Abstract
The mammalian RBC lacks de novo lipid synthesis but maintains its membrane composition by rapid turnover of acyl moieties at the sn-2 position of phospholipids. Plasma-derived fatty acids are esterified to acyl-CoA by acyl-CoA synthetases and transferred to lysophospholipids by acyl-CoA:lysophospholipid acyltransferases. We report the characterization of three lysophosphatidylcholine (lysoPC) acyltransferases (LPCATs), products of the AYTL1, -2, and -3 genes. These proteins are three members of a LPCAT family, of which all three genes are expressed in an erythroleukemic cell line. Aytl2 mRNA was detected in mouse reticulocytes, and the presence of the product of the human ortholog was confirmed in adult human RBCs. The three murine Aytl proteins generated phosphatidylcholine from long-chain acyl-CoA and lysoPC when expressed in Escherichia coli membranes. Spliced variants of Aytl1, affecting a conserved catalytic motif, were identified. Calcium and magnesium modulated LPCAT activity of both Aytl1 and -2 proteins that exhibit EF-hand motifs at the C terminus. Characterization of the product of the Aytl2 gene as the phosphatidylcholine reacylating enzyme in RBCs represents the identification of a plasma membrane lysophospholipid acyltransferase and establishes the function of a LPCAT protein.
Keywords: phosphatidylcholine, RBC, AYTL2
The complex plasma membrane lipid composition of the mature mammalian RBC is maintained throughout the lifespan of the cell. Its molecular composition comprises more than 250 different glycerophospholipid molecular species (1). This enucleated cell lacks internal organelles and de novo lipid synthesis. To maintain the integrity of its membrane, lipid molecules are continuously renewed. Different classes of fatty acids are found at the two positions of the glycerol backbone of phospholipids, and their distribution is heterogeneous (1). The sn-1 position of glycerophospholipids is mainly occupied by saturated acylchains 16–18 carbon atoms long, whereas a variety of unsaturated fatty acylchains are found at the sn-2 position. The oxygen-carrying function of the RBC leads to free radical damage of unsaturated acyl chains that need to be removed and replaced to maintain cellular integrity (2). Oxidized acyl chains are rapidly removed from membranes by (calcium-dependent) phospholipase A2 action (2). The reacylation of the resulting lysophospholipid (lysoPL) is achieved by a two-step CoA and ATP-dependent process. Fatty acids from the environment are activated to acyl-CoA by membrane-bound long-chain acyl-CoA synthetases (ACSL), after which the acyl group is transferred to lysophospholipid by acyl-CoA:lysophosphatidylcholine (lysoPC) acyltransferases (LPLAT). This repair mechanism is also known as the Lands cycle (3).
Phosphatidylcholine (PC) is a major component of RBC membranes and serum lipoproteins, and lysoPC is rapidly converted in RBC membranes to PC (4). We previously identified the acyl-CoA synthetase (ACSL6) catalyzing the formation of long-chain acyl-CoA esters at the expense of CoA and ATP (5), but no member of the LPLAT family has been reported in RBCs. We report the identification of an acyl-CoA:lysophosphatidylcholine acyltransferase (LPCAT) in the RBC. The murine form, Aytl2 (GeneID 210992; LPCAT1), was successfully expressed in Escherichia coli and generated PC from long-chain acyl-CoA and lysoPC. The product of the human ortholog, AYTL2, has been identified in the proteome of the human RBC (6). LPCAT activity was inhibited by divalent cations (Ca2+ and Mg2+), indicating that the predicted EF-hand motifs present in the C terminus regulate activity of Aytl2 and suggesting that not only deacylation (phospholipase action) but also reacylation in RBCs can be modulated by divalent cations.
In addition, we report the identification of two additional homolog proteins with LPCAT activity. The proteins coded by Aytl1 (GeneID 270084) and Agpat7 (previously named Aytl3; GeneID 99010) converted lysoPC to PC in the presence of acyl-CoA when expressed in the membrane of E. coli. Aytl1 contains a plsC domain and two EF-hand motifs and has been identified previously as a protein with unknown function. Agpat7 has been annotated previously by sequence similarity as member 7 (eta form) of the lysophosphatidic acid acyltransferase (LPAAT) family (7). Based on our findings, we propose that the products of the three Aytl genes are all three members of the LPCAT family, one of which is responsible for lysoPC reacylation in RBC membranes and is a LPCAT previously unidentified in plasma membranes.
Results
Characterization of the Aytl Family Members.
