Abstract
Tendons play a significant role in the modulation of forces transmitted between bones and skeletal muscles and consequently protect muscle fibers from contraction-induced, or high-strain, injuries. Myostatin (GDF-8) is a negative regulator of muscle mass. Inhibition of myostatin not only increases the mass and maximum isometric force of muscles, but also increases the susceptibility of muscle fibers to contraction-induced injury. We hypothesized that myostatin would regulate the morphology and mechanical properties of tendons. The expression of myostatin and the myostatin receptors ACVR2B and ACVRB was detectable in tendons. Surprisingly, compared with wild type (MSTN+/+) mice, the tendons of myostatin-null mice (MSTN−/−) were smaller and had a decrease in fibroblast density and a decrease in the expression of type I collagen. Tendons of MSTN−/− mice also had a decrease in the expression of two genes that promote tendon fibroblast proliferation: scleraxis and tenomodulin. Treatment of tendon fibroblasts with myostatin activated the p38 MAPK and Smad2/3 signaling cascades, increased cell proliferation, and increased the expression of type I collagen, scleraxis, and tenomodulin. Compared with the tendons of MSTN+/+ mice, the mechanical properties of tibialis anterior tendons from MSTN−/− mice had a greater peak stress, a lower peak strain, and increased stiffness. We conclude that, in addition to the regulation of muscle mass and force, myostatin regulates the structure and function of tendon tissues.
Keywords: scleraxis, tenomodulin, type I collagen, GDF-8
Tendons are a critical component of the musculoskeletal system. Situated between bones and skeletal muscles, tendons are in a position to transmit forces generated within skeletal muscle fibers to bone and conversely transmit to skeletal muscle external loads placed on bone. The extracellular matrix (ECM) of tendon tissue is composed primarily of type I collagen, as well as type III collagen, elastin, and various proteoglycans and mucopolysaccharides. Tendons are in series with both the contractile and noncontractile elements of skeletal muscles as well as with bone. Consequently, tendons are able to both store elastic energy during locomotion and protect muscle fibers from stretch-induced and contraction-induced injuries (1, 2). Whereas considerable research has been conducted on the effects of exercise, immobilization, and aging on the structure and function of tendons (3, 4), much less is known about the specific cytokines that regulate the structure and function of tendons.
During embryonic development of the limb, early tendon development can be categorized into three phases, with each phase corresponding to an up-regulation of the basic helix–loop–helix transcription factor scleraxis (5, 6). Scleraxis is a marker of the tendon cell lineage (6, 7), and mice deficient in scleraxis display severe tendon defects and have impaired locomotion and a complete inability to use their tails (8). Scleraxis promotes the formation of tendon ECM by inducing the expression of type I collagen (9) and increases tendon fibroblast proliferation by the up-regulation of the expression of the type II transmembrane protein tenomodulin (10). During the final stages of early tendon development FGF-4 and FGF-8 are secreted by adjacent myogenic cells and induce the expression of scleraxis (11, 12). Although FGF-4 and FGF-8 can induce scleraxis expression in the final phase of early tendon development, they do not appear to be responsible for the first and second phases of scleraxis expression (5). In fact, another member of the GDF family may be a candidate for the regulation of scleraxis expression during the first and second phases of early tendon development (5), but the specific member of the GDF family has not been identified.
Three members of the GDF family have been reported to influence the development of tendon tissue. The placement of matrices coated with GDF-5, GDF-6, and GDF-7 into skeletal muscle induces the ectopic formation of tendon-like tissue (13). The tendons of GDF5−/− mice are smaller and display decreases in type I collagen content, peak stress, stiffness, and energy absorption to yield (14). The GDF5−/− mice also have severe bone and joint defects (14), but whether these changes in tendon mechanical properties arise because of a direct effect of GDF-5 on tendon cells or the associated skeletal defects is not clear. Compared with wild-type mice, GDF7−/− mice have a minor tendon phenotype, with a decrease in proteoglycan content and smaller collagen fibrils, but no differences in Achilles tendon mechanical properties, type I collagen content, or gross morphology (15). Some evidence supports roles for GDF-5, GDF-6, and GDF-7 in tendon development, but whether other members of the GDF family influence tendon development has not been established.
Myostatin (GDF-8) is a member of the TGF-β superfamily of cytokines and is a negative regulator of skeletal muscle mass. Myostatin binds to the activin type IIB (ACVR2B) and type IB (ACVRB) receptors and activates the Smad2/3, p38 MAPK, and Erk1/2 signal transduction pathways (16–20). Myostatin regulates muscle mass in part by inhibiting the proliferation of myoblasts (21, 22). The treatment of myoblasts with myostatin induces the expression of the cyclin-dependent kinase inhibitor p21 and results in the subsequent arrest of myoblasts in the G1 phase of the cell cycle (21–23). Myostatin has a well established role in regulation of the structure and function of skeletal muscle, but the contribution of myostatin to the regulation of the structure and function of tendon has not been established.
In addition to the regulation of muscle mass, myostatin has a profound impact on the contractile properties of skeletal muscles. Inhibition of myostatin increases the maximum isometric force of skeletal muscles (24–26) and the susceptibility of muscles to contraction-induced injury (26). During a lengthening contraction, the series elastic component (aponeurosis and tendon) of a muscle protects muscle fibers from damage by reducing the strain on fibers (1). The MSTN−/− mice are much more susceptible to contraction-induced injury than the MSTN+/+ mice, an observation that is consistent with the possibility that myostatin might play a role in the regulation of the structural and functional properties of the tendons. Our prior study (26) focused on the mechanical and contractile properties of the muscle and aponeurosis but did not investigate the mechanical properties of the tendons of MSTN−/− mice directly. The overall aim of this investigation was to determine the role of myostatin in regulation of the mechanical and morphological properties of tendons. We hypothesized that a deficiency in myostatin results in smaller, stiffer, and more brittle tendons.
