably induced by residual [Ca2+]i after an initial EPSP (37–39). A similar effect on PPF was previously shown to occur in Snap25 Tkneo/Tkneo neurons, where the shift between the a and b isoforms is impaired (23). Moreover, in Snap25+/− with respect to WT mice, we found a significantly larger potentiation of the response induced by low-frequency stimulation, which also is considered a short-term plasticity phenomenon mainly dependent on presynaptic Ca2+ (39). It is noteworthy that the enhanced facilitation detected in Snap25 Tkneo/Tkneo hippocampi correlates with our finding that only SNAP-25b, and not SNAP-25a, is able to negatively modulate calcium dynamics.
In the present study, we demonstrate that in neurons the negative modulation of calcium dynamics by SNAP-25 occurs through the inhibition of VGCCs. The specific nature of the channels subtypes modulated by SNAP-25 is still to be defined, although L-type channels seem to be involved in such an effect (D.P., unpublished data). Calcium channel modulation is carried out by the SNAP-25b isoform by the phosphorylation of residue ser187. SNARE proteins are phosphorylated by different kinases, and this process plays an important role in modulating the molecular interactions between synaptic vesicles and the presynaptic membrane. In particular, phosphorylation of SNAP-25 by PKC affects the interaction of SNAP-25 with syntaxin and synaptotagmin (40, 41), potentiates the plasma membrane recruitment of dense-core vesicles in chromaffin cells (42, 43) and, in aplysia sensorimotor neuron synapses, regulates transmitter release, thereby decreasing the rate of synaptic depression (44). Our data now demonstrate that ser187 phosphorylation is required for the negative modulation of VGCCs. Notably, phosphorylation of SNAP-25 is dynamically regulated by neuronal activity, being transiently enhanced by neuronal firing and then returning to basal levels, or even below, both in vivo and in vitro. These data are in agreement with previous findings that, in the rat hippocampus, SNAP-25 phosphorylation is decreased in the long-term scale after seizure induction by KA (36). These data suggest that SNAP-25b provides a negative feedback mechanism for controlling neuronal excitability during intense network activation. The exact mechanism by which SNAP-25b phosphorylation controls neuronal calcium dynamics is still to be defined. It was shown previously that PKC or CaMKII phosphorylation of the N-type synprint peptide affects interactions with SNARE complexes containing syntaxin and SNAP-25 (45). Interestingly, however, when immobilized GST-SNAP-25 was phosphorylated by PKC, no effect on the SNARE interaction with calcium channels was detected (45). These data suggest that PKC phosphorylation of SNAP-25 does not directly influence the interaction between the SNARE and the channels. However, SNAP-25 phosphorylation at ser187 might control the intracellular distribution of the protein, favoring its localization at the plasmamembrane, where SNARE proteins and calcium channels are clustered in lipid microdomains (46). In line with this possibility, ser187 phosphorylation of SNAP-25 in PC12 cells has been reported to result in the protein relocating to the plasma membrane (47). Interestingly, SNAP-25a, which fails to modulate calcium dynamics, differs from the b isoform by only nine residues, which include palmitoylated cysteines necessary for proper membrane localization, therefore raising the possibility that SNAP-25a may be less efficiently targeted to the plasma membrane (2, 3, 48). Furthermore, SNAP-25a is phosphorylated to a lesser degree, compared with the b isoform (49). Although we cannot exclude additional scenarios, the possibility thus arises that both SNAP-25a and the nonphosphorylated mutant S187A are unable to be efficiently sorted to the plasma membrane and therefore fail to modulate calcium channels. In support of this possibility, the 1–197 SNAP-25 fragments, which modulate calcium dynamics, but not the 1–180 fragments, which are unable to do it, maintain an association to the plasma membrane (50). These data suggest that modulation of calcium channels by SNAP-25 is regulated at multiple levels by phosphorylation, which may intervene in both localizing the SNARE in suitable cellular compartments and directly modulating its interaction with the channel synprint. Consistent with the regulation of calcium responsiveness, heterozygous deletion of the SNAP-25 gene in the mouse mutant coloboma results in absence-like epilepsy (51) and a hyperactive phenotype, similar to attention deficit hyperactivity disorder (ADHD) (52). Notably, SNAP-25 polymorphisms in humans are associated with the disorder (53–55), thus suggesting SNAP-25 as a genetic susceptibility factor in ADHD. In agreement with our finding that the S187A mutant does not negatively modulate calcium responses to stimuli, homozygous S187A mutant mice show strong anxiety-related behavior and exhibit hyperlocomotor activity (M.T., unpublished data). These data indicate that mutations leading to either reduction of protein expression (e.g., coloboma mice) or expression of mutants that do not negatively modulate calcium (e.g., S187A homozygous mice) display a hyperactive phenotype, further supporting the crucial role of calcium regulation by SNAP-25 for correct brain functioning.