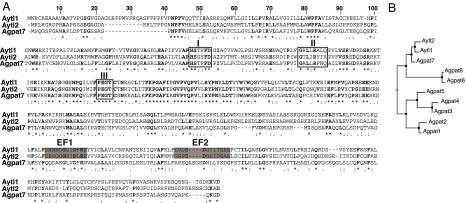
Until recently (8, 9), LPCAT has been an elusive protein not identified in any organism. Previous attempts for its isolation suggested that protein(s) of an apparent molecular mass of 50–60 kDa could be labeled with a photoreactive azido-lysoPC derivative in a partially purified microsomal fraction containing a LPCAT activity (10). To identify candidate LPCATs in RBCs, we searched databases for proteins carrying the E. coli plsC domain, which is characteristic of all known 1-acyl-sn-glycerol-3-phosphate acyltransferases (11–13), and excluded all known enzymes that have been described as lysophosphatidic acid acyltransferases (LPAATs). We focused particularly on two gene products of unknown function with a predicted molecular mass of 60 kDa, annotated in human as AYTL1 and AYTL2 and as Aytl1 and Aytl2 in mouse (Fig. 1). In addition, it became apparent that the product of member 7 of the postulated LPAAT family, defined as AGPAT7 in the human and Agpat7 in the mouse, shows more similarity to the AYTL proteins than to the members of the AGPAT family (Fig. 1). This gene was originally annotated as AYTL3 and although its activity was not determined, it was later reannotated as AGPAT7 (7). Orthologs of the human and mouse AYTL proteins are present in other mammals [supporting information (SI) Fig. 7]. The three isoforms Aytl1, -2, and -3 are more similar to each other than to the other known and predicted LPAAT enzymes (Fig. 1B). In Aytl1, -2, and -3, the acyltransferase domain was at the N terminus (1–250 aa), with the three motifs (I, II, and III) conserved among acyltransferase enzymes (Fig. 1A). In addition, two EF-hand motifs, EF1 and EF2, could be identified at the C terminus of Aytl1 and -2 (Fig. 1A). EF-hand motifs form a hairpin structure that constitutes a binding site for the cations calcium and magnesium and are often present in tandem or multicopy.
Fig. 1.
Sequence analysis of the Aytl family members. (A) Sequence alignment of mouse Aytl proteins. Note that Agpat7 was originally annotated as Aytl3. The conserved residues are in bold, the positions of the three conserved motifs are boxed, and the two predicted EF-hand hairpins (EF1 and EF2) are shaded in gray. Alignment was performed with CLUSTALW. The GenBank accessions numbers are as follows: Aytl1, NP_766602.1; Aytl2, NP_663351.2; Agpat7, NP_997089.1. (B) Phenogram illustrating the sequence relatedness of the putative mouse lysoPL acyltransferase members. Analysis was performed with MUTALIN.
Identification of an Acyl-CoA:1-Acyl-LysoPC Acyltransferase Family.
Full-length cDNA for mouse Aytl1, -2, and -3 was cloned and expressed in E. coli. Proteins were detected in the membrane fraction (SI Fig. 8 and data not shown). In addition, two recombinant truncated forms of Aytl2 were generated by removal of the plsC domain at the N terminus (construct ΔAT) and of the predicted EF-hand (ΔEF) at the C terminus. The expression level of the ΔAT construct was similar to the full-length protein, but the expression of the ΔEF truncated version was low.
Acyltransferase activities were measured with E. coli membranes in the presence of lysophospholipid and [14C]acyl-CoA.
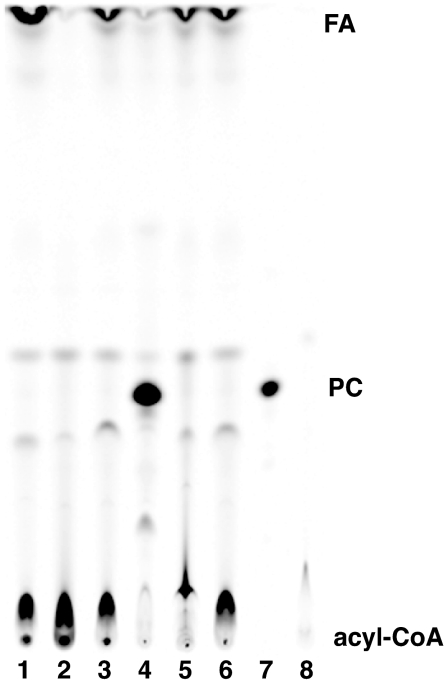
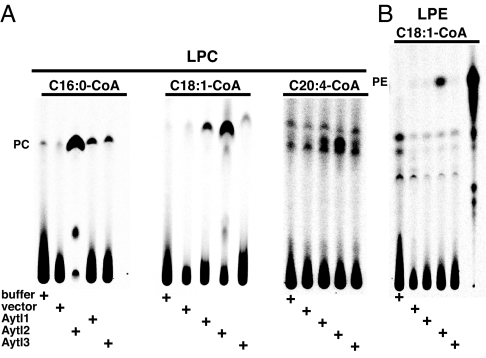
The full-length Aytl2 construct generated [14C]PC from [14C]C18:1-CoA and lysoPC (Fig. 2). The acyltransferase activity in the E. coli membrane appeared to be robust, because the reaction reached completion and all [14C]C18:1-CoA was ligated into PC in our conditions within 20 min. Different acyl-CoA species were accepted by full-length Aytl2 as substrates (Fig. 3A). The two recombinant Aytl1 and Aytl3 proteins, in the E. coli membrane fraction, were also able to acylate lysoPC in the presence of various acyl-CoA donors (Fig. 3A). Whereas the same amount of membranes was used in each reaction (10 μg), the PC formation by Aytl1 and -3 was found to be lower than for Aytl2. Activity was higher with increased amount of microsomes (data not shown). This result may reflect the difference in expression and/or activity of those enzymes in the lipid environment of E. coli. Nevertheless, Aytl1 and Aytl3 appeared to have a similar preference for the acyl-CoA donor presented. PC formation was higher with C16:0-CoA than with C18:1-CoA and lowest with C20:4-CoA. In the presence of lysophosphatidylethanolamine (lysoPE), full-length Aytl2 generated small amounts of radioactive PE from [14C]C18:1-CoA (Fig. 3B). This was not observed with Aytl1 and Aytl3. None of the other lysophospholipids tested (lysophosphatidic acid, lysophosphatidylserine, and lysophosphatidylinositol) resulted in the generation of the corresponding radioactive phospholipid by any of the three proteins (data not shown). Truncation of the N terminus of Aytl2 (construct ΔAT) or the C terminus (ΔEF) led to a loss in acyltransferase activity despite the presence of the catalytic motifs I, II, and III in ΔEF.
