Abstract
Prior to current studies on the emergence of drug resistance with the introduction of antiretroviral therapy (ART) in Cameroon, we performed genotypic analysis on samples from drug-naïve, human immunodeficiency virus (HIV)-infected individuals in this country. Of the 79 HIV type 1 (HIV-1) pol sequences analyzed from Cameroonian samples, 3 (3.8%) were identified as HIV-1 group O, 1 (1.2%) was identified as an HIV-2 intergroup B/A recombinant, and the remaining 75 (95.0%) were identified as HIV-1 group M. Group M isolates were further classified as subtypes A1 (n = 4), D (n = 4), F2 (n = 6), G (n = 12), H (n = 2), and K (n = 1) and as circulating recombinant forms CRF02_AG (n = 41), CRF11_cpx (n = 1), and CRF13_cpx (n = 2). Two pol sequences were identified as unique recombinant forms of CRF02_AG/F2 (n = 2). M46L (n = 2), a major resistance mutation associated with resistance to protease inhibitors, was observed in 2/75 (2.6%) group M samples. Single mutations associated with resistance to nucleoside reverse transcriptase inhibitors (T215Y/F [n = 3]) and nonnucleoside reverse transcriptase inhibitors (V108I [n = 1], L100I [n = 1], and Y181C [n = 2]) were observed in 7 of 75 (9.3%) group M samples. None of the patients had any history of ART exposure. Population surveillance of transmitted HIV drug resistance is required and should be included to aid in the development of appropriate guidelines.
The current standard for antiretroviral drug therapy (ART) in developed countries is the combination of two nucleoside reverse transcriptase (RT) inhibitors (NRTIs) plus a nonnucleoside RT inhibitor (NNRTI) or a protease inhibitor (PI). Since the successful trials in the late 1990s, combination ART has benefited and continues to aid many human immunodeficiency virus type 1 (HIV-1)-infected patients in developed countries, and it is becoming increasingly available in resource-constrained countries (17, 20, 24, 29, 30).
In countries with multiple antiretrovirals (ARVs) readily available, the prevalence of drug-resistant variants has ranged from 10 to 20% among drug-naïve patients (33), while in resource-constrained areas, resistance in the untreated HIV-infected population is rarely reported (23, 31). Recent interventions through such programs as the World Health Organization (WHO)'s 3 by 5 plan to treat 3 million people by the end of 2005 (33a) and the President's Emergency Plan for AIDS Relief have promoted significant access to ART in low- and middle-income countries. As of June 2005, about 500,000 people in sub-Saharan Africa were receiving ART, although the regional coverage rate was still 11% of the estimated number of patients with CD4 cell counts of ≤300/ml (2% of all HIV-infected patients in this region) (33a). Developing countries, including Cameroon, are moving towards universal access to HIV prevention, care, and treatment for those in need and at high risk of infection. This has led to the widespread use of antiretroviral drugs through structured national ART scale-up plans. Because of the complexity and open-ended duration of HIV treatments and the need to begin programs to treat many patients quickly, fears have been raised that emergence of ARV resistance may become a serious public health concern and render anti-HIV drugs useless. To assist ART programs and to minimize the emergence and transmission of HIV drug resistance strains and their public health consequences, WHO has developed a minimum-resource strategy for the surveillance and monitoring of HIV drug resistance in resource-limited countries. In Kenya, for example, where ART has been provided for 12 to 17% of the estimated need, the prevalence of resistant strains among drug-naïve patients has recently risen from 1% (2002) to more than 5% (2003) (WHO, personal communication). In Botswana, where treatment is available to all patients with <300 CD4 cells/ml, the prevalence of major mutations conferring PI resistance was estimated to be 4% among drug-naïve patients (4).
Unlike the case in southern and eastern African countries, where one or two HIV-1 subtypes dominate (22), all major groups and subtypes of HIV-1 cocirculate in Cameroon (1, 6, 14-19, 21, 22, 24, 28, 34-38). According to WHO/UNAIDS, as of the end of 2004, the prevalence of HIV-1 infection was estimated to be 4.8% overall and 9.8% for adults. To date, there have been several reports on the prevalence of ARV resistance mutations in the drug-naïve HIV-1-infected population of Cameroon (1, 6, 14-16, 19, 31). Baseline information on the frequency and types of ARV resistance mutations in Cameroon will help to inform optimal ART and enable the government to monitor the success of the national AIDS treatment program.
ART in Cameroon is based on the WHO guidelines, i.e., the combination of two NRTIs and one NNRTI. With the rapid introduction of ART and with limited health care infrastructure for care and monitoring, this country may face similar emergence rates of ARV resistance to those described for other developing countries (29, 30). With a higher prevalence of ARV resistance in the drug-naïve population (18, 32), resistance may emerge at an even higher rate.
