Abstract
Escherichia coli strains producing extended-spectrum β-lactamases (ESBLs) are a major problem in many different hospitals worldwide, causing outbreaks as well as sporadic infections. The prevalence of Escherichia coli ESBL producers was analyzed in a surveillance study performed on the population attending the Policlinico Umberto I, the largest university hospital in Rome, Italy. We also investigated genotypes, pathogenicity islands, and plasmids in the ESBL-positive E. coli isolates as further markers that are useful in describing the epidemiology of the infections. In this survey, 163 nonreplicate isolates of Escherichia coli were isolated from patients from 86 different wards, and 28 were confirmed as ESBL producers. A high prevalence (26/28) of CTX-M-15 producers was observed within the bacterial population circulating in this hospital, and the dissemination of this genetic trait was associated with the spread of related strains; however, these do not have the characteristics of a single epidemic clone spreading. The dissemination was also linked to horizontal transfer among the prevalent E. coli genotypes of multireplicon plasmids showing FIA, FIB, and FII replicons in various combinations, which are well adapted to the E. coli species. The analysis of related bacteria suggests a probable interpatient transmission occurring in several wards, causing small outbreaks.
A rapid dissemination of isolates producing CTX-M-type extended-spectrum β-lactamases (ESBLs) has recently been reported in some European countries, including Italy, and is a matter of major concern. The blaCTX-M genes have been captured on transferable plasmids from the chromosomes of Kluyvera spp., and their products are becoming the most prevalent ESBLs encountered in Enterobacteriaceae (19). The prevalent blaCTX-M-type genes in Europe have been identified as blaCTX-M-1, blaCTX-M-3, blaCTX-M-9, blaCTX-M-14, and blaCTX-M-15 (2, 19). Infections caused by enterobacteria producing ESBLs are associated with increased morbidity, mortality, and health care-associated costs (9, 16).
In Italy, the presence of CTX-M-type ESBLs in clinical isolates of Enterobacteriaceae examined in a Italian nationwide survey (23, 27), as well as in isolates from companion animals (7), was previously reported, and the results showed that CTX-M-type enzymes were common overall (around 20%) among ESBL producers (20). However, the local epidemiology of Escherichia coli producing ESBLs may differ from the national picture, with dominance of different strains. We therefore studied the prevalence and the clonality of the Escherichia coli ESBL producers in the population attending the Policlinico Umberto I, the largest university hospital in Rome, Italy, which had not been included in the nationwide survey. We also investigated virulence factors encoded on pathogenicity islands (PAIs) and plasmids carried by the E. coli ESBL producers as further markers that are useful in describing the epidemiology of nosocomially acquired infections.
MATERIALS AND METHODS
Clinical isolates and antibiotic susceptibilities.
A total of 4,769 various clinical samples from 86 different wards were sent for bacterial isolation to the microbiology laboratory of the Policlinico Umberto I hospital in Rome from 1 April to 15 September 2006. A total of 163 nonreplicate clinical isolates of Escherichia coli were isolated by routine procedures. In vitro antimicrobial activities were determined by the Vitek2 system with AST-N041 cards (BioMérieux, Mercy l'Etoile, France). Strains with suspected ESBL production were tested by the double-disk (BioMérieux, Mercy l'Etoile, France) synergy test between amoxicillin-clavulanic acid (20/10 μg) and cefotaxime (30 μg), ceftazidime (30 μg), aztreonam (30 μg), and cefepime (30 μg) (12). A cefpodoxime disk (10 μg) was used as an additional test for analyzing suspected ESBL production.
Extraction of β-lactamases and analysis by IEF.
Crude extracts of β-lactamase were obtained by ultrasonication. Analytical isoelectric focusing (IEF) of these extracts was performed on polyacrylamide gels at pH 3.5 to 11 (Amersham Pharmacia Biotech, Uppsala, Sweden). Enzymatic activity was assayed by the iodometric method with 250 mg/liter of ceftriaxone and 250 mg/liter of penicillin G (1). Crude extracts of the TEM-1 (pI 5.4), SHV-2 (pI 7.6), CTX-M-14 (pI 8.0), SHV-12 (pI 8.2), and CTX-M-15 (pI 8.6) plasmid β-lactamases were used as controls.
Molecular typing, phylogenetic groups, and PAI detection.