Fig. 2.
LysoPC acyltransferase activity of Aytl2. Formation of [14C]PC in the presence of [14C]C18:1-CoA (10 μM) and lysoPC (36 μM) with 20 μg of E. coli microsomes is shown. Lane 1, cloned rat LPAAT; lane 2, control no protein; lane 3, vector; lane 4, full-length Aytl2; lane 5, truncated Aytl2-ΔAT construct; lane 6, truncated Aytl2-ΔEF construct; lane 7, [14C]PC standard; lane 8, [14C]C18:1-CoA standard. Reactions were performed at 37°C for 20 min. Products were separated by TLC (see Materials and Methods). FA, fatty acid.
Fig. 3.
Acyltransferase activity of the Aytl proteins. (A) Formation of [14C]PC in the presence of lysoPC (LPC) (36 μM) and 10 μM different acyl-CoA {[14C]C16:0-CoA, [14C]C18:1-CoA, [14C]C20:4-CoA}, as indicated. (B) Formation of [14C]PE in the presence of lysoPE (LPE) (36 μM) was determined with [14C]C18:1-CoA (10 μM). Position of [14C]PC/PE was defined with pure standard. Reactions were performed at 37°C for 20 min with 10 μg of E. coli microsomes.
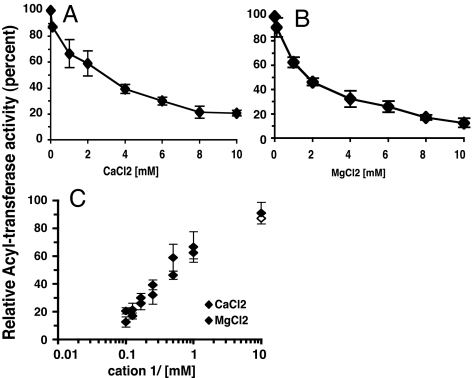
Formation of PC from lysoPC by Aytl2 (Fig. 4) and Aytl1 (data not shown) was inhibited by calcium and magnesium (Fig. 4 A and B). The concentration dependency of this inhibition was very similar for the two cations, achieving 50% inhibition of the enzyme activity rate at ≈2 mM (Fig. 4C). The addition of EDTA to the incubation mix in which the cations were present restored the activity of the enzymes (data not shown).
Fig. 4.
Inhibition of the lysoPC acyltransferase activity of Aytl2 by cations. (A) Rate of formation of [14C]PC as a function of concentration of calcium chloride. (B) Rate of formation of [14C]PC as a function of the concentration of magnesium chloride. (C) Comparison of the acyltransferase inhibition by both cations. Reactions were performed with [14C]C18:1-CoA (10 μM), lysoPC (36 μM), and 5 μg of Aytl2 microsomes in the absence or presence of the indicated cations. Under those conditions, formation of PC was linear during the first 10 min of the reaction, and rates were calculated from 1 to 6 min. The results are expressed as percentages of the relative rates of formation of PC in the presence of the cations to the rate in their absence (see Materials and Methods). The results are the average values of three experiments.
Expression of the Aytl Family Members.
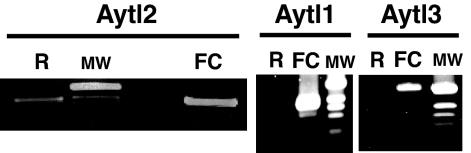
We tested the presence of mRNA for Aytl1, -2, and -3 in reticulocytes and erythroleukemic mouse Friend cells by RT-PCR (Fig. 5). Aytl1, -2, and -3 were detected in erythroleukemic mouse Friend cells. In reticulocytes, only mRNA for Aytl2 was found. Data analysis of the reported proteins in the human RBC proteome supported those findings (6). AYTL1 and AYTL3 (AGPAT7) were not detected, whereas AYTL2 (International Protein Index no. IPI00171626) was identified. Although, recombinant Aytl2 was detected in membranes of other cells (8, 9), AYTL2 was reported in the RBC fraction defined as cytosolic based on the fraction used in mass spectrometry analysis. It is important to note that this fraction also contains other proteins that are membrane proteins (6, 14). We identified three spliced isoforms of Aytl1 that differ in the conserved plsC domain (SI Fig. 9). The splicing events affected exon 5 and exon 6. Aytl1_v1 cDNA represents the correct full-length version predicted from the gene sequence. The Aytl1_v2 variant lacked both exons and Aytl1_v3 lacked exon 5. Exclusion of both exon 5 and exon 6 conserved the correct ORF and produced an isoform lacking 40 residues and the conserved motif III, PEGT. This motif is essential for catalytic activity and may participate in binding to the lysoPL acceptor (12). Splicing of exon 5 in Aytl1 variant 3 resulted in the loss of frame and early translational termination.