In this study, we evaluated the prevalence of drug-resistant HIV-1 strains in treatment-naïve HIV-1-infected individuals in a resource-limited country where ART is being scaled up rapidly to determine whether standard first-line regimens will continue to be effective. Samples were obtained prior to the rollout of significant ART programs in Yaoundé, the capital city of Cameroon. We examined the prevalence of ARV resistance mutations in 79 patient samples and found a low rate of major drug resistance mutations to RTIs and PIs.
MATERIALS AND METHODS
Study population.
Blood specimens were drawn in 2004 from newly diagnosed HIV-1 patients attending a clinic in Yaoundé, Cameroon. All participants provided written informed consent and were likely to be recently infected. Sera found to be reactive for HIV by enzyme-linked immunosorbent assay confirmed with Western blotting were included in this study to explore the prevalence of intrinsic resistance to ARV drugs from treatment-naïve patients. This study received ethical clearance from the National Ethics Committee of Cameroon. Exclusion criteria included any previous form of ARV treatment, including that given to women for prevention of mother-to-child transmission.
PCR and sequencing.
Peripheral blood mononuclear cells (PBMCs) from HIV-seroreactive blood donors were obtained by Ficoll-Hypaque density gradient centrifugation. Proviral DNA was extracted from uncultured PBMCs with a DNA extraction kit (Qiagen, Hilden, Germany). Nested PCR amplification was performed using AmpliTaq DNA polymerase (Roche Molecular Systems, Branchburg, NJ). A segment of the PR-RT region of the pol gene was first PCR amplified using the universal external primers univ-PS1 (TTTTTTAGGGAAAATTTGGCCTTC) and univ-RTA4 (CTGTATATCATTGACAGTCCAGCT), resulting in a 1.2-kbp product. Nested PCR was then performed with the universal primers univ-PS2 (5′-TCCCTCAAATCACTCTTTGGCAAC-3′) and univ-RTA3 (5′-TTCATAACCCATCCAAAGAAATGG-3′) to generate a fragment of 1.0 kbp. The PCR products were then purified with a QIAquick PCR purification kit (Qiagen, Valencia, CA) and sequenced in the sense and antisense directions with a set of nested primers (25). All sequencing reactions were performed using an ABI Prism Big Dye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA) and an ABI 3730 DNA sequencer by Davis Sequencing, Inc. The chromatogram files were read using the Chromas 1.6 program (Helensvale, Australia). All sequences were edited with the BioEdit program.
Phylogenetic analysis and subtyping.
Neighbor-joining phylogenetic trees including reference pol sequences were constructed using Clustal W and then drawn using Treeview PPC, version 1.6.6 (Institute of Biochemical and Life Sciences, Scotland, United Kingdom). Bootstrap resampling (1,000 data sets) of multiple alignments was performed to test the statistical robustness of the trees. Kimura-2 parameters were calculated with the DNADIST program in the PHYLIP package (13, 27).
Genotypic resistance analysis.
Genotypic resistance was defined as the presence of one or more resistance-related mutations, as specified by the consensus mutation figures of the International AIDS Society—USA (11). The emergence of amino acid substitutions associated with resistance to RTIs and PIs has been characterized extensively, and these substitutions can be classified into major and accessory/minor (modifying) mutations. Major mutations lead to severalfold decreases in sensitivity to one or more ART drug. Accessory mutations may not result in a significant decrease in sensitivity but are associated with an increase in viral fitness (replication capacity) (9). Although resistance testing was performed retrospectively, for ethical reasons these results were fed back to the clinicians at the study site regarding the relative merits of change in therapy.
Nucleotide sequence accession numbers.
The DNA sequences of HIV-1 pol PR-RT regions determined as part of this study were submitted to GenBank under the following accession numbers: DQ990400 to DQ990455.
RESULTS
HIV-1 subtype distribution.