Materials and Methods
Cultures of Hippocampal Neurons.
Primary cultures of rat or mouse hippocampal neurons were prepared from the hippocampi of 18- or 16-day-old fetuses, respectively, after the procedure described by Banker and Cowan (56) as described in ref. 35. Cells were maintained in MEM (Invitrogen) without sera, supplemented with 1% N2 (Invitrogen) and 1 mg/ml BSA (neuronal medium), or in neurobasal medium supplemented with B27 (57). Neurons were transfected at 5–6 days in vitro (DIV) by calcium phosphate precipitation. cDNA for SNAP-47 and SNAP-29 were a gift of R. Jahn and cDNA for SNAP-25a and SNAP-25b were provided by J. Sorensen (both of Max Planck Institute for Göttingen, Germany).
In Vivo Kainate Treatment.
Three-month-old male C57BL/6 mice received a 30 mg/kg convulsant dose of KA i.p. (Ocean Produce International). Saline-injected animals were used as controls. Occurrence of behavioral seizures was scored according to a previously defined rating scale (58) for a maximum of 3 h after KA administration. For phospho-SNAP-25 immunoblotting, the hippocampi, parietal cortices, and entorhinal cortices were dissected at 15, 30, 60, and 180 min. For in situ hybridization, brains were dissected at 15, 30, and 180 min and embedded in OCT-Tisue Tek as previously indicated (59).
Single-Cell Calcium Imaging.
Calcium imaging experiments were performed as described in ref. 21. Polychrome IV (TILLPhotonics) was used as a light source, and images were collected with a PCO Sensicam EM (TILL Photonics) and analyzed with the TILLVISION software (TILL Photonics). Thirty mM KCl stimulation was carried out in KRH in the presence of 1 μM TTX (Tocris), 100 μM APV (Tocris), and 20 μM CNQX (Tocris). Retrospective immunofluorescence analysis was carried out with antibodies against GABA (Sigma–Aldrich).
Western Blotting and RT-PCR.
Homogenates from hippocampal cultures and from different brain regions were analyzed by Western blot by using monoclonal antibodies against SNAP-25 (Synaptic System or Sternberger). Antibodies against SNAP-47 and ribophorin were a gift of R. Jahn and G. Kreibich (New York University, New York), respectively. Immunoreactive bands were analyzed with Image J software. RT-PCR analysis was performed by using specific primers (59) as described in ref. 60.
Statistical Analysis.
Results are presented as means ± SE. Data were statistically compared using Student's t test. Differences were considered significant if P < 0.05 and are indicated by an asterisk in all figures; those differences at P < 0.01 are indicated by double asterisks.
Electrophysiology.
Primary Cultures.