Fig. 5.
Expression of the Aytl members in erythroid cells. Reverse transcription of Aytl1, Aytl2, and Aytl3 from total RNA of isolated mouse reticulocytes (lane R) and Friend cells (lane FC) (see Materials and Methods). RT-PCR products were separated on agarose gel and stained with ethidium bromide. In each gel, the 1-kbp DNA molecular weight (MW) standard (Invitrogen) is shown. Note that the two faint bands smaller than the major products of Aytl1 (lane FC) correspond to alternatively spliced variants 2 and 3, shown in SI Fig. 9.
Characterization of Acyl-CoA:1-Acyl-LysoPC Acyltransferase in Isolated Human RBC Membranes.
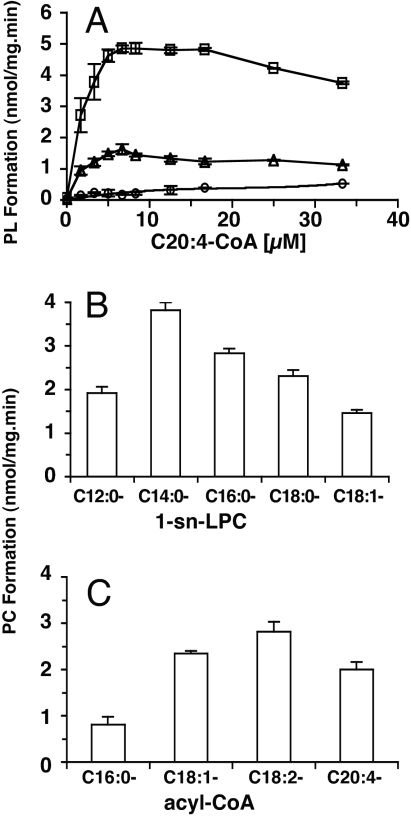
LPLAT activities for the most abundant glycerophospholipids, PC (28% of total phospholipid), phosphatidylethanolamine (25%) and phosphatidylserine (15%) (1), were detected in the membrane of RBCs in the presence of [14C]C20:4-CoA and different 1-acyl-sn-lysoPL. The rate of incorporation of radioactive arachidonoyl-CoA into lysoPC was greater than in lysoPS and lysoPE (Fig. 6A). Under the conditions tested, formation of PC by LPCAT was maximal with acyl-CoA in the 10–20 μM range and was inhibited at higher concentrations. The presence of other lysophospholipid species did not compete with PC formation (data not shown). All of the radioactivity in the PC formed could be released as radioactive fatty acid by phospholipase A2 treatment, indicating that incorporation occurred at the sn-2 position of lysoPC.
Fig. 6.
Characterization of the RBC lysoPC acyltransferase. (A) Rate of formation of [14C]PL (PC, square; PS, triangle; PE, circle) as a function of concentration of [14C]C20:4-CoA. All lysoPLs were 25 μM. The results are the average values of three independent experiments. (B) Rate of formation of [14C]PC as a function of the sn-1-acyl moiety of the lysoPC acceptor. Reactions were performed with [14C]C20:4-CoA (10 μM) in the presence of the different 1-acyl-sn-lysoPCs at 25 μM. (C) Rate of formation of [14C]PC as a function of the acyl species of the acyl-CoA donor. Reactions were performed with 1-[14C](C16:0)-lysoPC (50 μM) in the presence of the different acyl-CoA species at 16.6 μM. The results are the average values of three (A and B) and six (C) independent experiments.