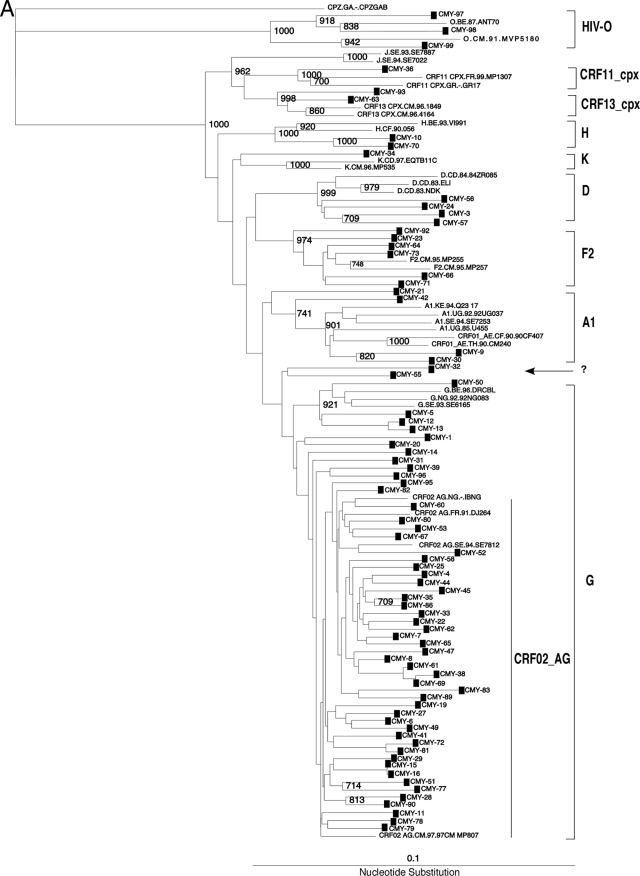
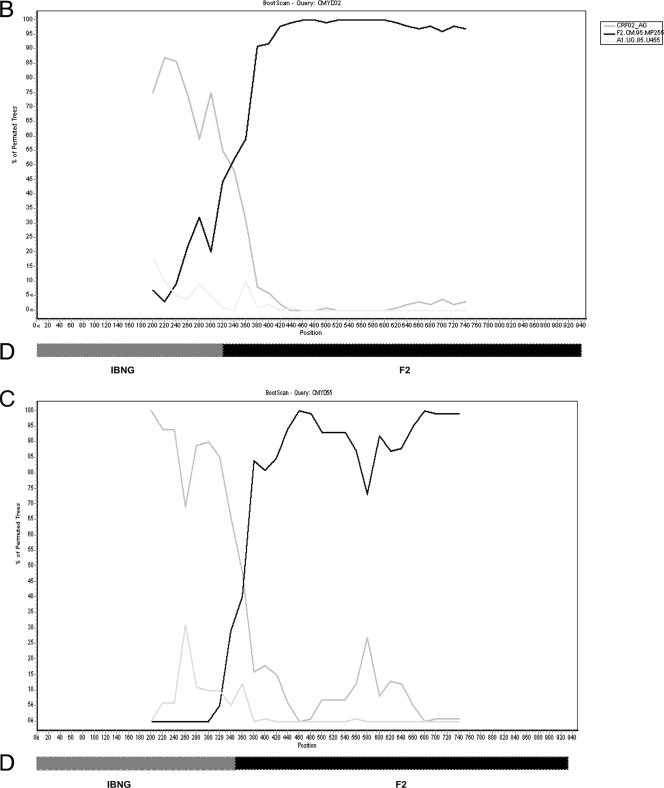
Seventy-nine HIV-infected samples from drug-naïve patients were obtained in 2004. A 1.0-kbp fragment encompassing amino acids 1 to 99 of PR and 1 to 234 of RT was PCR amplified and sequenced as described above. Sequences were then aligned and phylogenetic trees constructed to classify the different HIV sequences into groups, subtypes, and recombinant forms (Fig. 1A). Three sequences (3.8%; 95% confidence interval [CI], 1.6 to 5.9%) belonged to HIV-1 group O, and 75 sequences (94.9%; CI, 94.8 to 95.0%) were identified as HIV-1 group M. Group M isolates were further classified into the following six subtypes and three circulating recombinant forms (CRFs): subtypes A1 (n = 4), D (n = 4), F2 (n = 6), G (n = 12), H (n = 2), and K (n = 1) and CRF02_AG (n = 41), CRF11_cpx (n = 1), and CRF13_cpx (n = 2), with an intersubtype recombinant, CRF02_AG/F2 (n = 2). The two CRF02_AG/F2 isolates were identified using SimPlot for bootscanning analysis (Fig. 1B and C), with a 400-nucleotide (nt) rolling window and a significance threshold of 95% over the 1,000-bp PR-RT gene. Figure 1D shows the SimPlot output and a schematic representative plot and indicates that samples CMYD-32 and CMYD-55 have different breakpoints in the PR-RT gene, at 350 nt and 425 nt, respectively.
FIG. 1.