Whole-cell calcium currents were recorded from 9–13 DIV neurons bathed in an external solution containing 115 mM NaCl, 4 mM KCl, 10 mM BaCl2, 1 mM MgCl2, 10 mM TEACl, 10 mM Glucose, 10 mM HEPES, and 1 μM TTX (pH 7.4). In some experiments, 10 μM Bisindolymaleimide I or 500 nM PMA were added to the bathing solution before recording. Individual transfected neurons were identified by positive GFP fluoresence by using an Axiovert 200-inverted fluorescence microscope (Carl Zeiss). Patch pipettes with resistances measuring 2–4 MΩ were filled with internal recording solution [120 mM CsMeSO4−, 4 mM MgCl2, 10 mM EGTA, 10 mM HEPES, 4 mM Mg-ATP, and 3 mM Tris-GTP (pH 7.2) with CsOH], and peak whole-cell Ba2+ currents were measured by using an Axon Axopatch 200Bamplifier (Axon Instruments) interfaced to a PC by a Digidata 1320 (Axon Instruments). Data were acquired at 5 kHz, with leak and capacitative transients subtracted online with a P/4 protocol by using pClamp 8.0 software. Access resistance was continually monitored during experiments, and cells with calculated voltage errors of >5 mV were excluded from analysis.
Hippocampal Slices.
Three- to six-month-old WT or heterozygous mice were deeply anesthetized with ether and decapitated. Transverse slices of 450 μm were cut with a vibratome, transferred to an interface chamber, and kept at a constant temperature of 35°C. The perfusing artificial cerebrospinal fluid contained 124 mM NaCl, 4 mM KCl, 2 mM MgSO4, 2 mM CaCl2, 1.25 mM KH2PO4, 26 mM NaHCO3, and 10 mM glucose (pH 7.4) equilibrated with 95% O2 and 5% CO2. Orthodromic field potentials, PSs, and fEPSPs were recorded simultaneously from the cell layer and dendritic area in the CA1 subfields by using glass microelectrodes filled with NaCl 0.9% (resistance 1–2 MΩ) amplified with a high-impedance amplifier (Neurodata) and digitized with a Labview interface. Bipolar stimulation of Schaffer collateral was performed by 50-μm-diameter stainless steel wire electrodes. PPF stimulus protocol was performed with stimuli giving barely maximal responses to an unconditioned stimulus and included IPIs from 5 to 500 ms; low-frequency stimulation protocol was performed with 2-Hz stimuli for 15 sec by using stimuli capable of evoking either maximum or half-maximal responses. Paired pulse ratio (PPR) of PSs was calculated as 2nd PS/1st PS; PPR of EPSPs was calculated as 2nd EPSP 10–90% slope/1st EPSP 10–90% slope. The response to 2-Hz stimuli was calculated as (fEPSPn/fEPSP1−1)×100(%). Statistic evaluation was made by using a nonparametric Mann–Whitney U test.
ACKNOWLEDGMENTS.
We thank Dr. M. Wilson (University of New Mexico, Albuquerque, NM) for the SNAP-25 mutant mice; Dr. J. Sorensen for the cDNAs for SNAP-25a and SNAP-25b; M. Holt and M. Druminski (both of Max Planck Institute for Biophysical Chemistry, Göttingen, Germany) for SNAP-47 cDNA and antibody and SNAP-29 cDNA; Dr. F. Bianco (University of Milan, Milan) for mice genotyping; Drs. M. Caleo and F. Antonucci (both of Consiglio Nazionale delle Richerche–Institute of Neuroscience, Pisa, Italy) for help in some experiments; and Dr. T. Galli (Centre National de la Recherche Scientifique, Paris) and C. Frassoni (Istituto Besta, Milan, Italy) for support and discussions. This work was supported by EU Synapse Integrated Project (LSHM-CT-2005-019055), Telethon GGP04196, and Italian Ministry of University and Research Grant PRIN2005 2005051177_002 (to M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Bark IC. Mol Biol. 1993;5:67–76. doi: 10.1006/jmbi.1993.1485. [DOI] [PubMed] [Google Scholar]