The LPCAT activity was detected over a broad pH range, with a maximum rate observed between pH 5.5 and 7.5 (data not shown). The preference of the enzyme for chain length and degree of unsaturation of the acyl moiety of the lysoPC acceptor was tested with different 1-acyl-sn-lysoPC species. The rate of PC formation from saturated lysoPC was highest for the lysoPC species, with a 14-carbon chain acyl [1-(C14:0)-lysoPC] (Fig. 6B). The increase or reduction of the chain length decreased the rate. This decrease was more pronounced with a reduction in length of the acyl moiety. The decrease in rate from C14 to C12 [1-(C12:0)-lysoPC] was twice as much compared with the addition of 2 carbon atoms [1-(C16:0)-lysoPC]. PC formation from 1-(C18:0)-lysoPC was slower compared with 1-(C16:0)-lysoPC but faster compared with 1-(C12:0)-lysoPC. Together, these data indicate that the chain length of the fatty acyl chain at the sn-1 position of lysoPC affects the incorporation at the sn-2 position. An additional decrease in rate was observed with the presence of one unsaturated bound. The incorporation into [1-(C18:1)-lysoPC] was 40% less compared with the incorporation into 1-(C18:0)-lysoPC (Fig. 6B). Because the predominant fatty acid found at the sn-1 position of PC of human RBCs is C16:0 (1), we chose 1-(C16:0)-lysoPC as the lysoPC acceptor for additional experiments. The diacyl subclass of PC accounts for more than 93% of the PC molecular species in RBCs, and the major acyl species found at the sn-2 position of diacyl-PC in RBC membranes are C16:0 (5%), C18:1 (29%), C18:2 (42%), and C20:4 (9%) (1). At a concentration of 16 μM acyl-CoA, transfer rates were highest for the precursor of the two most abundant acyl species (C18:1-CoA and C18:2-CoA), followed by the polyunsaturated C20:4-CoA and the saturated C16:0-CoA (Fig. 6C). The apparent kinetic parameters of LPCAT were determined by varying the concentrations of the four acyl-CoA donors in the presence of 1-(C16:0)-lysoPC and by changing the concentration of 1-(C16:0)-lysoPC in the presence of different acyl-CoA species (Table 1). It should be noted that RBC membranes prepared from freshly collected RBCs contain lysoPC species up to concentrations of 2% of the total phospholipid pool. The variable amount of this substrate is expected to interfere with the determination of the apparent enzyme kinetics for LPCAT (15). As previously observed for the LPCAT present in rat liver microsomes (15), the nature of the acyl-CoA donor affected the affinity of the enzyme for the lysoPC acceptor. The apparent affinity was at least 3 times lower with the donors C18:1 and C18:2 than with C16:0 and C20:4. The apparent velocity of the enzyme for the different acyl-CoA species confirmed the assays performed at a single concentration of 16 μM (Fig. 6C). Maximum velocity was highest for C18:2-CoA and lowest for C16:0-CoA. Interestingly, the apparent affinity for the two less abundant species found at the sn-2 position (C16:0 and C20:4) was nearly one order of magnitude apart. There was only a 2-fold difference between the two abundant species C18:1 and C18:2. A similar preference was detected for the LPCAT of liver microsomes (16).
Table 1.
Characteristics of the lysoPC acyltransferase activity in RBC membrane
| Species | C16:0-CoA |
C18:1-CoA |
C18:2-CoA |
C20:4-CoA |
||||
|---|---|---|---|---|---|---|---|---|
| Km | Vmax | Km | Vmax | Km | Vmax | Km | Vmax | |
| Acyl-CoA | 0.6 | 2.8 | 4.0 | 5.6 | 7.8 | 11.3 | 4.3 | 4.6 |
| 1-(C16:0)-sn-lysoPC | 6.8 | 1.4 | 27 | 6.8 | 44.5 | 11.8 | 9.2 | 3.6 |
Values represent the kinetic parameters (Km, μM; Vmax, nmol/mg·min) for the indicated acyl-CoA species in the presence of 1-(C16:0)-sn-lysoPC (first row) and for 1-(C16:0)-sn-lysoPC in their presence (second row). Six independent experiments were performed for each combination set.
In summary, acyl-CoA is incorporated into different lysophospholipids by distinct proteins in the red cell membrane. LPCAT activity exhibits specificity for both lysophospholipid and acyl-CoA species. We identified three members of the Aytl family, Aytl1, -2, and -3 (Agpat7), that when expressed as recombinant proteins in E. coli, are LPCATs that generate PC from lysoPC and acyl-CoA. Aytl2 was the only form detected in RBCs.
Discussion
Lysophospholipid acyltransferases play an important role in lipid metabolism because they provide a mechanism for the generation of new phospholipid molecules independent of de novo synthesis (3, 17). Phospholipase-mediated deacylation, followed by reacylation of the lysophospholipid by using acyl-CoA, provides a pathway to remodel phospholipid molecular species in membranes. LPLAT activity, first described in the microsomal fraction of various tissues, has been characterized in many organisms (3, 18, 19).
The eight members of a mammalian protein family, annotated as 1-acylglycerol-3-phosphate-O-acyltransferase (AGPAT), are postulated to represent lysophosphatidic acid (lysoPA) acyltransferases and their products are annotated as LPAAT (alpha, beta, etc.). Five proteins have been identified in human and mouse containing the so-called plsC domain of the E. coli PlsC protein and have been reported to acylate lysoPA in the presence of acyl-CoA (12, 13). However, systematic annotation of any plsC-containing protein homologous to AGPAT as an additional member of this family has led to some confusion, because not all appear to use lysoPA as substrate. No acyltransferase activity could be detected for AGPAT6 (zeta isoform) with lysoPA or any other lysophospholipid (11). We report that member 7 (eta form) acylates lysoPC rather than lysoPA and therefore belongs to the LPCAT family. AGPAT7 was annotated originally as AYTL3 and although its activity has not been determined, unfortunately, it has been reannotated as AGPAT7 (7). Based on our results, this reannotation should be reversed. Activity of AGPAT8 (theta form) has not been determined (20). A protein (GenBank accession no. NP_001002257) that has been proposed to be the product of the AGPAT8 gene (GeneID 84803; MAG1) has been shown to acylate lysoPA (21), but this enzyme was erroneously identified because it is the product of the LYCAT gene (Gene ID:253558, ALCAT1). ALCAT1 has been shown previously to be a lysocardiolipin acyltransferase, and no lysoPA-AT activity could be detected (22). In this report, we establish that each product of the three genes currently or formerly annotated as AYTL (acyltransferase-like member 1, 2, and 3) has a lysoPC acyltransferase activity. In addition, mouse Aytl2 has been reported previously as LPCAT1 in lungs (8, 9). Therefore, these proteins could be identified as members of the to be defined AGPCAT (1-acylglycerol-3-phosphatidyl-choline-O-acyltransferase) family, rather than “acyltransferase-like” members.