Results
MSTN−/− Mice Have Greater Muscle Masses but Smaller Tendons.
We first determined the impact of myostatin deficiency on the mass of tendons. Although the mass of the tibialis anterior (TA) muscles of MSTN−/− mice was 72% greater than that of MSTN+/+ mice, the TA tendons of the MSTN−/− mice were 40% smaller (Table 1). When the tendon mass was normalized by the muscle mass, the MSTN−/− mice had a 64% decrease in the tendon/muscle mass ratio. Similar results were observed for soleus muscles and Achilles tendons. The mass of the soleus muscles of MSTN−/− mice was 82% greater than that of MSTN+/+ mice, but the Achilles tendons of the soleus muscles of MSTN−/− mice were 44% smaller than those of MSTN+/+ mice. Consequently, for MSTN−/− mice the Achilles tendon/soleus muscle mass ratio was decreased by 69%. Furthermore, for the MSTN−/− mice, the cross-sectional area (CSA) of the TA tendons was 50% smaller than that of the MSTN+/+ mice (Table 2). The lengths and densities of the TA tendons of MSTN−/− and MSTN+/+ mice were not different. CSA and the lengths and densities of Achilles tendons were not determined, because the Achilles tendons were not used in testing of mechanical properties.
Table 1.
Whole animal, muscle, and tendon masses
| Mouse | Mouse mass, g | TA tendon mass, mg | TA muscle mass, mg | TA tendon/muscle mass ratio | Achilles tendon mass, mg | Soleus mass, mg | Achilles tendon/soleus muscle mass ratio |
|---|---|---|---|---|---|---|---|
| MSTN+/+ | 33.8 ± 1.1 | 1.30 ± 0.10 | 51.84 ± 1.58 | 0.025 ± 0.002 | 2.34 ± 0.10 | 8.10 ± 0.29 | 0.291 ± 0.005 |
| MSTN−/− | 36.8 ± 1.1 | 0.78 ± 0.05* | 89.34 ± 3.92* | 0.009 ± 0.001* | 1.32 ± 0.10* | 14.80 ± 0.21* | 0.090 ± 0.008* |
Values are means ± SE. n = 5 for each genotype.
*, significantly different from MSTN+/+ at P < 0.05.
Table 2.
Morphological and mechanical properties of tibialis anterior tendons
| Mouse | Lo, mm | CSA, mm2 | Density, mg/mm3 | Peak strain, ΔL/Lo | Peak stress, kPa | Peak stiffness, mN/mm2 | Average stiffness, mN/mm2 | Energy absorption, mJ/mg |
|---|---|---|---|---|---|---|---|---|
| MSTN+/+ | 6.78 ± 0.28 | 0.16 ± 0.03 | 1.54 ± 0.42 | 0.75 ± 0.02 | 8,250 ± 1,223 | 95.0 ± 17.1 | 49.6 ± 8.3 | 1.93 ± 0.56 |
| MSTN−/− | 6.12 ± 0.10 | 0.08 ± 0.01* | 1.63 ± 0.06 | 0.33 ± 0.04* | 20,300 ± 1,067* | 1,386 ± 212* | 711 ± 253* | 1.89 ± 0.27 |
Values are means ± SE. n = 5 for each genotype.
*, significantly different from MSTN+/+ at P < 0.05.
Tendon Fibroblasts Express the Myostatin Receptors and Activate the p38 MAPK and Smad2/3 Signaling Pathways in Response to Myostatin Treatment.
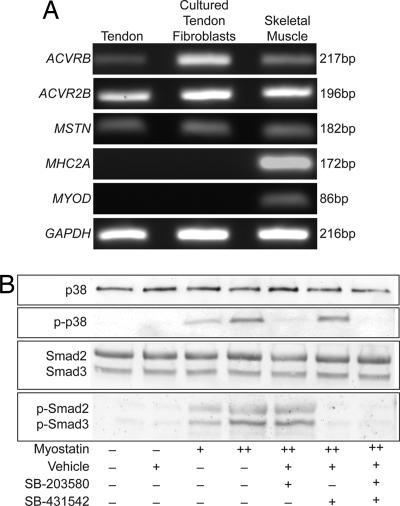
Subsequently, the expression of the myostatin receptors ACVR2B and ACVRB was examined in tendon fibroblasts. Transcripts for both ACVR2B and ACVRB were identified in whole tendon tissue as well as in cultured tendon fibroblasts (Fig. 1A). The myostatin transcript was also detected in tendon tissue and in cultured fibroblasts. Because of the close anatomical proximity between muscle and tendon tissue, the purity of tendon samples was verified by probing for the presence of type IIa myosin heavy chain and MyoD. Neither muscle-specific gene was detected in tendon samples.
Fig. 1.
Tendon fibroblasts expressed the myostatin receptors and activated signal transduction cascades in the presence of myostatin. (A) PCR analysis of cDNA libraries from whole tendon tissue and cultured tendon fibroblasts indicated that tendon fibroblasts expressed both of the myostatin receptors (ACVRB and ACVR2B) as well as myostatin (MSTN) itself. MHC2A and MyoD were used as negative controls to indicate purity of tendon samples. (B) Treatment of tendon fibroblasts with myostatin for 2 h results in the phosphorylation of both p38 MAPK and Smad2/3. The p38 MAPK inhibitor SB-203580 was able to specifically block the myostatin-mediated phosphorylation of p38 MAPK, and the Smad2/3 inhibitor SB-431542 was able to specifically block the myostatin-mediated phosphorylation of Smad2/3. +, 500 ng/ml myostatin; ++, 1,000 ng/ml myostatin.