- 2.Bark IC, Hahn KM, Ryabinin AE, Wilson MC. Proc Natl Acad Sci USA. 1995;28:1510–1514. doi: 10.1073/pnas.92.5.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bark IC, Wilson MC. Gene. 1994;139:291–292. doi: 10.1016/0378-1119(94)90773-0. [DOI] [PubMed] [Google Scholar]
- 4.Jahn R, Lang T, Sudhof TC. Cell. 2003;21:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 5.Sudhof TC. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 6.Schiavo G, Stenbeck G, Rothman JE, Sollner TH. Proc Natl Acad Sci USA. 1997;4:997–1001. doi: 10.1073/pnas.94.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Augustine GJ. Curr Opin Neurobiol. 2001;11:320–326. doi: 10.1016/s0959-4388(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 8.Chapman ER. Nat Rev Mol Cell Biol. 2002;3:498–508. doi: 10.1038/nrm855. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Kim-Miller MJ, Fukuda M, Kowalchyk JA, Martin T. Neuron. 2002;16:599–611. doi: 10.1016/s0896-6273(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 10.Bai J, Wang CT, Richards DA, Jackson MB, Chapman ER. Neuron. 2004;25:929–942. doi: 10.1016/s0896-6273(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 11.Gerachshenko T, Blackmer T, Yoon EJ, Bartleson C, Hamm HE, Alford S. Nat Neurosci. 2005;8:597–605. doi: 10.1038/nn1439. [DOI] [PubMed] [Google Scholar]
- 12.He Y, Kang Y, Leung YM, Xia F, Gao X, Xie H, Gaisano HY, Tsushima RG. Biochem J. 2006;1:363–369. doi: 10.1042/BJ20051478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvis SE, Zamponi GW. Cell Calcium. 2005;37:483–488. doi: 10.1016/j.ceca.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Sheng ZH, Rettig J, Cook T, Catterall WA. Nature. 1996;1:451–454. doi: 10.1038/379451a0. [DOI] [PubMed] [Google Scholar]
- 15.Rettig J, Sheng ZH, Kim DK, Hodson CD, Snutch TP, Catterall WA. Proc Natl Acad Sci USA. 1996;9:7363–7368. doi: 10.1073/pnas.93.14.7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Moutot N, Charvin N, Leveque C, Sato K, Nishiki T, Kozaki S, Takahashi M, Seagar M. J Biol Chem. 1996;22:6567–6570. doi: 10.1074/jbc.271.12.6567. [DOI] [PubMed] [Google Scholar]
- 17.Wiser O, Trus M, Hernandez A, Renstrom E, Barg S, Rorsman P, Atlas D. Proc Natl Acad Sci USA. 1999;5:248–253. doi: 10.1073/pnas.96.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiser O, Bennett MK, Atlas D. EMBO J. 1996;15:4100–4110. [PMC free article] [PubMed] [Google Scholar]
- 19.Ji J, Yang SN, Huang X, Li X, Sheu L, Diamant N, Berggren PO, Gaisano HY. Diabetes. 2002;51:1425–1436. doi: 10.2337/diabetes.51.5.1425. [DOI] [PubMed] [Google Scholar]
- 20.Zhong H, Yokoyama CT, Scheuer T, Catterall WA. Nat Neurosci. 1999;2:939–941. doi: 10.1038/14721. [DOI] [PubMed] [Google Scholar]
- 21.Verderio C, Pozzi D, Pravettoni E, Inverardi F, Schenk U, Coco S, Proux-Gillardeaux V, Galli T, Rossetto O, Frassoni C, Matteoli M. Neuron. 2004;19:599–610. doi: 10.1016/s0896-6273(04)00077-7. [DOI] [PubMed] [Google Scholar]