Remodeling and repair of plasma membrane phospholipids, including the RBC membrane, are essential to maintain cellular viability. In the RBC, fatty acids from plasma are rapidly incorporated into phospholipids, and lipid turnover occurs continuously (23). The substrate pool of plasma fatty acids changes depending on diet, but the phospholipid molecular species composition of the RBC membrane is remarkably well maintained, suggesting a highly specific system. An ATP- and CoA-dependent acyl-CoA synthetase (ACSL6) catalyzes the activation of fatty acids to acyl-CoA esters (5, 24). LysoPL acyltransferases transfer the activated fatty acid to lysoPL. Fatty acids are taken up into phospholipids at different rates (4), and acyl-CoA specificity of LPCAT can be established in isolated RBC membranes. We show that formation of PC from lysoPC and acyl-CoA follows in a general sense the abundance of the different PC molecular species in the RBC (1). Although such studies are informative with respect to the relative activity toward different substrates, the enzyme kinetics for LPCAT can only be described as “apparent”. Several factors, including supporting proteins such as acyl-CoA-binding proteins (25) and the complex protein lipid environment, will affect the enzyme kinetics of any of the LPLATs. The specificity of the reacylation of lysophospholipids in the RBC plasma membrane may be typical for all plasma membranes.
We characterized the product of Aytl2 as a candidate for the RBC LPCAT. Detection of Aytl2 mRNA in reticulocytes was confirmed by reported proteomic data (6). When expressed in the lipid environment of E. coli, the Aytl2 protein preferred acyl-CoA ester with a C16:0 and C18:1 species over C20:4. In the RBC membrane, apparent affinity of the enzyme was highest for C16:0-CoA, but enzyme activity was higher for longer and unsaturated acyl species (Fig. 6C). The poor utilization of C20:4-CoA by Aytl2 in E. coli and other cells (8) is different from the high incorporation rate of arachidonoyl-CoA by isolated RBC membranes. As argued above, a different membrane environment will influence the apparent enzyme activity and will likely lead to the observed differences. Despite these considerations, preference for lysoPC species with a short and saturated acyl moiety at the sn-1 position is very similar in RBCs and other cells (8). The weak, but measurable lysoPE acyltransferase activity of mouse Aytl2, was detected in E. coli and reported in CHO-K1 microsomes (8). Enzymes with a similar specificity for lysoPC and lysoPE have also been reported in rat liver Golgi membranes (26) and other tissues (19).
In contrast to Aytl2, the two other homologs, Aytl1 and Aytl3 (Agapt7), were not expressed in reticulocytes and could not be identified in adult RBCs. Expression of these proteins in E. coli established their LPCAT activity. Because RBCs generate all glycerophospholipids, other yet to be determined LPLATs must be present in the RBC membrane.
Increased calcium concentrations above the normal submicromolar level in the cytosol of RBCs affect lipid turnover. Permeabilization and increase in cytosolic calcium will trigger many pathways, including those that lead to a loss in phospholipid asymmetry and the activation of phospholipases including phospholipase D (27). Under such conditions, it is difficult to determine the direct effect of calcium on the LPCAT enzyme in the RBC membranes. We found that the acyltransferase activity of Aytl2 in E. coli was reversibly inhibited by Ca2+ and Mg2+. Inhibition of LPCAT activity by Ca2+ and Mg2+ has been reported previously in the microsomal fraction of rat submandibular glands (50 and 67% inhibition at 10 mM, respectively) (28) and heart microsomes (50% inhibition at 10 mM) (18). Whereas no effect of calcium (at an unspecified concentration) has been observed on mouse Aytl2 activity expressed in CHO-K1 cells (8), both magnesium (≥5 mM) and manganese (≥2 mM) have been reported to stimulate rat Aytl2 activity expressed in HEK293 cells (9). The reason for these discrepancies is unknown. Importantly, our results indicate that 50% inhibition was only observed in the 1–2 mM range, the range found in plasma. This is several orders of magnitude higher than that found under normal physiological conditions in RBC cytosol (29). One could argue that under conditions in which the RBC is not able to maintain its calcium homeostasis and phospholipases are activated, reacylation would be a futile activity. In fact, failure to repair their damaged membrane would ensure their rapid destruction.
In summary, we identified a family of three LPCAT enzymes and proposed that AYTL2 is the reacylating enzyme for PC in RBC membrane. To our knowledge, this would represent the first established biological function of a LPCAT in plasma membranes.