The next step was to determine whether cultured tendon fibroblasts activate intracellular signaling cascades in response to myostatin treatment. Myostatin induced the phosphorylation of both p38 MAPK and Smad2/3 (Fig. 1B). When cells were treated with myostatin in the presence of the p38 MAPK inhibitor SB-203580 (27), the phosphorylation of p38 MAPK did not occur. Consequently, SB-203580 was specific to the p38 MAPK pathway, because this inhibitor did not block the phosphorylation of Smad2/3. For cells that were treated with myostatin in the presence of the Smad2/3 inhibitor SB-431542 (28), the phosphorylation of Smad2/3 was blocked, with no effect on the phosphorylation of p38 MAPK. These results indicated that tendon fibroblasts expressed the myostatin receptors and were responsive to myostatin treatment and that this response could be blocked by the use of SB-203580 and SB-431542.
Myostatin Induces the Proliferation of Tendon Fibroblasts.
To determine whether myostatin induced the proliferation of tendon fibroblasts, fibroblasts were pulsed with BrdU and the cells were subsequently treated with myostatin, SB-203580, and SB-431542 for 24 h. Treatment with 1,000 ng/ml myostatin increased tendon cell proliferation by 37% over controls (Fig. 2A). Whereas the inhibition of the p38 MAPK pathway decreased the myostatin-mediated increase in fibroblast proliferation, the inhibition of the Smad2/3 pathway did not block the myostatin-mediated increase in fibroblast proliferation. Consistent with the increase in proliferation, myostatin decreased the levels of p21 in fibroblasts (Fig. 2B). Compared with MSTN+/+ mice, the density of fibroblasts in whole tendon tissue was 47% less than that of the MSTN−/− mice (Fig. 2C). These results indicated that myostatin was a potent regulator of tendon cell proliferation.
Fig. 2.
Myostatin induced the proliferation of tendon fibroblasts. (A) Treatment of tendon fibroblasts for 24 h with myostatin increases cell proliferation as measured by the relative incorporation of the thymidine analog BrdU. n = 3 independent experiments. +, 500 ng/ml myostatin; ++, 1,000 ng/ml myostatin; *, significantly different from control group at P < 0.05. (B) Treatment of tendon fibroblasts for 24 h with myostatin decreases p21 protein levels. β-Tubulin is shown as a loading control. (C) Cell density data from tibialis anterior tendon sections stained with H&E. MSTN−/− mice have a lower fibroblast density than MSTN+/+ mice. n = 6 tendons per genotype. *, significantly different from MSTN+/+ at P < 0.05.
Myostatin Induces the Expression of Scleraxis, Tenomodulin, and Type I Collagen in Tendon Fibroblasts.
The next step was to determine whether myostatin regulated the expression of scleraxis and tenomodulin, the two genes that induce the proliferation of tendon fibroblasts. Treatment with 1,000 ng/ml myostatin resulted in a >2-fold increase in scleraxis expression (Fig. 3A). Inhibiting the p38 MAPK pathway resulted in a 70% decrease in scleraxis expression, whereas inhibition of the Smad2/3 pathway resulted in a 50% increase in scleraxis expression. Myostatin treatment doubled the expression of tenomodulin, and inhibition of both the p38 MAPK and Smad2/3 pathways blocked the myostatin-mediated increase in tenomodulin expression (Fig. 3B). Compared with MSTN+/+ mice, MSTN−/− mice had a 64% decrease in scleraxis expression (Fig. 3D) and a 63% decrease in tenomodulin expression (Fig. 3E). These results indicated that the mechanisms responsible for the myostatin-mediated increase in fibroblast proliferation were due to an up-regulation of scleraxis and tenomodulin.
Fig. 3.
Myostatin induces the expression of scleraxis, tenomodulin, and collagen Iα2 genes in tendon fibroblasts. Treatment of cells with myostatin for 24 h increases the relative expression of scleraxis (A), tenomodulin (B), and collagen Iα2 (C) normalized to GAPDH. +, 500 ng/ml myostatin; ++, 1,000 ng/ml myostatin; *, significantly different from control group at P < 0.05. There is a decrease in the expression of scleraxis (D), tenomodulin (E), and collagen Iα2 (F) normalized to GAPDH in tendons of MSTN−/− mice. n = 4 tendons per genotype. *, significantly different from MSTN+/+ at P < 0.05.
Because of the smaller mass and CSA of the tendons of MSTN−/− mice, the impact of myostatin on the major structural protein of tendon, type I collagen, was determined. Treatment with 1,000 ng/ml myostatin resulted in a 67% increase in the expression of type I collagen (Fig. 3C). The inhibition of either the p38 MAPK pathway or the Smad2/3 pathway was sufficient to block this increase in type I collagen expression. Compared with MSTN+/+ mice, a 54% decrease in type I collagen expression was observed in the tendons of MSTN−/− mice (Fig. 3F). Taken together, the cell proliferation and gene expression data indicated that the smaller tendons of the MSTN−/− mice depended on a decrease in tendon fibroblast proliferation and on the production of the constituents of the ECM.
MSTN−/− Mice Have Stiff, Brittle Tendons.
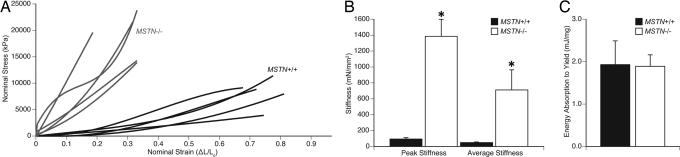
The profound impact of myostatin on the structure of tendons indicated that myostatin deficiency likely influenced the mechanical properties of tendons. Consequently, the stress–strain relationships of tendons from MSTN+/+ and MSTN−/− mice were measured (Fig. 4A). Compared with the tendons of MSTN+/+ mice, those of MSTN−/− mice reached a >2-fold higher peak stress before yielding but reached less than half of the peak strain before yielding (Table 2). Tendons of MSTN−/− mice also demonstrated a 14-fold-greater peak stiffness and average stiffness values than those of MSTN+/+ mice (Fig. 4B and Table 2). Despite the different stress–strain and stiffness properties of tendons from MSTN+/+ and MSTN−/− mice, the tendons absorbed the same amount of energy before reaching the yield point (Fig. 4C and Table 2). These results indicated that the loss of myostatin had a major impact on the mechanical properties of tendons.