(A) Phylogenetic tree of HIV-1 PR-RT sequences from 78 HIV-1 group M and O isolates. “CMY” refers to PR-RT sequences from the cross-sectional analysis and indicates the country (Cameroon) and location (Yaoundé) of sample collection. The bootstrap value at each node represents the number among 1,000 bootstrap replicates that supported the branching order. Bootstrap resampling values of 70% or higher are shown. Brackets on the right represent the major group M subtypes. Newly derived sequences are marked with filled squares, and the novel unique recombinant form CRF02_AG/F2 is shown by an arrow. A 950-nt segment of the PR-RT coding region was used to construct this tree by the neighbor-joining method. PR-RT genetic subtypes A, D, F, G, H, and K and recombinants CRF02_AG, CRF11.cpx, and CRF13.cpx, as well as HIV-1 group O, are indicated. GenBank accession numbers for the reference sequences are as follows: A1.KE.93.Q23-17, AF004885; A1.UG.85.U455, M62320; A1.UG.92.92UG037, U51190; D.CD.83.ELI, K03454; D.CD.83.NDK, M27323; DCD.84.84ZR085, U88822; F2.CM.95.MP257, AJ249237; G.NG.92.92NG083, U88826; G.SE.93.SE6165, AF061642; G.BE.96.DRCBL, AF084936; H.BE.93.VI991, AF190127; J.SE.93SE7887, AF082394; J.SE.94.SE7022, AF082395; K.CM.96.MP535, AJ249239; K.CD.97.EQTB11C, AJ249234; 01_AE.TH.90.90CM240, U54771; 01_AE.CF.90.90CF4071, AF197341; 02_AG.NG.-.IBNG, L39106; 02_AG.FR91.DJ264, AF063224; 02_AG.SE.94.SE7812, AF107770; 02_AG.CM.97.97CM.MP807, AJ251056; 11_CPX.CM.97.MP818, AJ291718; 13 CPX.CM.96.1849, AF460972; 13 CPX.CM.96.4164, AF460974; O.CM.-.ANT70, L20587; O.CM.91.MVP5180, L20571; and CPZ.GA.-.CPZGAB, X52154. (B and C) SimPlot analyses of unclassifiable Cameroonian PR-RT (approximately 1,000 nt) sequences 04CMY-32 (B) and 04CMY-55 (C), showing the recombination between subtype F2 and CRF02_AG (A). The bootscan analysis was performed against reference strains from clades A (strain A1.UG.85.U455), B (strain B.US.83.RF), D (strain D,CD.84.84ZR085), F1 (strain F1.FI.93.FIN9363), F2 (strain F2.CM.95,MP255), G (strain G.SE.93.SE6155), and 02_AG (strain AG.NG.-.IBNG). (D) Segments derived from an IBNG-like strain and subtype F2 are shown.
HIV-2 intersubtype B/A recombinant.
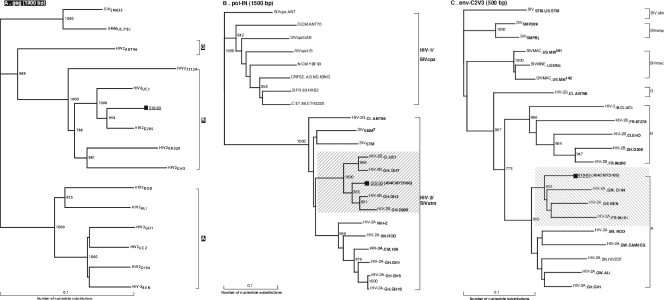
One sample was seropositive for HIV infection but could not be PCR amplified by our set of primers. PCR amplification and subsequent DNA sequencing with a set of HIV-2-specific primers confirmed the identity of this isolate as not only HIV-2 but also the first documented case of an HIV-2 intersubtype B/A recombinant, based on gag-p17/pol-IN/env-C2V3 sequence analyses, with three breakpoints in the env-nef gene (Fig. 2). The HIV-2 isolate (510-03) was subtype B based on gag and pol sequences, while the env-nef region is an intersubtype recombinant of subtypes A and B, with three recombination breakpoints identified. Further data analyses are in progress (N. Ndembi and C. Brennan, unpublished data).
FIG. 2.
Phylogenetic trees based on gag-p17/p24 gene (1,900 bp) of HIV-2 (510-03/04CMYD-100) subtype B/A recombinant strain (A) and the pol-IN (1,500 bp) gene (B) and env-C2V3 (500 bp) gene (C) from the Cameroonian HIV-2 strain. The bootstrap value at each node represents the number among 1,000 replicates that supported the branching order. Bootstrap values of >70% are shown. The brackets on the right represent the major HIV-2 subtypes. The newly analyzed sequence (510-03) is marked with a filled square.
PI resistance-associated mutations.
The amino acid sequence of each strain was compared to the subtype B consensus amino acid sequence, using the published HIV drug resistance algorithm from the International AIDS Society (10, 11) for mutations associated with resistance to PIs and RTIs. Based on subtype B sequences, drug resistance mutations in the protease region at positions 10, 13, 16, 20, 24, 30, 32, 33, 34, 36, 43, 46, 47, 48, 50, 53, 54, 58, 60, 62, 63, 64, 71, 73, 76, 77, 82, 84, 85, 88, 89, 90, and 93 (11), i.e., 33 mutations in total, have been shown to be associated with resistance to PIs.
Primary PI resistance-associated mutations were found in 2 of 75 cases (2.6%). These two patients harbored a CRF02_AG or CRF13_cpx HIV-1 isolate with an M46L amino acid substitution in the protease coding region. The M46L mutation in subtype B is associated with resistance to amprenavir, atazanavir (ATV), indinavir, and nelfinavir. The CRF02_AG-infected patient CMY-72 also contained a G48R mutation linked to the M46L mutation in the protease gene. The G48V mutation in subtype B is responsible for saquinavir, ritonavir, and ATV resistance (9). A V82I mutation was detected in the protease sequences of three patients, but the V82I mutation is a minor/accessory mutation and confers only minimal resistance to ATV and ritonavir (10). An alanine, threonine, phenylalanine, or serine at this position, however, is responsible for resistance to all PIs. Isoleucine at position 82 is also a naturally occurring polymorphism in subtype strains (9, 23) and was observed in 3 of 12 (25%; CI, 5.5 to 57.2%) of our G isolates. Minor or accessory PI resistance mutations were also found as wild-type sequences in Cameroonian isolates at the following positions, in order of decreasing frequency: M36I (74/75 isolates; 98.7%), K20I/M/R (67/75 isolates; 89.3%), L10V (5/75 isolates; 6.7%), L63P (4/75 isolates; 5.3%), and D60E (4/75 isolates; 5.3%).