Chromosomal DNA was prepared for pulsed-field gel electrophoresis (PFGE) as previously described (28). After digestion with XbaI (Roche Diagnostics, Monza, Italy), the DNA fragments were separated by electrophoresis in 1% agarose gels (pulsed-field agarose certified; Bio-Rad) and 0.5× Tris-borate-EDTA buffer by using a contour-clamped homogeneous electric field (CHEF-Mapper system; Bio-Rad). Electrophoresis conditions were 14°C at 6 V/cm (200 V) for 22 h with switch times ranging from 2 to 64 s of linear ramping. To perform phylogenetic analysis of restriction profiles, TIFF files were analyzed with BioNumerics software (v.3.5; Applied Maths, Belgium). Strains showing >80% similarity were classified as genetically related and assigned to the same lineage, identified as PFGE types A to G (33). Different numbers were assigned to more closely related strains to differentiate members of the two prevalent A and B PFGE types as subtypes (e.g., A1 to A3).
Total DNA of higher purity was prepared with the Wizard genomic DNA purification kit (Promega, Milan, Italy) according to the manufacturer's procedure. The phylogenetic groups of these isolates were determined by a previously described PCR-based method (8). Two multiplex PCR assays were applied to detect eight PAI markers as previously described (29), and uropathogenic E. coli (UPEC) strains 536 and J96 were used as positive controls.
ESBL gene identification.
ESBL genes (blaTEM, blaSHV, and blaCTX-M type) were characterized by PCR and sequencing using specific primers previously published (11, 21, 24, 34). New primers were designed to amplify and sequence the complete blaCTX-M-15 gene: CTX-M-1g Fw (5′-CCCATGGTTAAAAAATCACTGC-3′) and CTX-M-1g Rv (5′-CAGCGCTTTTGCCGTCTAAG-3′).
Plasmid analysis.
Plasmids were analyzed by the extraction method described by Kado and Liu (13), separated by 0.8% agarose gel electrophoresis and blotted onto positively charged nylon membrane (Roche Diagnostics, Monza, Italy) by standard procedures (30), and cross-linked to the membrane with UV light. Filters were consecutively hybridized with the repF amplicon (6) used as a probe for the FII replicon and with the amplicon obtained with the CTX-M-1g FW and CTX-M-1g RV primers mentioned above, recognizing the blaCTX-M-15 gene. DNA probes were digoxigenin labeled, using the digoxigenin PCR DIG probe synthesis kit (Roche Diagnostics GmbH, Mannheim, Germany). Hybridization and detection were performed according to the manufacturer's instructions.
For transformation and restriction analysis of the plasmids, plasmid DNA was purified with the Qiagen plasmid midikit (Qiagen Inc., Milan, Italy). Purified plasmids were used to transform competent E. coli DH5α cells (MAX Efficiency DH5α chemically competent cells; Invitrogen, Milan, Italy). Selection of the transformants was performed on LB agar plates containing ampicillin (100 μg/ml). Plasmids were typed by the PCR-based replicon typing (PBRT) method as previously described (6).
RESULTS
Identification and epidemiology of ESBL producers.
A prospective study was carried out from April to September 2006 at the Policlinico Umberto I Hospital in Rome. In that survey, 163 nonreplicate clinical isolates of Escherichia coli were isolated from 162 patients; two E. coli strains showing different resistance profiles were isolated from the same patient, and both were included within the collection. Twenty-five patients were from emergency units, seven patients were recovering in the intensive care unit of the hospital and the remaining were recovering in medical, surgical, or geriatric units. The majority of E. coli strains was isolated from urine specimens (53%); others were from blood, central vein catheters, tracheal/bronchial aspirates, wound swabs, and other various clinical samples. All the 163 E. coli isolates were analyzed for antimicrobial resistance by Vitek2 automatic screening and 32 were found to be suspected ESBL producers. Of these, 28 were confirmed using the double-disk diffusion assay, indicating a prevalence of 17.2% (28/163) in this study. Eight (28%) E. coli ESBL producers were from medical wards, seven (25%) were from surgical wards, seven (25%) were from the geriatric ward, three (11%) were from the intensive care unit, and three (11%) were from emergency units (Table 1). The clinical sites of isolation were as follows: 13 (46%) were from urine specimens, 2 (7%) were from blood specimens, 2 (7%) were from the respiratory tract, 1 (4%) was from a central vein catheter, and the remaining 10 (36%) were from wound swabs and various other clinical samples (Table 1). For 10 patients the ESBL-producing strain was isolated in the first 24 h after admission. Six of these patients had been admitted for first aid at the surgical, emergency, or intensive care unit, suggesting that these infections were incubating prior to admittance. However, past clinical information for these patients is lacking, and thus we cannot exclude hospitalization in the previous months. The remaining patients presented the infection from 5 days up to 6 months after admission, suggesting that these were hospital-acquired infections (Table 1).