Fig. 4.
Mechanical properties of tibialis anterior tendons of MSTN+/+ and MSTN−/− mice. (A) The stress–strain relationship of tendons from MSTN+/+ and MSTN−/− mice indicates that MSTN−/− mice develop a higher peak stress but have a lower peak strain. (B) MSTN−/− mice have a greater peak stiffness and average stiffness than MSTN+/+ mice. (C) The energy absorbed to the yield point is not different between MSTN+/+ and MSTN−/− mice. n = 5 tendons per genotype. *, significantly different from MSTN+/+ at P < 0.05.
Discussion
Myostatin has a well characterized role in the regulation of the structure and function of skeletal muscles. Although the myostatin transcript has been detected previously in tendons (29), to our knowledge our results provide the first evidence that myostatin regulates directly the structure and function of tendons. For the MSTN−/− mice, the deficiency in myostatin resulted in small, brittle, hypocellular tendons. The difference in tendon phenotypes between the MSTN−/− and MSTN+/+ mice is clearly not attributable to any indirect influence of a greater muscle mass of the MSTN−/− mice, because both genotypes were limited to cage sedentary activity levels and the two genotypes showed no differences in body masses. Consequently, the tendons of both groups of mice experienced very similar mechanical loads throughout their lifespan, and the dramatic change in the structure and function of tendons could be attributed directly to the effect of myostatin on tendon fibroblasts. In addition, this conclusion is supported further by the concurrent cell culture experiments.
Although myostatin promotes the synthesis of intramuscular collagen content (26), whether myostatin regulated the collagen content of tendon was not immediately evident. The inhibition of myostatin in mdx mice, a murine model of Duchenne muscular dystrophy, decreased fibrosis and increased maximum isometric force production (24, 25, 30). Compared with MSTN+/+ mice, a decrease in the type I collagen content of extensor digitorum longus muscles of MSTN−/− mice was observed (26). Myostatin also induced the expression of type I collagen in skeletal muscle myotubes (26) and muscle-derived fibroblasts (31). The current investigation indicated that myostatin also promotes type I collagen synthesis in tendon fibroblasts both in vivo and in vitro.
Myostatin plays very different roles in the control of the proliferation of tendon cells and muscle cells. This difference is evident in the regulation of the expression of p21. In the present study myostatin decreased the expression of p21 and subsequently increased the proliferation of tendon fibroblasts. A cytokine closely related to myostatin, TGF-β, decreased the expression of p21 and increased the proliferation of tendon fibroblasts (32) and NIH/3T3 fibroblasts (33). In contrast to tendon cells, myostatin decreased the proliferation of myoblasts by increasing the expression of the muscle-specific basic helix–loop–helix transcription factor MyoD and subsequently increased the expression of p21, resulting in arrest in the G1 phase of the cell cycle (21–23). Although MyoD appears to be a key transcription factor in the regulation of p21 in myoblasts (34, 35), the transcription factors that regulate the expression of p21 in tendon cells have yet to be determined.
A rapidly growing body of literature supports the role of scleraxis and tenomodulin in the embryonic development of tendons (5–12, 36), but little is known regarding their function in adult tendons. FGF-4 and FGF-8 directly induced the expression of scleraxis in tendon fibroblasts (11, 12, 37, 38). Furthermore, TGF-β induced the expression of scleraxis in osteosarcoma cells (39). Overexpression of scleraxis in tendon fibroblasts either directly or indirectly resulted in the up-regulation of tenomodulin (10). The relative expression of scleraxis and tenomodulin in the tendons of MSTN+/+ and MSTN−/− mice in this study was in good agreement with the data on tendon mass and cell density. Similar to tenomodulin-deficient mice, the tendons of MSTN−/− mice were hypocellular (36). The results from the current study indicate that scleraxis and tenomodulin are expressed in adult tendons and that both genes are downstream targets of myostatin, which activates signal transduction cascades similar to TGF-β and FGF.
During several stages of embryonic development, myogenic cells and tendon precursor cells interact with each other to ensure proper spatial alignment and proper timing of differentiation events (5, 40). While myostatin was expressed in myogenic cells and decreased the expression of the myogenic genes MyoD, Myf-5, and Pax3 in these cells, myostatin was also expressed in nonmyogenic cells of the ectoderm (41), but the reason for the expression of myostatin in nonmuscle cells was not known. Scleraxis expression occurred in three phases of tendon development (5, 6), with the final phase initiated by FGF-4 and FGF-8 (11, 12) produced in adjacent myogenic cells. The signal that initiated the first phase of scleraxis expression in tendon progenitor cells is not known, but this signal does not come from muscle cells, because removal of myogenic cells does not alter scleraxis expression (12). The ectoderm appeared critical to the induction of the first stage of scleraxis expression, because ablation of the ectoderm abolished the first phase of scleraxis expression (6). Myostatin was expressed in the ectoderm around the time of the first phase of scleraxis expression (41), and removal of the ectoderm resulted in a down-regulation of scleraxis. Consequently, myostatin may play a direct role in the development of tendons by regulating the initial expression of scleraxis.