RTI resistance-associated mutations.
Based on subtype B consensus sequences, mutations leading to resistance to NRTIs and NNRTIs are well defined and differ between the two classes of inhibitors. The most common major RT mutations leading to NRTI resistance occur at positions 41, 62, 65, 67, 69, 70, 74, 75, 77, 115, 116, 151, 184, 210, 215, and 219 (16 in total), and major mutations leading to NNRTI resistance are known to occur at positions 100, 103, 106, 108, 181, 188, 190, 225, (11), and 236 (9 in total).
Of the 79 cases analyzed, 7 (9.3%) showed major mutations associated with resistance to RTIs (zidovudine [ZDV], nevirapine [NVP], delavirdine [DLV], and efavirenz [EFV]). A V108I mutation was found in a CRF02_AG-infected patient, a Y181C mutation was found in a CRF13_cpx-infected patient, and V118C and V179E mutations were found in subtype G isolates. The subtype B mutations V118C and V179E result in moderate NNRTI resistance, whereas Y181C and V108I mutations are responsible for high-level NNRTI resistance (DLV, EFV, and NVP resistance and EFV and NVP resistance, respectively). The L210W mutation in subtype B (ZDV resistance) and the Y181C mutation (in subtype B [NNRTI resistance]) are found as the wild-type sequences in most HIV-1 group O isolates, including the three group O samples from this cohort, i.e., CMYD-97, -98, and -99 (5, 19, 26). Possible accessory amino acid mutations R211K and G333E in subtype B isolates were also observed in the RT genes of viruses from 54 patients (Table 1).
TABLE 1.
Overview of epidemiologic and genetic information for acutely HIV-1-infected subjects in central Cameroon
| Patient no. | Age (yr) | Sexd | Genetic subtypea
|
Drug resistance-associated mutation(s)c
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GenBank accession no. | Pol-PR | Pol-RT | Unique recombinant form | PR
|
RT
|
|||||
| Primary | Secondary | Primary | Secondary | |||||||
| 04CMYD1 | 50 | F | DQ990377 | G | G | K20I, M36I | ||||
| 04CMYD3 | 21 | M | DQ990378 | D | D | M36I | ||||
| 04CMYD4 | 29 | F | DQ990379 | CRF02_AG | CRF02_AG | K20R, M36I | ||||
| 04CMYD5 | 28 | F | DQ990380 | G | G | K20I, M36I | ||||
| 04CMYD6 | 25 | F | DQ990381 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD7 | 45 | M | DQ990382 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD8 | 27 | F | DQ990383 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD9 | 23 | F | DQ990384 | A1 | A1 | M36I, D60E, V77I | ||||
| 04CMYD10 | 33 | M | DQ990385 | H | H | K20R, M36I, D60E | ||||
| 04CMYD11 | 23 | F | DQ990386 | CRF02_AG | CRF02_AG | L10I, K20I, M36I | ||||
| 04CMYD12 | 21 | F | DQ990387 | G | G | K20I, M36I, (V82I) | ||||
| 04CMYD13 | 23 | F | DQ990388 | G | G | K20I, M36I | ||||
| 04CMYD14 | 40 | F | DQ990389 | G | G | K20I, M36I | ||||
| 04CMYD15 | 34 | M | DQ990390 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD16 | 25 | F | DQ990391 | CRF02_AG | CRF02_AG | K20I, M36I | V100I | |||
| 04CMYD19 | 29 | F | DQ990392 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD20 | 33 | F | DQ990393 | G | G | K20I, M36I | ||||
| 04CMYD21 | 47 | M | DQ990394 | A1 | A1 | M36I | ||||
| 04CMYD22 | 43 | M | DQ990395 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD23 | 54 | M | DQ990396 | F2 | F2 | M36I | ||||
| 04CMYD24 | 28 | M | DQ990397 | D | D | K20I, M36I | ||||
| 04CMYD25 | 14 | M | DQ990398 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD27 | 35 | F | DQ990455 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD28 | 56 | M | DQ990399 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD29 | 45 | F | DQ990400 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD30 | 26 | M | DQ990401 | A1 | A1 | M36I | ||||
| 04CMYD31 | 46 | M | DQ990402 | G | G | K20I, M36I, (V82I) | ||||
| 04CMYD32 | 30 | F | DQ990403 | CRF02_AG | F2 | CRF02_AG)/F2b | K20I, M36I, V77I | |||
| 04CMYD33 | 40 | M | DQ990404 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD34 | 35 | F | DQ990405 | K | K | K20R, M36I | ||||