TABLE 1.
ESBL-producing E. coli isolates analyzed in this study
| Isolate | Date (day/mo/yr) of:
|
Warda | Site of isolation | PFGE type | Phylogenetic group | PAI
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Patient admission | Isolation | IV | II | I | |||||
| 45545 | 30/01/06 | 06/04/06 | Cardiology (MED 1) | Wound swab | A1 | B2 | 536 | CFT073 | CFT073 |
| 45606 | 30/03/06 | 07/04/06 | Oncology (SUR 1) | Bile | B1 | B2 | 536 | CFT073 | CFT073 |
| 45852 | 12/04/06 | 13/04/06 | General surgery (SUR 2) | Blood | B1 | B2 | 536 | CFT073 | CFT073 |
| 45913 | 06/04/06 | 15/04/06 | Internal medicine (MED 2) | Urine | B1 | B2 | 536 | CFT073 | Negative |
| 46214 | 14/04/06 | 26/04/06 | General surgery (SUR 2) | Venous catheter | A1 | B2 | 536 | CFT073 | CFT073 |
| 46427 | 12/04/06 | 03/05/06 | GER | Wound swab | C | B2 | 536 | CFT073 | CFT073 |
| 46595 | 01/03/06 | 06/05/06 | Angiology (MED 3) | Wound swab | A1 | B2 | 536 | CFT073 | CFT073 |
| 46644 | 07/05/06 | 08/05/06 | Neurology (MED 4) | Urine | NDb | B2 | 536 | CFT073 | CFT073 |
| 46688 | 10/05/06 | 10/05/06 | Surgery day hospital (SUR 3) | Abdominal abscess | B1 | B2 | 536 | CFT073 | CFT073 |
| 46733 | 11/05/06 | 11/05/06 | ICU | Tracheal aspirate | D | B2 | 536 | Negative | Negative |
| 46844 | 14/05/06 | 15/05/06 | General surgery (SUR 4) | Wound swab | A1 | B2 | 536 | CFT073 | CFT073 |
| 47242 | 10/03/06 | 24/05/06 | EME 1 | Blood | G | B2 | 536 | Negative | Negative |
| 47731 | 15/05/06 | 09/06/06 | General surgery (SUR 2) | Bile | B1 | B2 | 536 | CFT073 | CFT073 |
| 47736 | 08/06/06 | 09/06/06 | Angiology (MED 3) | Wound swab | A2 | B2 | 536 | CFT073 | CFT073 |
| 47849 | 11/05/06 | 13/06/06 | ICU | Urine | B2 | B2 | 536 | CFT073 | CFT073 |
| 47856 | 30/05/06 | 13/06/06 | GER | Urine/blood | F | D | 536 | Negative | CFT073 |
| 48112 | 20/06/06 | 20/06/06 | EME 1 | Wound swab | A3 | B2 | 536 | CFT073 | CFT073 |
| 48115 | 19/06/06 | 20/06/06 | Internal medicine (MED 5) | Urine | A2 | B2 | 536 | CFT073 | CFT073 |
| 48165 | 20/06/06 | 21/06/06 | Internal medicine (MED 6) | Urine | A3 | B2 | 536 | CFT073 | CFT073 |
| 49107 | 18/07/06 | 18/07/06 | ICU | Tracheal aspirate | A3 | B2 | 536 | CFT073 | CFT073 |
| 49403 | 14/01/06 | 27/07/06 | GER | Urine | E | B2 | 536 | Negative | CFT073 |
| 50055 | 09/05/06 | 18/08/06 | GER | Urine | A2 | B2 | 536 | CFT073 | CFT073 |
| 50068 | 22/05/06 | 18/08/06 | General surgery (SUR 2) | Urine | B2 | B2 | 536 | CFT073 | CFT073 |
| 50252 | 14/07/06 | 24/08/06 | GER | Urine | A1 | B2 | 536 | CFT073 | CFT073 |
| 50419 | 09/08/06 | 30/08/06 | GER | Urine | A2 | B2 | 536 | CFT073 | CFT073 |
| 50552 | 29/08/06 | 04/09/06 | EME 2 | Wound swab | A1 | B2 | 536 | CFT073 | CFT073 |
| 50811 | 01/09/06 | 09/09/06 | Internal medicine (MED 2) | Urine | A1 | B2 | 536 | CFT073 | CFT073 |
| 50814 | 08/08/06 | 09/09/06 | GER | Urine | A1 | B2 | 536 | CFT073 | CFT073 |
MED, medical; SUR, surgical; GER, geriatric unit; ICU, intensive care unit; EME, emergency unit.