One of the most striking differences between the mechanical properties of MSTN+/+ and MSTN−/− mice was the 14-fold increase in the stiffness of tendons in MSTN−/− mice. The stiffness of tendons is a critical factor in determining the damage to muscle fibers during lengthening contractions. Immediately after a two-stretch lengthening contraction protocol, the extensor digitorum longus muscles of MSTN−/− mice had a 15% greater force deficit than MSTN+/+ mice (26). The force deficit after a contraction-induced injury is directly related to the strain on the muscle fibers during the lengthening contraction (42). Consequently, the series elastic component of a muscle protects the sarcomeres from damage by limiting the strain of the muscle fibers during the contraction (1). The increased stiffness of tendons from MSTN−/− mice is likely an underlying mechanism behind the greater force deficits of muscles from MSTN−/− mice after contraction-induced injury. Although the mechanisms responsible for the increased stiffness of tendons from MSTN−/− mice are not known, the stiffening of tendons is thought to arise as a result of the increased cross-linking between type I collagen molecules (43). Future studies that evaluate the biochemical and molecular differences between the tendons of MSTN+/+ and MSTN−/− mice are necessary to determine the mechanisms behind the regulation of tendon mechanical properties by myostatin.
Considerable interest has focused on the potential use of myostatin inhibitors in the treatment of muscle wasting diseases such as Duchenne muscular dystrophy (44–46). Although myostatin inhibition has the potential to produce a greater muscle mass and decrease fibrosis, muscles of mdx mice (47–51) and patients with Duchenne muscular dystrophy (52) are highly susceptible to contraction-induced injury. An increase in the stiffness of tendons would increase the susceptibility of dystrophic muscles to contraction-induced injury and exacerbate the symptoms of muscular dystrophy. Whether the stiff, brittle, hypocellular phenotype of tendons of adult MSTN−/− mice arises because of prenatal or postnatal mechanisms, or a combination of both, is not clear. However, the spatiotemporal expression of myostatin during development, in conjunction with the current results that tendon fibroblasts continue to express both the myostatin cytokine and receptors into adulthood and that scleraxis is a direct downstream target of myostatin signaling, strongly suggest that myostatin plays a role in both the prenatal and postnatal development and regulation of tendons.
Methods
Animals.
All experiments were conducted in accordance with the guidelines of the University of Michigan Committee on the Use and Care of Animals. Mice were housed in specific-pathogen-free conditions and were provided food and water ad libitum. The MSTN−/− mice used in this study are of a C57BL/6 background and were a kind gift of Se-Jin Lee (The Johns Hopkins University, Baltimore). The null MSTN allele was generated by replacing a portion of the third exon of the MSTN gene that encodes the C-terminal region of the mature myostatin protein with a neo cassette (53). The wild-type (MSTN+/+) littermates of the MSTN−/− mice served as controls. The genotype of mice was determined by PCR-based analysis of DNA samples obtained via tail biopsy.
Mechanical Testing of Tendons.
To evaluate the mechanical properties of the tibialis anterior tendon, the entire tendon unit (from the myotendinous junction to the base of the first metatarsal bone) was used. The tibialis anterior tendon was chosen based on its relative uniformity of diameter, minimal aponeurosis, and long gauge length (54). Six-month-old male mice were anesthetized with i.p. injection of Avertin (400 mg/kg). Braded silk sutures were tied around the distal end of the tibialis anterior muscle just superior to the myotendinous junction and at the very distal end of the tendon just superior to the first metatarsal. The length (Lo) of the tibialis anterior tendon was measured by using digital calipers while the ankle was placed in maximal plantarflexion. The tendon was removed by cutting the muscle just superior to the proximal suture and by removing the first metatarsal bone that was inferior to the distal suture. The tendon was immediately submerged in PBS maintained at 25°C. The tendon was held at Lo, and CSA was calculated from 10 evenly spaced width and depth measurements from high-resolution digital photographs of both top and side views of the tendon. Side views were obtained by using a 90° prism embedded in the side of the bath. These measurements were fitted to an ellipse, and the ellipse area was used as the tendon CSA.
The proximal end of the tendon was attached to a dual-mode servo motor/force transducer (model 305C; Aurora Scientific), and the distal end was attached to a fixed post. Custom-designed software (LabVIEW 7.1; National Instruments) controlled the servo motor motion and recorded force and strain data at a sampling rate of 20 kHz. The tendon was stretched to a 100% strain relative to Lo at a velocity of 1 Lo × s−1. Peak stress was defined as the stress at which further increases in length resulted in a rupture of the tendon or the point at which yield strength had been reached without a frank rupture of the tendon. Peak strain was defined as the strain at which peak stress was reached. The data were fitted by either a fourth- or fifth-order polynomial function with an R2 ≥ 0.9995. Peak tendon stiffness was calculated by differentiating the fitted polynomial and then determining its maximum value between Lo and peak strain. Average tendon stiffness was calculated as the mean value of the differentiated polynomial between Lo and peak strain. The energy absorption to the yield point was calculated by integrating the force-displacement function from Lo through peak strain and normalizing this value by the mass of the tendon.
Histology.
To determine the density of fibroblasts in tendon tissue, tibialis anterior tendons were removed from 6-month-old anesthetized mice, placed in embedding media, and snap-frozen in isopentane cooled with dry ice. Sections were obtained from the proximal, middle, and distal thirds of the tendon and stained with H&E. Fibroblast density for the entire tendon was calculated by taking the mean value of the counts from each of the three regions of the tendon.
Tendon Fibroblast Isolation, Culture, and Treatment with Myostatin.