| 04CMYD35 | 35 | M | DQ990406 | CRF02_AG | CRF02_AG | K20I, M36I | Y188C | |||
| 04CMYD36 | 31 | F | DQ990407 | CRF11_cpx | CRF11_cpx | D60E, V77I | ||||
| 04CMYD38 | 49 | M | DQ990408 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD39 | 34 | F | DQ990409 | G | G | K20I, M36I | ||||
| 04CMYD41 | 32 | M | DQ990410 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD42 | 43 | F | DQ990411 | A1 | A1 | K20I, M36I, L63P | ||||
| 04CMYD44 | 33 | M | DQ990412 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD45 | 36 | F | DQ990413 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD47 | 29 | F | DQ990414 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD49 | 24 | M | DQ990415 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD50 | 35 | M | DQ990416 | G | G | K20I, M36I | ||||
| 04CMYD51 | 28 | F | DQ990417 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD52 | 26 | M | DQ990418 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD53 | 50 | M | DQ990419 | CRF02_AG | CRF02_AG | K20I, M36I, L63P | ||||
| 04CMYD55 | 20 | M | DQ990420 | CRF02_AG | F2 | CRF02_AG/F2b | K20I, M36I | |||
| 04CMYD56 | 26 | M | DQ990421 | D | D | L10V, K20R, M36I | ||||
| 04CMYD57 | 35 | F | DQ990422 | D | D | M36I | ||||
| 04CMYD58 | 60 | F | DQ990423 | CRF02_AG | CRF02_AG | K20I, M36I | V108I | |||
| 04CMYD60 | 43 | M | DQ990424 | CRF02_AG | CRF02_AG | L10V, K20R, M36I | ||||
| 04CMYD61 | 21 | F | DQ990425 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD62 | 32 | M | DQ990426 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD63 | 30 | F | DQ990427 | CRF13_cpx | CRF13_cpx | K20I, M36I, V77I | ||||
| 04CMYD64 | 36 | M | DQ990428 | F2 | F2 | L10V, K20R, M36I | ||||
| 04CMYD65 | 42 | F | DQ990429 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD66 | 28 | M | DQ990430 | F2 | F2 | L10V, K20R, M36I | ||||
| 04CMYD67 | 35 | M | DQ990431 | G | G | K20I, M36I, (V82I) | ||||
| 04CMYD69 | 43 | M | DQ990432 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD70 | 38 | F | DQ990433 | H | H | K20R, M36I, D60E | ||||
| 04CMYD71 | 44 | F | DQ990434 | F2 | F2 | K20R, M36I, D60E | ||||
| 04CMYD72 | 48 | M | DQ990435 | CRF02_AG | CRF02_AG | M46L | K20I, M36I | |||
| 04CMYD73 | 39 | F | DQ990436 | F2 | F2 | K20R, M36I | ||||
| 04CMYD77 | 22 | F | DQ990437 | CRF02_AG | CRF02_AG | K20I, M36I | T215Y | |||
| 04CMYD78 | 36 | F | DQ990438 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD79 | 33 | F | DQ990439 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD80 | 55 | M | DQ990440 | CRF02_AG | CRF02_AG | K20I, M36I, L63P | T215F | |||
| 04CMYD81 | 33 | F | DQ990441 | CRF02_AG | CRF02_AG | K20I, M36I | T215Y | |||
| 04CMYD82 | 37 | M | DQ990442 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD83 | 47 | M | DQ990443 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD86 | 45 | F | DQ990444 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD89 | 40 | F | DQ990445 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD90 | 24 | M | DQ990446 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD92 | 32 | M | DQ990447 | F2 | F2 | M36I, L63P | ||||
| 04CMYD93 | 42 | M | DQ990448 | CRF13_cpx | CRF13_cpx | M46L | K20I, M36I | Y181C | ||
| 04CMYD95 | 31 | M | DQ990449 | CRF02_AG | CRF02_AG | K20I, M36I | ||||
| 04CMYD96 | 37 | F | DQ990450 | G | G | K20I, M36I, L63P | ||||
| 04CMYD97 | 23 | M | DQ990451 | HIV-1 group O | HIV-1 group O | M36L, I93L | Y181C, L210W | |||
| 04CMYD98 | 38 | M | DQ990452 | HIV-1 group O | HIV-1 group O | M36L, I93L | Y181C, L210W | |||
| 04CMYD99 | 25 | F | DQ990453 | HIV-1 group O | HIV-1 group O | M36L, I93L | Y181C, L210W | |||
| 04CMYD100 | 28 | F | DQ990454 | HIV-2 group A | HIV-2 group A | HIV2.B/A | NA | NA | ||
Typing of the pol gene (approximately 1,000 bp), encoding the Pol protease (Pol-PR) and Pol reverse transcriptase (Pol-RT) regions.
Possible recombination between subtype F and CRF_02 within the region.