ND, not determined because of DNA self-degradation.
All 28 identified ESBL producers were resistant to cefpodoxime. All strains but two were resistant to ceftazidime, cefotaxime, and cefepime; strain 47242 was susceptible to ceftazidime and showed intermediate resistance to aztreonam, and strain 46733 was susceptible to cefotaxime and cefepime. All the isolates except five showed resistance to amoxicillin-clavulanic acid, suggesting the presence of an oxacillinase gene.
Genotyping of the ESBL producers.
Twenty-seven of the 28 ESBL producers were assigned to the group B2, which has been more frequently associated with extraintestinal E. coli isolates (22), and one strain was group D (Table 1).
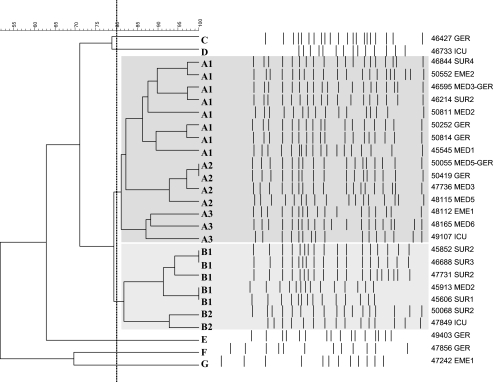
Strains were genotyped by PFGE of genomic DNA restricted by XbaI (Fig. 1). Seven different clonal lineages were identified and classified as PFGE types A to G. PFGE types A and B were prevalent within this collection (Table 1; Fig. 1). Three and two subtypes were further discerned within the A and B PFGE types; these were named A1, A2, and A3 and B1 and B2, respectively, with each subtype including the strains showing higher levels of similarity (85 to 100%).
FIG. 1.
Dendrogram (unweighted-pair group method using average linkages and Dice coefficient) showing the relatedness of PFGE patterns for the 28 E. coli ESBL producers. The dotted line indicates 80% similarity. Related A and B clusters showing >80% similarity are shaded dark gray and light gray, respectively. The isolate numbers and the wards where the patients recovered are indicated and refer to Table 1.
Distribution of PAIs in the E. coli ESBL producers.
Strains were tested by multiplex PCR (29) for the presence of specific markers recognizing the major PAIs associated with UPEC.
Only three types of PAIs were detected, with the most prevalent being PAI IV536, which was detected in all of the isolates. The majority of the strains also carried PAI ICFT073 and PAI IICFT073; only two strains lacked ICTF073, and four strains lacked IICFT073. The strains showing one or two PAIs belong to rare PFGE types, while those showing the three PAIs were mainly of the prevalent A and B genotypes (Table 1).
β-Lactamase gene identification.
The 28 ESBL producers were first tested by IEF to determine the ESBLs contained in each strain. Most of the strains showed enzymes migrating at pI 8.6 and pI 5.4; a few strains showed different β-lactamases migrating at pI 8.0, 8.2, or 7.6 (Table 2). All of the strains were then screened for the blaSHV-, blaTEM-, and blaCTX-M-type ESBL genes and for the blaOXA-1 gene, and amplicons were sequenced. The most widespread class A ESBL in this collection was CTX-M-15 (pI 8.6), which was detected in 26/28 E. coli strains. The two strains lacking CTX-M-15 produced CTX-M-14 (isolate 47242, showing a β-lactamase at pI 8.0) and SHV-12 (isolate 46733, showing a β-lactamase at pI 8.2) (Table 2). Two strains carried both SHV-11 (pI 7.6) and CTX-M-15. Most of the strains (79%) were also positive for the blaTEM gene (pI 5.4), and 75% were positive for the blaOXA-1 gene; the latter was found in all isolates showing amoxicillin-clavulanic resistance. The blaCTX-M-15 gene was found in different PFGE genetic lineages. Strains carrying the blaSHV-12 or blaCTX-14 gene were of the rare D and G PFGE types, respectively.