Hindlimb and forelimb tendons were isolated from anesthetized 4-month-old male MSTN+/+ mice, carefully trimmed of muscle and fat tissue, finely minced, and placed in DMEM plus 0.05% type II collagenase (Invitrogen) in a shaking water bath for 2 h at 37°C. After dissociation, fibroblasts were pelleted by centrifugation, resuspended in DMEM plus 2% FBS plus 1% antibiotic-antimycotic (AbAm), and expanded in 100-mm culture dishes coated with type I collagen (BD Biosciences). Fibroblasts were passaged twice upon reaching 70% confluence. After the last passage, 2 × 104 fibroblasts were plated in 35-mm culture dishes coated with type I collagen and expanded until reaching 80% confluence. Fibroblasts were then starved of serum for 24 h before treatment by replacing serum-containing media with DMEM plus 1% AbAm plus 1× insulin–transferrin–selenium supplement (Invitrogen). Recombinant murine myostatin, produced in NS0 mouse myeloma cells (R & D Systems), was dissolved into the serum-free media at a final concentration of 500 or 1,000 ng/ml. Stock solutions of the p38 MAPK inhibitor SB-203580 (27) and the Smad2/3 inhibitor SB-431542 (28) were prepared by dissolving these solid anhydrous compounds in DMSO at a concentration of 10 mM. These stock solutions were then added to serum-free media containing 1% DMSO at a final concentration of 10 μM for SB-203580 and 5 μM for SB-431542. Fibroblasts were pretreated with SB-203580 or SB-431542 for 1 h before treatment with myostatin.
Cell Proliferation and Immunocytochemistry.
After serum starvation, fibroblasts were incubated in serum-free media containing 20 μM of the thymidine analog BrdU (Sigma) for 3 h. Fibroblasts were rinsed twice with serum-free media and treated with myostatin, SB-203580, and SB-431542 as described above. After 24 h of treatment, fibroblasts were rinsed with PBS, fixed in ice-cold methanol, and permeabilized with 0.5% Triton X-100. The BrdU epitope was exposed by digesting DNA with 200 units/ml EcoRI and denaturing DNA with 2 M HCl. BrdU was visualized by using an anti-BrdU antibody (G3G4; S. J. Kaufman, Developmental Studies Hybridoma Bank) and a Cy3-conjugated secondary antibody (Jackson ImmunoResearch). DAPI (Sigma) was used as a nonspecific nuclear stain. Twenty-five random fields were counted per dish. Cell proliferation data presented are the mean ± SE values of three independent experiments.
RT-PCR.
RNA was isolated from samples by using an RNeasy Mini Kit (Qiagen). When isolating RNA from whole tendons, the tissue was treated with type II collagenase and proteinase K before vigorous homogenization in guanidine thiocyanate buffer. Because of the smaller size of the tibialis anterior tendons, we were unable to consistently obtain adequate quantities of RNA with an A260/A280 ratio between 1.8 and 2.0. We instead used RNA from Achilles tendons because we were able to obtain RNA with A260/A280 ratios between 1.8 and 2.0 from these tendons. RNA was treated with DNase I and reverse-transcribed by using an Omniscript RT kit (Qiagen) and oligo(dT)15 primers. Primers for PCRs (Table 3) were designed to generate amplicons that span multiple exons. For standard PCR, 250 ng of cDNA underwent 42 rounds of amplification with GoTaq Green (Promega). PCR products were separated by using a 2% agarose gel. For real-time PCR, cDNA was amplified by using a QuantiTect SYBR Green I PCR system (Qiagen) with uracil-N-glycosylase (Invitrogen) in an Opticon 2 real-time thermal cycler (Bio-Rad). Quantitative PCRs (qPCRs) were conducted in quadruplicate for each sample. We used the methods of Livak and Schmittgen (55) to determine optimal loading quantities of cDNA and to validate the use of GAPDH as a housekeeping gene. Gene expression was normalized to GAPDH expression by using the 2-ΔΔC(t) method (55). The presence of single amplicons from qPCRs was verified by melting curve analysis as well as electrophoresis using a 2% agarose gel. qPCR data presented are the combined means ± SE of three independent experiments.
Table 3.
PCR primer sequences
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| ACVRB | GCTGGAAAGCCCTTCTACTG | TGATGACCAGGAAGACGATG |
| ACVR2B | GAAGATGAGGCCCACGATTA | GGAGGTCACCAGAGAGACGA |
| COL1A2 | CCAGCGAAGAACTCATACAGC | GGACACCCCTTCTACGTTGT |
| GAPDH | TGGAAAGCTGTGGCGTGAT | TGCTTCACCACCTTCTTGAT |
| MHC2A | CCAAGTCAGAGGCAAAGAGG | TCTTTGATTTTGGCCTCCAG |
| MSTN | TGCAAAATTGGCTCAAACAG | GCAGTCAAGCCCAAAGTCTC |
| MYOD | CGCTCCAACTGCTCTGATG | TAGTAGGCGGTGTCGTAGCC |
| TNMD | TGTACTGGATCAATCCCACTCT | GCTCATTCTGGTCAATCCCCT |
| SCX | CCTTCTGCCTCAGCAACCAG | GGTCCAAAGTGGGGCTCTCCGTGACT |
Primers for RT-PCR and quantitative RT-PCR. Primers are designed to generate an amplicon that spans two or more exons and can discriminate cDNA from genomic DNA.
Immunoblot.