- 22.Washbourne P, Thompson PM, Carta M, Costa ET, Mathews JR, Lopez-Bendito G, Molnar Z, Becher MW, Valenzuela CF, Partridge LD, Wilson MC. Nat Neurosci. 2002;5:19–26. doi: 10.1038/nn783. [DOI] [PubMed] [Google Scholar]
- 23.Bark C, Bellinger FP, Kaushal A, Mathews JR, Partridge LD, Wilson MC. J Neurosci. 2004;6:8796–8805. doi: 10.1523/JNEUROSCI.1940-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan PY, Cai Q, Lin L, Lu PH, Duan S, Sheng ZH. J Biol Chem. 2005;8:25769–25779. doi: 10.1074/jbc.M502356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holt M, Varoqueaux F, Wiederhold K, Takamori S, Urlaub H, Fasshauer D, Jahn R. J Biol Chem. 2006;23:17076–17083. doi: 10.1074/jbc.M513838200. [DOI] [PubMed] [Google Scholar]
- 26.Sorensen JB, Matti U, Wei SH, Nehring RB, Voets T, Ashery U, Binz T, Neher E, Rettig J. Proc Natl Acad Sci USA. 2002;5:1627–1632. doi: 10.1073/pnas.251673298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimazaki Y, Nishiki T, Omori A, Sekiguchi M, Kamata Y, Kozaki S, Takahashi MY. J Biol Chem. 1996;14:14548–14553. doi: 10.1074/jbc.271.24.14548. [DOI] [PubMed] [Google Scholar]
- 28.Genoud S, Pralong W, Riederer BM, Eder L, Catsicas S, Muller D. J Neurochem. 1999;72:1699–1706. doi: 10.1046/j.1471-4159.1999.721699.x. [DOI] [PubMed] [Google Scholar]
- 29.Hepp R, Cabaniols JP, Roche PA. FEBS Lett. 2002;4:52–56. doi: 10.1016/s0014-5793(02)03629-3. [DOI] [PubMed] [Google Scholar]
- 30.Yokoyama CT, Sheng ZH, Catterall WA. J Neurosci. 1997;15:6929–6938. doi: 10.1523/JNEUROSCI.17-18-06929.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levitan IB. Annu Rev Physiol. 1994;56:193–212. doi: 10.1146/annurev.ph.56.030194.001205. [DOI] [PubMed] [Google Scholar]
- 32.Ahlijanian MK, Striessnig J, Catterall WA. J Biol Chem. 1991;266:20192–20197. [PubMed] [Google Scholar]
- 33.Hell JW, Appleyard SM, Yokoyama CT, Warner C, Catterall WA. J Biol Chem. 1994;269:7390–7396. [PubMed] [Google Scholar]
- 34.Iwasaki S, Kataoka M, Sekiguchi M, Shimazaki Y, Sato K, Takahashi M. J Biochem. 2000;128:407–414. doi: 10.1093/oxfordjournals.jbchem.a022768. [DOI] [PubMed] [Google Scholar]
- 35.Verderio C, Bacci A, Coco S, Pravettoni E, Fumagalli G, Matteoli M. Eur J Neurosci. 1999;11:2793–2800. doi: 10.1046/j.1460-9568.1999.00697.x. [DOI] [PubMed] [Google Scholar]
- 36.Kataoka M, Kuwahara R, Matsuo R, Sekiguchi M, Inokuchi K, Takahashi M. Neurosci Lett. 2006;30:258–262. doi: 10.1016/j.neulet.2006.08.055. [DOI] [PubMed] [Google Scholar]
- 37.Katz B, Miledi R. J Physiol. 1968;195:481–492. doi: 10.1113/jphysiol.1968.sp008469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zucker RS. Annu Rev Neurosci. 1989;12:13–31. doi: 10.1146/annurev.ne.12.030189.000305. [DOI] [PubMed] [Google Scholar]
- 39.Zucker RS, Regehr WG. Annu Rev Physiol. 2002;64:355–405. doi: 10.1146/annurev.physiol.64.092501.114547. [DOI] [PubMed] [Google Scholar]