TABLE 2.
Characterization of ESBL genes and PBRT of plasmids
| Isolate | PFGE type | ESBL pI by IEF | β-Lactamase genes
|
PBRT | |||
|---|---|---|---|---|---|---|---|
| CTX-M | TEM | SHV | OXA | ||||
| 45545 | A1 | 8.6 | CTX-M-15 | Negative | Negative | OXA-1 | FIA, FIB, FII[2] |
| 46214 | A1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | Negative | FIA, FII[1] |
| 46595 | A1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FII[1] |
| 46644 | A1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FIB, FII[2], A/C, I1 |
| 46844 | A1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | Negative | FIA, FIB, FII[1], I1 |
| 50252a | A1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FII[1] |
| 50552 | A1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FIB, FII[2] |
| 50811 | A1 | 8.6, 7.6, 5.4 | CTX-M-15 | TEM-1 | SHV-11 | OXA-1 | FIA, FII[1] |
| 50814a | A1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FII[1] |
| 47736 | A2 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | Negative | FIA, FII[1], A/C, T |
| 48115 | A2 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FII[1] |
| 50055a | A2 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FII[1] |
| 50419a | A2 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FII[1] |
| 48112 | A3 | 8.6 | CTX-M-15 | Negative | Negative | OXA-1 | FIA, FIB, FII[2] |
| 48165 | A3 | 8.6 | CTX-M-15 | Negative | Negative | OXA-1 | FII[1] |
| 49107 | A3 | 8.6 | CTX-M-15 | Negative | Negative | OXA-1 | FII[2] |
| 45606 | B1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FIB, FII[2] |
| 45852 | B1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FIB, FII[2] |
| 45913 | B1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FIB, FII[2] |
| 46688 | B1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FIB, FII[2] |
| 47731 | B1 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FIB, FII[2], A/C, I1 |
| 47849 | B2 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FIB, FII[2], A/C, I1 |
| 50068 | B2 | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | Negative | FIA, FII[1] |
| 46427a | C | 8.6, 5.4 | CTX-M-15 | TEM-1 | Negative | OXA-1 | FIA, FII[1] |
| 46733 | D | 8.2 | Negative | Negative | SHV-12 | Negative | FII[1], A/C |
| 49403a | E | 8.6, 7.6, 5.4 | CTX-M-15 | TEM-1 | SHV-11 | OXA-1 | FIA, FIB, FII[2] |
| 47856a | F | 8.6 | CTX-M-15 | Negative | Negative | Negative | FIA, FIB, FII[2] |
| 47242 | G | 8.0, 5.4 | CTX-M-14 | TEM-1 | Negative | Negative | FIA, FIB, FII[2], A/C, I1 |
Strain isolated in the geriatric unit (see text).
Plasmid analysis and PBRT.
The blaCTX-M-15 gene was successfully transferred by transformation from several PFGE type A and B prototypic strains, demonstrating a plasmid localization of this ESBL gene (data not shown). Plasmids obtained from all the strains and from the transformants were classified by PBRT (Table 2). All the strains were positive for plasmids belonging to the IncF incompatibility group, since all carried the FII replicon. The blaCTX-M-15 gene was invariantly associated with the FII plasmids by Southern blot hybridization experiments performed on crude plasmid extracts obtained from the 26 positive strains (data not shown). The DNA sequence of the FII replicon showed the IncF plasmids in two types, identified as FII[1] and FII[2], as previously described (10). The FII[2] plasmid group was characterized by association with the FIA and FIB replicons in multireplicon plasmids (32). The FII[1] plasmid group was more frequently (80%) associated with the FIA replicon only (Table 2).
Seven strains were also positive for the simultaneous presence of additional plasmids, more frequently of the IncA/C or IncI1 group (Table 2).