Tendon fibroblasts were prepared as described above and starved of serum for 24 h. To determine the phosphorylation of p38 MAPK and Smad2/3, cells were treated for 2 h with myostatin and pathway inhibitors. To determine p21 protein content, cells were treated for 24 h with myostatin and pathway inhibitors. After treatment, cells were rinsed with PBS and scraped and homogenized in Laemmli's sample buffer with 1:20 2-mercaptoethanol, 1:20 protease inhibitor mixture (Sigma), and 1:40 phosphatase inhibitor mixture (Sigma) and then placed in boiling water for 5 min. Protein concentration of the samples was determined by using an RC DC Protein Assay (Bio-Rad). Equal amounts of protein were loaded into polyacrylamide gels and subjected to electrophoresis using a 4% stacking, 10% resolving gel for p38 MAPK and Smad2/3 and a 4% stacking, 15% resolving gel for p21 and β-tubulin. Proteins were transferred to a 0.45-μm nitrocellulose membrane, stained with Ponceau S to verify equal protein transfer, and blocked by using casein (Vector Laboratories). Primary antibodies against p38 MAPK and phospho-p38 MAPK were purchased from Cell Signaling Technology, antibodies against Smad2/3 and phospho-Smad2/3 were purchased from Millipore, and antibodies against p21 and β-tubulin were purchased from AbCam. Biotinylated secondary antibodies were purchased from Pierce. Avidin–HRPO conjugates were purchased from Vector Laboratories. Membranes were developed by using SuperSignal West Dura enhanced chemiluminescent reagents (Pierce) and visualized by using a FluorChem SP chemiluminescent documentation system (Alpha Innotech).
Statistical Analysis.
Results are presented as means ± SE. KaleidaGraph 4.02 software was used to conduct statistical tests. For gene expression and cell proliferation data from cell culture experiments, differences between groups were tested with a one-way ANOVA with α = 0.05. Fisher's least significant post hoc test was used to identify specific differences when significance was tested. For all other data, differences between MSTN+/+ and MSTN−/− mice were tested with Student's t test with α = 0.05.
ACKNOWLEDGMENTS.
We thank Dennis Claflin, Keith Baar, Cheryl Hassett, and Kimberly Gates for providing technical assistance. This work was supported by National Institute on Aging Grants AG-13283 and AG-20591 and National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-070071.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Griffiths RI. J Physiol. 1991;436:219–236. doi: 10.1113/jphysiol.1991.sp018547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McHugh M, Pasiakos S. Eur J Appl Physiol. 2004;93:286–293. doi: 10.1007/s00421-004-1196-0. [DOI] [PubMed] [Google Scholar]
- 3.Kjaer M. Physiol Rev. 2004;84:649–698. doi: 10.1152/physrev.00031.2003. [DOI] [PubMed] [Google Scholar]
- 4.Kjaer M, Magnusson P, Krogsgaard M, Boysen Møller J, Olesen J, Heinemeier K, Hansen M, Haraldsson B, Koskinen S, Esmarck B, et al. J Anat. 2006;208:445–450. doi: 10.1111/j.1469-7580.2006.00549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edom-Vovard F, Duprez D. Dev Dyn. 2004;229:449–457. doi: 10.1002/dvdy.10481. [DOI] [PubMed] [Google Scholar]
- 6.Schweitzer R, Chyung J, Murtaugh L, Brent A, Rosen V, Olson E, Lassar A, Tabin C. Development (Cambridge, UK) 2001;128:3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- 7.Pryce B, Brent A, Murchison N, Tabin C, Schweitzer R. Dev Dyn. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- 8.Murchison N, Price B, Conner D, Keene D, Olson E, Tabin C, Schweitzer R. Development (Cambridge, UK) 2007;134:2697–2708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- 9.Léjard V, Brideau G, Blais F, Salingcarnboriboon R, Wagner G, Roehrl M, Noda M, Duprez D, Houillier P, Rossert J. J Biol Chem. 2007;282:17665–17675. doi: 10.1074/jbc.M610113200. [DOI] [PubMed] [Google Scholar]
- 10.Shukunami C, Takimoto A, Oro M, Hiraki Y. Dev Biol. 2006;298:234–247. doi: 10.1016/j.ydbio.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 11.Brent A, Schweitzer R, Tabin C. Cell. 2003;113:235–248. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
- 12.Edom-Vovard F, Schuler B, Bonnin M, Teillet M, Duprez D. Dev Biol. 2002;247:351–366. doi: 10.1006/dbio.2002.0707. [DOI] [PubMed] [Google Scholar]
- 13.Wolfman N, Hattersley G, Cox K, Celeste A, Nelson R, Yamaji N, Dube J, DiBlasio-Smith E, Nove J, Song J, et al. J Clin Invest. 1997;100:321–330. doi: 10.1172/JCI119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikic B, Schalet B, Clark R, Gaschen V, Hunziker E. J Orthop Res. 2001;19:365–371. doi: 10.1016/S0736-0266(00)90018-4. [DOI] [PubMed] [Google Scholar]
- 15.Mikic B, Bierwert L, Tsou D. J Orthop Res. 2006;24:831–841. doi: 10.1002/jor.20092. [DOI] [PubMed] [Google Scholar]
- 16.Lee S, McPherron A. Proc Natl Acad Sci USA. 2001;98:9306–9311. doi: 10.1073/pnas.151270098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philip B, Lu Z, Gao Y. Cell Signalling. 2005;17:365–375. doi: 10.1016/j.cellsig.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Rebbapragada A, Benchabane H, Wrana J, Celeste A, Attisano L. Mol Cell Biol. 2003;23:7230–7242. doi: 10.1128/MCB.23.20.7230-7242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, Chen Y, Zhang Y, Wang X, Yang N, Zhu D. Cancer Res. 2006;66:1320–1326. doi: 10.1158/0008-5472.CAN-05-3060. [DOI] [PubMed] [Google Scholar]