Materials and Methods
Cloning of Mouse Aytl1, Aytl2, and Aytl3 cDNA.
cDNA clones encoding the full-length product of mouse Aytl1 (GeneID 270084) and Aytl2 (GeneID 210992) were identified in the RIKEN database (Kabushiki Kaisha DNAForm) as Mouse Fantom clone ID A330042H22 [DNA Data Bank of Japan (DDBJ) accession no. AK039431; 2,800 bp] and clone ID F630214O14 (DDBJ accession no. AK155286; 3,645 bp), respectively. Full-length mouse AGPAT7 (Aytl3; GeneID 99010) cDNA, represented by IMAGE clone 6414152 (GenBank accession no. BC068131; 1,919 bp), was obtained from the American Type Collection Center (ATCC no. 10698833). PCR fragments encoding the full-length ORF were cloned into the expression vector pET28a (Novagen), to yield plasmid pFK221 (Aytl1), pFK192 (Aytl2), and pFK225 (Aytl3), respectively, with a unique in-frame hexahistidine tag at the N terminus. Deletions of the plsC domain (N terminus) and of the EF-hand motifs (C terminus) were performed by restriction digests of the cloned full-length recombinant Aytl2 construct (549 aa, 61.1 kDa) carried by pFK192. The plsC domain was removed with PasI restriction, followed by self-ligation of the large fragment to yield pFK207 (Aytl2-ΔAT) (324 aa, 35.5 kDa). Deletion of the region containing the two EF-hand motifs was performed by restriction with MscI, filling in with a Klenow fragment, and self-ligation of the large fragment to yield pFK208 (Aytl2-ΔEF) (316 aa, 35.4 kDa).
Protein Expression and Membrane Preparation.
Rosetta 2(DE3) was used as the host strain in all experiments. Cells were grown at 37°C in LB liquid medium to an OD600 nm of ≈0.6, followed by 3 h in the presence of 500 μM isopropyl β-d-thiogalactoside. Microsomes were made in 0.2 M Tris·HCl, pH 7.4/0.5 mM PMSF, as described in ref. 5. After obtaining informed consent, heparinized blood was collected from healthy human donors by venipuncture. RBC ghost membranes were prepared according to Dodge et al. (30).
Measurement of Acyl-CoA:1-Acyl-LysoPL Acyltransferase in Isolated Human RBC Membranes.
The activity of LPCAT activity in isolated human RBC membranes was determined by measuring the formation of [14C]PC from either 1-[14C](C16:0)-lysoPC and acyl-CoA or lysoPC and [14C]acyl-CoA. All reactions were performed in 100 mM Bistris, pH 6.5, at 37°C with 15 μg of RBC membrane proteins in a total volume of 600 μl. The reaction was stopped by adding 0.5 ml of the incubation mixture to 2 ml of chloroform/methanol (2:1, vol/vol). Lipids were extracted and separated by TLC with chloroform/methanol/acetic acid/0.9% NaCl (100:50:16:5, vol/vol). Lipid fractions were visualized by exposure to I2 vapor. PC was scraped off after I2 evaporation, and radioactivity was measured in a scintillation counter. Measurement of acyl-CoA:1-acyl-lysoPL acyltransferase in isolated human RBC membranes was performed as described for LPCAT activity in the presence of the respective lysophospholipid precursors and [14C]C20:4-CoA.
Measurement of LPCAT Activity in Isolated E. coli Membranes.
Incorporation of [14C]acyl-CoA into egg lysoPC by recombinant Aytl proteins in E. coli membranes was determined as follows. Reactions were performed at 37°C in 100 μl of 20 mM Tris·HCl, pH 7.4/0.8 mg/ml Tween-20 containing 36 μM lysoPC and 10 μM [14C]acyl-CoA. The amount of E. coli membrane protein (5 to 20 μg) used in each assay is indicated in the legend of the figures. Reactions were stopped by the addition of 200 μl of chloroform/methanol/12 M HCl (40:40:0.26, vol/vol). Samples were applied to TLC silica plates and developed with the solvent system described above. TLC plates were exposed to a PhosphorImager screen (Storm 840, Molecular Dynamics). Quantification of PC formation was performed with ImageQuant software, subtracting the plate background. The effect of divalent cations on LPCAT activity was obtained by measuring the rate of [14C]PC formation in the absence or presence of various concentrations (up to 10 mM) of calcium chloride or magnesium chloride in the incubation mixture. Each assay was performed in triplicate, with samples to be compared applied and developed on the same plate. The relative activity rate is expressed as the ratio of the rates in the presence compared with the rate in the absence of the divalent cation.
RT-PCRs.
Total RNA isolation and reverse transcription of total RNA isolated from mouse reticulocytes and Friend erythroleukemia cells D1B (ATCC no. TIB-56) were performed as described in ref. 14.
Supplementary Material
ACKNOWLEDGMENTS.
This work was supported by National Institutes of Health Grant HL070583.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF442783 and EF442784).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709737104/DC1.