DISCUSSION
E. coli strains with the CTX-M-15 ESBL are a major problem in many different hospitals worldwide, causing outbreaks as well as many sporadic infections. The impressive prevalence of this genetic trait in the hospital analyzed in this study was associated with the spread of related strains; these, however, did not have the characteristic of the spread of a single epidemic clone, because the bacterial population was not perfectly homogeneous and very few identical patterns were observed by PFGE analysis. The majority of the strains in our study belonged to two closely related clusters that probably originate from a common clone that was widely distributed in the hospital. Multireplicon plasmids showing FIA, FIB, and FII replicons in various combinations were identified by replicon typing in these isolates as the horizontal vehicle for blaCTX-M-15 gene dissemination. IncF plasmids have been previously identified as the prevalent carriers of the blaCTX-M-15 gene in other countries also, suggesting an impressive worldwide distribution of these plasmids, prevalently associated with the E. coli species (3, 10, 14, 18, 25). The first evidence of the association of the FII plasmid with the blaCTX-M-15 gene was obtained by the determination of the full sequence of the pC15-1a plasmid from epidemic E. coli isolated in Canada in 2000 (EMBL no. NC_005327) (3). The pC15-1a plasmid did not contain additional FIA or FIB replicons. IncF plasmids carrying the FII replicon in association with the FIA or FIB replicon were later identified as associated with the blaCTX-M-15 gene in several studies performed on E. coli strains from Tunisia, France, Spain, and the United Kingdom, demonstrating that these multireplicon plasmids were also important vehicles for the dissemination of this gene variant (10, 14, 18, 25). A recent study performed on a nonbiased Enterobacteriaceae population demonstrated that plasmids belonging to the IncFII group are unevenly distributed between the different species, being more prevalent in E. coli (58%) than in other genera (31). These data could suggest that the blaCTX-M-15 gene has been acquired on a plasmid type that frequently occurs in the E. coli species and is now spreading in association with it.
It is interesting to note the role played by IncFII plasmids in intestinal bacterial pathogens. There are very few biochemical properties that distinguish intestinal pathogens from normal inhabitants of the human gastrointestinal tract. Diseases are often due to the presence of specific virulence genes harbored on the so-called virulence plasmids or on chromosomally located PAIs (15). Salmonella enterica, Shigella spp., Yersinia spp., and enteropathogenic, enteroinvasive, and enterohemorrhagic O157:H7 E. coli all show virulence plasmids, and these virulence plasmids are in the range of 60 to 200 kb in size and carry replicons belonging to the IncFII group (4). The current information on the blaCTX-M-15 gene-carrying plasmids seems to exclude the possibility that these IncFII plasmids are related to the virulence plasmids associated with intestinal pathogens, since they are more related to the R100 IncFII reference plasmid (EMBL no. AP000342) (3) or to the pRSB107 IncF multireplicon plasmid isolated from a sewage treatment plant in Germany (EMBL no. AJ851089) (18, 32). These plasmids could be characteristic of UPEC E. coli, carrying important but still unknown functions promoting their large prevalence within this kind of E. coli strain.
From this and previous studies, the role of PAIs is not completely clear. In a previous study (29) PAI IV536 was one of the most frequently encountered in both commensal and UPEC E. coli strains, indicating that it is the first to be acquired and fixed on the chromosome, making it the most stable PAI. PAI IV536 was frequently found with the PAI I and PAI II of the CFT073 locus, and these two considered the second and third to be acquired, respectively. This study for the first time describes uropathogenic PAIs associated with resistant E. coli carrying the worldwide distributed blaCTX-M-15 gene. Since the strains analyzed in this study belong to phylogenetic group B2 and show the combination of these three PAIs, they probably cause urinary tract infections, and in fact the majority of our ESBL producers were isolated from urine specimens. However, several isolates, which were indistinguishable by PFGE type, phylogenetic group, plasmid type, and PAI profile, were from invasive infections and isolated from blood or other sterile sites, thus indicating that these strains can cause more serious infections probably affecting immunocompromised hosts.
Among the epidemiological results obtained in this study, we noted that the ESBL producers isolated from May to July in the geriatric unit belonged to the rare C (isolate 46427), E (isolate 49403), and F (isolate 47856, phylogenetic group D) PFGE types, while those isolated in the same ward in August belonged to the A1 (isolates 50252 and 50814) and A2 (isolates 50055 and 50419) PFGE types, which largely spread in the other wards of the hospital. Irrespective of the PFGE type, all these isolates showed the blaCTX-M-15 gene, and this gene was linked to IncFII multireplicon plasmids. The E and F type strains were associated with two PAIs (IV536 and ICFT073), while the four cases observed in August also carried the third PAI IICFT073. The epidemiological investigation of these cases showed that the introduction of the prevalent A2 PFGE type E. coli within the geriatric unit correlated with the admission of patient 50055, who was previously recovering in June in a medical ward where this A2 clone was isolated. The patient was transferred to the geriatric unit in August and there developed a urinary tract infection caused by the E. coli ESBL producer. The introduction of the A1 PFGE type in the geriatric unit could also be due to the transfer of patient 46595 to this unit: a wound swab taken in May from the patient was found to be positive for a CTX-M-15 producer of this genotype.