Amino acid changes denote International AIDS Society (30) recognized mutations, while amino acid changes in parentheses stand for the presence of resistance mutations as minor mutations and subtype G naturally occurring polymorphisms. Primary drug resistance-associated mutations, shown in boldface type, lead to severalfold decreases in sensitivity to one or more ARTs. NA, not analyzed. All HIV group O samples contained Y181C as a natural occurring polymorphism. HIV-2B/A is a new recombinant strain, based on its gag-p17/24 (1,500 bp), pol-IN (1,500 bp), and env-C2V3 (500 bp) sequences.
F, female; M, male.
Dual-class resistance-associated mutations.
In one of the CRF13_cpx isolates (1.2%; CI, 0.93 to 1.46%), we identified primary amino acid sites associated with resistance to PIs (M46L mutation [resistance to amprenavir, indinavir, ATV, and nelfinavir]) and NRTIs (Y181C mutation [resistance to DLV, EFV, and NVP). Further phenotypic resistance would be needed to confirm these genotypic analyses.
DISCUSSION
In the current study, we found 2.6% PI resistance and 9.3% major RTI resistance mutations in HIV-1-infected drug-naïve individuals in Yaoundé, Cameroon. Unlike the case in developed countries, where antiretroviral regimens containing PIs are readily available, the first line of ART in Cameroon is the combination of two NRTIs plus one NNRTI. Very few patients in Cameroon are currently being or have been treated with PIs (1, 16-18). Konings et al. (16) reported that only the minor mutations associated with PI resistance were detected among HIV-1-infected drug-naïve patients in Cameroon during the period of 2000 to 2002. Our study confirmed previous reports and describes a high frequency of minor mutations (isoleucine or valine at position 10 in CRF02_AG; K20I and M36I mutations), which were found in all sequences except one, i.e., the CMY-36 isolate classified as CRF11_cpx. Of greater concern is the appearance of the major PI resistance mutation M46L in two infected patients (one with CRF02_AG and another with CRF13_cpx). The identification of this amino acid mutation in the protease warrants a more thorough screen of CRF02_AG and CRF13_cpx protease sequences, which is currently under way.
Three major NRTI resistance mutations were observed as wild-type sequences in three CRF02_AG (T215Y/F) and one CRF13_cpx (Y118C) virus. The T215Y/F mutation confers resistance to ZDV in nearly all HIV-1 isolates, whereas Y118C is a mutation related to native versus nucleoside analog discrimination but confers only low-level resistance (10, 11, 30). Limited studies on ART drug resistance in Africa, especially for non-B subtypes in Europe, have shown a strong correlation between the presence of major mutations and phenotypic resistance, similar to the case for mutations seen in subtype B infections with similar treatment regimens (31, 33). However, studies have also documented some salient differences among patients infected with non-B subtypes. A study of single-dose NVP to prevent mother-to-child transmission of HIV-1, conducted in Uganda, showed that selection of genotypic mutations associated with resistance to NVP occurred more frequently in women infected with subtype D than in women infected with subtype A viruses (23, 24). In addition, there has been identification of new mutational patterns conferring high-level drug resistance, previously not characterized for subtype B isolates (3, 23, 25, 26). For example, the V106M mutation in subtype C, as opposed to the V106A mutation of subtype B, is generally selected and confers resistance to EFV (3, 4). In addition, a combination of three mutations (I135L, T139V, and V245T) found as “wild-type” sequences in a subtype D HIV-1 isolate in Uganda conferred over 1,000-fold resistance to NVP and DLV and some cross-resistance to EFV (8). We are currently examining the phenotypic resistance of the PR-RT coding regions of Cameroonian HIV-1 isolates with or without any ARV resistance sequences. Although resistance testing was performed on PBMCs, this is a more sensitive method for detection of archived resistant mutants in persons lacking evidence of resistance by conventional assays.
This study provides the most recent data on molecular characterization of HIV-1 isolates in treatment-naïve individuals in Yaoundé, Cameroon. Overall, there is clear documentation of cocirculating HIV-1 group M and O strains as well as evidence for HIV-2 B/A recombinants, which are the subject of further investigation. At least six genetic subtypes (A, D, F2, G, H, and K) and three CRFs (CRF02_AG, CRF11_cpx, and CRF13_cpx) have been identified in HIV-1-infected patients in Yaoundé. Subtype CRF02_AG was responsible for 51.89% of the infections and was previously identified as predominant in west and west-central Africa (1, 6, 14-19, 21, 22, 24, 28). HIV-2 has been observed with a very low prevalence (0.06% of total HIV infections) in Douala but at a higher frequency in Yaoundé (0.2% to 1.2% of total HIV infections), based on independent epidemiological surveys (28, 36). A higher prevalence of HIV-2 infections was observed in commercial sex workers and tuberculosis patients, with no apparent link to other West African countries (36). However, the origin of the HIV-2 infection in our study was not available (7).
An obvious challenge in resource-limited settings such as Yaoundé, Cameroon, is maintaining a balance between rapid introduction of ART and continual surveillance of drug resistance to prevent treatment failures and to avoid a public health crisis. Expansion of molecular characterization on a nationwide basis would be useful to scientists developing prevention strategies based on vaccines and microbicides. Although there may be a cost factor involved, ART should be accompanied by testing for resistance before the choice of a particular ART regimen is made. This will reduce the selection pressure of resistance types, thus making first-line therapy more effective.