References
- 1.Myher JJ, Kuksis A, Pind S. Lipids. 1989;24:396–407. doi: 10.1007/BF02535147. [DOI] [PubMed] [Google Scholar]
- 2.van den Berg JJ, Op den Kamp JA, Lubin BH, Kuypers FA. Biochemistry. 1993;32:4962–4967. doi: 10.1021/bi00069a035. [DOI] [PubMed] [Google Scholar]
- 3.Lands WE. J Biol Chem. 1960;235:2233–2237. [PubMed] [Google Scholar]
- 4.Renooij W, Van Golde LMG, Zwaal RFA, Roelofsen B, van Deenen L. Biochim Biophys Acta. 1974;363:287–292. doi: 10.1016/0005-2736(74)90069-8. [DOI] [PubMed] [Google Scholar]
- 5.Soupene E, Kuypers FA. BMC Mol Biol. 2006;7:21. doi: 10.1186/1471-2199-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, Thomas AW, Mann M. Blood. 2006;108:791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- 7.Ye GM, Chen C, Huang S, Han DD, Guo JH, Wan B, Yu L. DNA Seq. 2005;16:386–390. doi: 10.1080/10425170500213712. [DOI] [PubMed] [Google Scholar]
- 8.Nakanishi H, Shindou H, Hishikawa D, Harayama T, Ogasawara R, Suwabe A, Taguchi R, Shimizu T. J Biol Chem. 2006;281:20140–20147. doi: 10.1074/jbc.M600225200. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Hyatt BA, Mucenski ML, Mason RJ, Shannon JM. Proc Natl Acad Sci USA. 2006;103:11724–11729. doi: 10.1073/pnas.0604946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser T, Stobart K. Biochem Soc Trans. 2000;28:715–718. [PubMed] [Google Scholar]
- 11.Beigneux AP, Vergnes L, Qiao X, Quatela S, Davis R, Watkins SM, Coleman RA, Walzem RL, Philips M, Reue K, Young SG. J Lipid Res. 2006;47:734–744. doi: 10.1194/jlr.M500556-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung DW. Front Biosci. 2001;6:D944–D953. doi: 10.2741/leung. [DOI] [PubMed] [Google Scholar]
- 13.Lu B, Jiang YJ, Zhou Y, Xu FY, Hatch GM, Choy PC. Biochem J. 2005;385:469–477. doi: 10.1042/BJ20041348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soupene E, Kuypers FA. Br J Haematol. 2006;133:436–438. doi: 10.1111/j.1365-2141.2006.06051.x. [DOI] [PubMed] [Google Scholar]
- 15.Lands WE, Hart P. J Biol Chem. 1965;240:1905–1911. [PubMed] [Google Scholar]
- 16.Jezyk P, Lands WE. J Lipid Res. 1968;9:525–531. [PubMed] [Google Scholar]
- 17.Lands WE. Annu Rev Biochem. 1965;34:313–346. doi: 10.1146/annurev.bi.34.070165.001525. [DOI] [PubMed] [Google Scholar]
- 18.Arthur G, Choy PC. Biochem J. 1986;236:481–487. doi: 10.1042/bj2360481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita A, Sugiura T, Waku K. J Biochem (Tokyo) 1997;122:1–16. doi: 10.1093/oxfordjournals.jbchem.a021715. [DOI] [PubMed] [Google Scholar]
- 20.Tang W, Yuan J, Chen X, Gu X, Luo K, Li J, Wan B, Wang Y, Yu L. J Biochem Mol Biol. 2006;39:626–635. doi: 10.5483/bmbrep.2006.39.5.626. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal AK, Barnes RI, Garg A. Arch Biochem Biophys. 2006;449:64–76. doi: 10.1016/j.abb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Cao J, Liu Y, Lockwood J, Burn P, Shi Y. J Biol Chem. 2004;279:31727–31734. doi: 10.1074/jbc.M402930200. [DOI] [PubMed] [Google Scholar]
- 23.Oliveira MM, Vaughan M. J Lipid Res. 1964;5:156–162. [PubMed] [Google Scholar]
- 24.Davidson BC, Cantrill RC. FEBS Lett. 1985;193:69–74. doi: 10.1016/0014-5793(85)80081-8. [DOI] [PubMed] [Google Scholar]
- 25.Fyrst H, Knudsen J, Schott MA, Lubin BH, Kuypers FA. Biochem J. 1995;306:793–799. doi: 10.1042/bj3060793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chambers K, Brown WJ. Biochem Biophys Res Commun. 2004;313:681–686. doi: 10.1016/j.bbrc.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Butikofer P, Yee MC, Schott MA, Lubin BH, Kuypers FA. Eur J Biochem. 1993;213:367–375. doi: 10.1111/j.1432-1033.1993.tb17770.x. [DOI] [PubMed] [Google Scholar]
- 28.Yashiro K, Kameyama Y, Mizuno M, Hayashi S, Sakashita Y, Yokota Y. Biochim Biophys Acta. 1989;1005:56–64. doi: 10.1016/0005-2760(89)90031-3. [DOI] [PubMed] [Google Scholar]
- 29.Tiffert T, Lew VL. J Physiol. 1997;505:403–410. doi: 10.1111/j.1469-7793.1997.403bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dodge JT, Mitchell C, Hanahan DJ. Arch Biochem Biophys. 1963;100:119–130. doi: 10.1016/0003-9861(63)90042-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.