The two unique ESBL producers not positive for the blaCTX-M-15 gene in our collection were of the D (isolate 46733) and G (isolate 47242) PFGE types, showing only PAI IV536 and coming from patients recovering in emergency units; thus, they were not related to the prevalent strains circulating within the hospital.
Analyzing the time of admission to the hospital and the date of the first isolation of the ESBL-producing strain (Table 1), we observed that a large proportion of the infections occurred after a long period of hospitalization, indicating that these infections were acquired within the hospital. The sparse dissemination of related bacteria suggests a probable contamination of foreign materials (water, catheter, or disinfectants) and an interpatient transmission occurring in the wards, in several cases causing small outbreaks involving two or three patients. The dissemination of relevant ESBL genes is linked to the spread of specific plasmids, well adapted to the E. coli species, by horizontal genetic exchange among bacteria of different genotypes.
The eruptive dissemination of the ESBL producers in this hospital is comparable to that observed in other hospitals in Italy and worldwide (5, 17, 19, 26) and emphasizes the necessity of continuous epidemiological monitoring followed by immediate intervention to prevent severe nosocomial infections.
Acknowledgments
We are grateful to Anna Maria Dionisi for her work on the PFGE pattern analysis, to Tonino Sofia for manuscript revision, and to G. Prats for kindly providing us with positive control UPEC strains 536 and J96.
This work was supported by the DRESP2 (LSHM-CT-2005-018705) contract with the European Commission and by the FIRB project of the Italian Ministry of Research and University.
Footnotes
Published ahead of print on 24 October 2007.
REFERENCES
- 1.Barthelemy, M., M. Guionie, and R. Labia. 1978. β-Lactamases: determination of their isoelectric points. Antimicrob. Agents Chemother. 13695-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, D. A., S. Tyler, S. Christianson, A. McGeer, M. P. Muller, B. M. Willey, E. Bryce, M. Gardam, P. Nordmann, and M. R. Mulvey. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum β-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 483758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 264196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caccamo, M., M. Perilli, G. Celenza, G. Bonfiglio, G. Tempera, and G. Amicosante. 2006. Occurrence of extended spectrum β-lactamases among isolates of Enterobacteriaceae from urinary tract infections in southern Italy. Microb. Drug Resist. 12257-264. [DOI] [PubMed] [Google Scholar]
- 6.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63219-228. [DOI] [PubMed] [Google Scholar]
- 7.Carattoli, A., S. Lovari, A. Franco, G. Cordaro, P. Di Matteo, and A. Battisti. 2005. Extended-spectrum β-lactamases in Escherichia coli isolated from dogs and cats in Rome, Italy, from 2001 to 2003. Antimicrob. Agents Chemother. 49833-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 664555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du, B., Y. Long, H. Liu, D. Chen, D. Liu, Y. Xu, and X. Xie. 2002. Extended spectrum-β-lactamase-producing Escherichia coli and Klebsiella pneumoniae bloodstream infection: risk factors and clinical outcome. Intensive Care Med. 281718-1723. [DOI] [PubMed] [Google Scholar]
- 10.Hopkins, K. L., E. Liebana, L. Villa, M. Batchelor, E. J. Threlfall, and A. Carattoli. 2006. Replicon typing of plasmids carrying CTX-M or CMY β-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob. Agents Chemother. 503203-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hujer, A. M., M. G. P. Page, M. S. Helfand, B. Yeiser, and R. A. Bonomo. 2002. Development of a sensitive and specific enzyme-linked immunosorbent assay for detecting and quantifying CMY-2 and SHV-12 β-lactamases. J. Clin. Microbiol. 401947-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility pattern. Rev. Infect. Dis. 10867-878. [DOI] [PubMed] [Google Scholar]
- 13.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 1451365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karisik, E., M. J. Ellington, R. Pike, R. E. Warren, D. M. Livermore, and N. Woodford. 2006. Molecular characterization of plasmids encoding CTX-M-15 β-lactamases from Escherichia coli strains in the United Kingdom. J. Antimicrob. Chemother. 58665-668. [DOI] [PubMed] [Google Scholar]