- 40.Risinger C, Bennett MK. J Neurochem. 1999;72:614–624. doi: 10.1046/j.1471-4159.1999.0720614.x. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, Craig TJ, Chen X, Ciufo LF, Takahashi M, Morgan A, Gillis KD. J Gen Physiol. 2007;129:233–244. doi: 10.1085/jgp.200609685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy G, Matti U, Nehring RB, Binz T, Rettig J, Neher E, Sorensen JB. J Neurosci. 2002;22:9278–9286. doi: 10.1523/JNEUROSCI.22-21-09278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoji-Kasai Y, Itakura M, Kataoka M, Yamamori S, Takahashi M. Eur J Neurosci. 2002;15:1390–1394. doi: 10.1046/j.1460-9568.2002.01972.x. [DOI] [PubMed] [Google Scholar]
- 44.Houeland G, Nakhost A, Sossin WS, Castellucci VF. J Neurophysiol. 2007;97:134–143. doi: 10.1152/jn.00122.2006. [DOI] [PubMed] [Google Scholar]
- 45.Yokoyama CT, Myers SJ, Fu J, Mockus SM, Scheuer T, Catterall WA. Mol Cell Neurosci. 2005;28:1–17. doi: 10.1016/j.mcn.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 46.Taverna E, Saba E, Rowe J, Francolini M, Clementi F, Rosa P. J Biol Chem. 2004;13:5127–5134. doi: 10.1074/jbc.M308798200. [DOI] [PubMed] [Google Scholar]
- 47.Kataoka M, Kuwahara R, Iwasaki S, Shoji-Kasai Y, Takahashi M. J Neurochem. 2000;74:2058–2066. doi: 10.1046/j.1471-4159.2000.0742058.x. [DOI] [PubMed] [Google Scholar]
- 48.Andersson J, Fried G, Lilja L, Meister B, Bark C. Eur J Cell Biol. 2000;79:781–789. doi: 10.1078/0171-9335-00106. [DOI] [PubMed] [Google Scholar]
- 49.Gonelle-Gispert C, Costa M, Takahashi M, Sadoul K, Halban P. Biochem J. 2002;15:223–232. doi: 10.1042/BJ20020896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez-Salas E, Steward LE, Ho H, Garay PE, Sun SW, Gilmore MA, Ordas JV, Wang J, Francis J, Aoki KR. Proc Natl Acad Sci USA. 2004;2:3208–3213. doi: 10.1073/pnas.0400229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Vilaythong AP, Yoshor D, Noebels JL. J Neurosci. 2004;2:5239–5248. doi: 10.1523/JNEUROSCI.0992-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hess EJ, Collins KA, Wilson MC. J Neurosci. 1996;1:3104–3111. doi: 10.1523/JNEUROSCI.16-09-03104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barr CL, Feng Y, Wigg K, Bloom S, Roberts W, Malone M, Schachar R, Tannock R, Kennedy JL. Mol Psychiatry. 2000;5:405–409. doi: 10.1038/sj.mp.4000733. [DOI] [PubMed] [Google Scholar]
- 54.Mill J, Curran S, Kent L, Gould A, Huckett L, Richards S, Taylor E, Asherson P. Am J Med Genet. 2002;114:269–271. doi: 10.1002/ajmg.10253. [DOI] [PubMed] [Google Scholar]
- 55.Kim JW, Biederman J, Arbeitman L, Fagerness J, Doyle AE, Petty C, Perlis RH, Purcell S, Smoller JW, Faraone SV, Sklar P. Am J Med Genet B Neuropsychiatr Genet. 2007;144:776–780. doi: 10.1002/ajmg.b.30522. [DOI] [PubMed] [Google Scholar]
- 56.Banker GA, Cowan WM. Brain Res. 1977;126:397–442. doi: 10.1016/0006-8993(77)90594-7. [DOI] [PubMed] [Google Scholar]
- 57.Brewer GJ, Torricelli JR, Evege EK, Price PJ. J Neurosci Res. 1993;1:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- 58.Bozzi Y, Vallone D, Borrelli E. J Neurosci. 2000;20:8643–8649. doi: 10.1523/JNEUROSCI.20-22-08643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grant NJ, Hepp R, Krause W, Aunis D, Oehme P, Langley K. J Neurochem. 1999;72:363–372. doi: 10.1046/j.1471-4159.1999.0720363.x. [DOI] [PubMed] [Google Scholar]
- 60.Frassoni C, Inverardi F, Coco S, Ortino B, Grumelli C, Pozzi D, Verderio C, Matteoli M. Neuroscience. 2005;131:813–823. doi: 10.1016/j.neuroscience.2004.11.042. [DOI] [PubMed] [Google Scholar]