- 20.Zhu X, Topouzis S, Liang L, Stotish R. Cytokine. 2004;26:262–272. doi: 10.1016/j.cyto.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 21.McCroskery S, Thomas M, Maxwell L, Sharma M, Kambadur R. J Cell Biol. 2003;162:1135–1147. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas M, Langley B, Berry C, Sharma M, Kirk S, Bass J, Kambadur R. J Biol Chem. 2000;275:40235–40243. doi: 10.1074/jbc.M004356200. [DOI] [PubMed] [Google Scholar]
- 23.Yang W, Zhang Y, Li Y, Wu Z, Zhu D. J Biol Chem. 2007;282:3799–3808. doi: 10.1074/jbc.M610185200. [DOI] [PubMed] [Google Scholar]
- 24.Bogdanovich S, Krag T, Barton E, Morris L, Whittemore L, Ahima R, Khurana T. Nature. 2002;420:418–421. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- 25.Bogdanovich S, Perkins K, Krag T, Whittemore L, Khurana T. FASEB J. 2005;19:543–549. doi: 10.1096/fj.04-2796com. [DOI] [PubMed] [Google Scholar]
- 26.Mendias C, Marcin J, Calerdon D, Faulkner J. J Appl Physiol. 2006;101:898–905. doi: 10.1152/japplphysiol.00126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuenda A, Rouse J, Doza Y, Meier R, Cohen P, Gallagher T, Young P, Lee J. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 28.Inman G, Nicolás F, Callahan J, Harling J, Gaster L, Reith A, Laping N, Hill C. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 29.Heinemeier K, Olesen J, Schjerling P, Haddad F, Langberg H, Baldwin K, Kjaer M. J Appl Physiol. 2007;102:573–581. doi: 10.1152/japplphysiol.00866.2006. [DOI] [PubMed] [Google Scholar]
- 30.Wagner K, McPherron A, Winik N, Lee S. Ann Neurol. 2002;52:832–836. doi: 10.1002/ana.10385. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, Li Y, Shen W, Qiao C, Ambrosio F, Lavasani M, Nozaki M, Branca M, Huard J. J Biol Chem. 2007;282:25852–25863. doi: 10.1074/jbc.M704146200. [DOI] [PubMed] [Google Scholar]
- 32.Salingcarnboriboon R, Yoshitake H, Tsuji K, Obinata M, Amagasa T, Nifuji A, Noda M. Exp Cell Res. 2003;287:289–300. doi: 10.1016/s0014-4827(03)00107-1. [DOI] [PubMed] [Google Scholar]
- 33.Dkhissi F, Raynal S, Jullien P, Lawrence DA. Oncogene. 1999;18:703–711. doi: 10.1038/sj.onc.1202341. [DOI] [PubMed] [Google Scholar]
- 34.Guo K, Wang J, Andres V, Smith RC, Walsh K. Mol Cell Biol. 1995;15:3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 36.Docheva D, Hunziker E, Fässler R, Brandau O. Mol Cell Biol. 2005;25:699–705. doi: 10.1128/MCB.25.2.699-705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brent A, Tabin C. Development (Cambridge, UK) 2004;131:3885–3896. doi: 10.1242/dev.01275. [DOI] [PubMed] [Google Scholar]
- 38.Smith T, Sweetman D, Patterson M, Keyse S, Münsterberg A. Development (Cambridge, UK) 2005;132:1305–1314. doi: 10.1242/dev.01699. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Cserjesi P, Nifuji A, Olson E, Noda M. J Endocrinol. 1996;151:491–499. doi: 10.1677/joe.0.1510491. [DOI] [PubMed] [Google Scholar]
- 40.Kardon G. Development (Cambridge, UK) 1998;125:4019–4032. doi: 10.1242/dev.125.20.4019. [DOI] [PubMed] [Google Scholar]
- 41.Amthor H, Huang R, McKinnell I, Christ B, Kambadur R, Sharma M, Patel K. Dev Biol. 2002;251:241–257. doi: 10.1006/dbio.2002.0812. [DOI] [PubMed] [Google Scholar]
- 42.Brooks SV, Zerba E, Faulkner JA. J Physiol. 1995;488:459–469. doi: 10.1113/jphysiol.1995.sp020980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tuite D, Renström P, O'Brien M. Scand J Med Sci Sports. 1997;7:72–77. doi: 10.1111/j.1600-0838.1997.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 44.Chakkalakal JV, Thompson J, Parks RJ, Jasmin BJ. FASEB J. 2005;19:880–891. doi: 10.1096/fj.04-1956rev. [DOI] [PubMed] [Google Scholar]
- 45.Patel K, Macharia R, Amthor H. Acta Myol. 2005;24:230–241. [PubMed] [Google Scholar]
- 46.Tobin J, Celeste A. Curr Opin Pharmacol. 2005;5:328–332. doi: 10.1016/j.coph.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Consolino CM, Brooks SV. J Appl Physiol. 2004;96:633–638. doi: 10.1152/japplphysiol.00587.2003. [DOI] [PubMed] [Google Scholar]
- 48.Dellorusso C, Crawford RW, Chamberlain JS, Brooks SV. J Muscle Res Cell Motil. 2001;22:467–475. doi: 10.1023/a:1014587918367. [DOI] [PubMed] [Google Scholar]
- 49.Moens P, Baatsen PH, Marechal G. J Muscle Res Cell Motil. 1993;14:446–451. doi: 10.1007/BF00121296. [DOI] [PubMed] [Google Scholar]
- 50.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Proc Natl Acad Sci USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stevens ED, Faulkner JA. J Physiol. 2000;522:457–466. doi: 10.1111/j.1469-7793.2000.t01-3-00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel IM. Muscle Nerve. 1978;1:453–460. doi: 10.1002/mus.880010603. [DOI] [PubMed] [Google Scholar]
- 53.McPherron A, Lawler A, Lee S. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 54.Arruda E, Calve S, Dennis R, Mundy K, Baar K. J Appl Physiol. 2006;101:1113–1117. doi: 10.1152/japplphysiol.00612.2005. [DOI] [PubMed] [Google Scholar]
- 55.Livak K, Schmittgen T. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]