- 15.Lan, R., and P. R. Reeves. 2002. Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 41125-1132. [DOI] [PubMed] [Google Scholar]
- 16.Lautenbach, E., J. B. Patel, W. B. Bilker, P. H. Edelstein, and N. O. Fishman. 2001. Extended-spectrum-β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin. Infect. Dis. 321162-1171. [DOI] [PubMed] [Google Scholar]
- 17.Lavigne, J. P., H. Marchandin, J. Delmas, J. Moreau, N. Bouziges, E. Lecaillon, L. Cavalie, H. Jean-Pierre, R. Bonnet, and A. Sotto. 2007. CTX-M β-lactamase-producing Escherichia coli in French hospitals: prevalence, molecular epidemiology, and risk factors. J. Clin. Microbiol. 45620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavollay, M., K. Mamlouk, T. Frank, A. Akpabie, B. Burghoffer, R. Ben Redjeb, S., R. Bercion, V. Gautier, and G. Arlet. 2006. Clonal dissemination of a CTX-M-15 β-lactamase-producing Escherichia coli strain in the Paris area, Tunis, and Bangui. Antimicrob. Agents Chemother. 502433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livermore, D. M., R. Canton, M. Gniadkowski, P. Nordmann, G. M. Rossolini, G. Arlet, J. Ayala, T. M. Coque, I. Kern-Zdanowicz, F. Luzzaro, L. Poirel, and N. Woodford. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59165-174. [DOI] [PubMed] [Google Scholar]
- 20.Luzzaro, F., M. Mezzatesta, C. Mugnaioli, M. Perilli, S. Stefani, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2006. Trends in the production of extended-spectrum-β-lactamases among enterobacteria of medical interest: report of the second Italian survey. J. Clin. Microbiol. 441659-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mabilat, C., and S. Goussard. 1995. PCR detection and identification of genes for extended-spectrum β-lactamases, p. 553-557. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, DC.
- 22.Moreno, E., I. Planells, G. Prats, A. M. Planes, G. Moreno, and A. Andreu. 2005. A comparative study of Escherichia coli virulence determinants in strains causing urinary tract bacteremia versus strains causing pyelonephritis and other sources of bacteremia. Diagn. Microbiol. Infect. Dis. 5393-99. [DOI] [PubMed] [Google Scholar]
- 23.Mugnaioli, C., F. Luzzaro, F. De Luca, G. Brigante, M. Perilli, G. Amicosante, S. Stefani, A. Toniolo, and G. M. Rossolini. 2006. CTX-M-type extended-spectrum β-lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob. Agents Chemother. 502700-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navarro, F., R. J. Mesa, E. Miró, L. Gómez, B. Mirelis, and P. Coll. 2007. Evidence for convergent evolution of CTX-M-14 ESBL in Escherichia coli and its prevalence. FEMS Microbiol. Lett. 273120-123. [DOI] [PubMed] [Google Scholar]
- 25.Novais, A., R. Canton, R. Moreira, L. Peixe, F. Baquero, and T. M. Coque. 2007. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob. Agents Chemother. 51796-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagani, L., E. Dell'Amico, R. Migliavacca, M. M. D'Andrea, E. Giacobone, G. Amicosante, E. Romero, and G. M. Rossolini. 2003. Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J. Clin. Microbiol. 414264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perilli, M., E. Dell'Amico, B. Segatore, M. R. de Massis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante. 2002. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 359-67. [DOI] [PubMed] [Google Scholar]
- 29.Sabaté, M., E. Moreno, T. Pérez, A. Andreu, and G. Prats. 2006. Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin. Microbiol. Infect. 12880-886. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J. E., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Sherley, M., D. M. Gordon, and P. J. Collignon. 2003. Species differences in plasmid carriage in the Enterobacteriaceae. Plasmid. 4979-85. [DOI] [PubMed] [Google Scholar]
- 32.Szczepanowski, R., S. Braun, V. Riedel, S. Schneiker, I. Krahn, A. Puhler, and A. Schluter. 2005. The 120 592 bp IncF plasmid pRSB107 isolated from a sewage-treatment plant encodes nine different antibiotic-resistance determinants, two iron acquisition systems and other putative virulence-associated functions. Microbiology 1511095-1111. [DOI] [PubMed] [Google Scholar]
- 33.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodford, N., E. J. Fagan, and M. J. Ellington. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 57154-155. [DOI] [PubMed] [Google Scholar]