Acknowledgments
We thank the Director of Jamot Hospital and the Dean of the Faculty of Medicine and Biomedical Sciences, P. Ndumbe, for administrative support and the use of their research facilities and clinics.
N.N. was supported by funds from the Ministry of Education of Japan. E.J.A. was supported by an NIAID/NIH grant (AI-49170). Additional support was provided by a Case University Center for AIDS research grant (AI36219).
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Aghokeng, A. F., L. Ewane, B. Awazi, A. Nanfack, E. Delaporte, L. Zekeng, and M. Peeters. 2005. Enfuvirtide binding domain is highly conserved in non-B HIV type 1 strains from Cameroon, West Central Africa. AIDS Res. Hum. Retrovir. 21430-433. [DOI] [PubMed] [Google Scholar]
- 2.Apetrei, C., D. Descamps, G. Collin, I. Loussert-Ajaka, F. Damond, M. Duca, F. Simon, and F. Brun-Vezinet. 1998. Human immunodeficiency virus type 1 subtype F reverse transcriptase sequence and drug susceptibility. J. Virol. 723534-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner, B., D. Turner, M. Oliveira, D. Moisi, M. Detorio, M. Carobene, R. G. Marlink, J. Schapiro, M. Roger, and M. A. Wainberg. 2003. A V106 mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non nucleoside reverse transcriptase inhibitors. AIDS 17F1-F5. [DOI] [PubMed] [Google Scholar]
- 4.Bussmann, H., V. Novitsky, and W. Wester. 2002. Frequency of drug resistant mutations among HIV-1 C infected drug-naïve patients in Botswana, abstr. TuPeB4607. Abstr. 14th Int. AIDS Conf., Barcelona, Spain.
- 5.Descamps, D., G. Collin, F. Letourneur, C. Apetrei, F. Damon, I. Loussert-Ajaka, F. Simon, S. Saragosti, and F. Brun-Vezinet. 1997. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J. Virol. 718893-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fonjungo, P., E. N. Mpoudi, J. N. Torimiro, G. A. Alemnji, L. T. Eno, E. J. Lyonga, J. N. Nkengasong, R. B. Lal, M. Rayfield, M. L. Kalish, T. M. Folks, and D. Pieniazek. 2002. Human immunodeficiency virus type 1 group M protease in Cameroon: genetic diversity and protease inhibitor mutational features. J. Clin. Microbiol. 40837-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, G. M. Shaw, et al. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 687433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao, Y., E. Paxinos, J. Galovich, R. Troyer, H. Baird, M. Abreha, C. Kityo, P. Mugyenyi, C. Petropoulos, and E. J. Arts. 2004. Characterization of a subtype D human immunodeficiency virus type 1 isolate that was obtained from an untreated individual and that is highly resistant to nonnucleoside reverse transcriptase inhibitors. J. Virol. 785390-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch, M. S., B. Conway, R. T. D'Aquila, V. A. Johnson, F. Brun-Vezinet, B. Clotet, L. M. Demeter, S. M. Hammer, D. M. Jacobsen, D. R. Kuritzkes, C. Loveday, J. W. Mellors, S. Vella, and D. D. Richman. 1998. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. JAMA 2792000-2002. [DOI] [PubMed] [Google Scholar]
- 10.Holguin, A., A. Alvarez, and V. Soriano. 2002. High prevalence of HIV-1 subtype G and natural polymorphisms at the protease gene among HIV-infected immigrants in Madrid. AIDS 161163-1170. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, V. A., F. Brun-Vezinet, B. Clotet, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2006. Update of the drug resistance mutations in HIV-1: fall 2006. Top. HIV Med. 14125-130. [PubMed] [Google Scholar]
- 12.Kantor, R., D. A. Katzenstein, B. Effron, A. P. Carvalho, B. Wynhoven, P. Cane, J. Clarke, S. Sirivichayakul, M. A. Soares, J. Snoeck, C. Pillay, H. Rudich, R. Rodrigues, A. Holguin, K. Ariyoshi, M. B. Bouzas, P. Cahn, W. Sugiura, V. Soriano, L. F. Brigido, Z. Grossman, L. Morris, A. M. Vandamme, A. Tanuri, P. Phanuphak, J. N. Weber, D. Pillay, P. R. Harrigan, R. Camacho, J. M. Schapiro, and R. W. Shafer. 2005. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2325-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16111-120. [DOI] [PubMed] [Google Scholar]
- 14.Koizumi, Y., N. Ndembi, M. Miyashita, R. Lwembe, S. Kageyama, D. Mbanya, L. Kaptue, K. Numazaki, Y. Fujiyama, and H. Ichimura. 2006. Emergence of antiretroviral therapy resistance-associated primary mutations among drug-naive HIV-1-infected individuals in rural western Cameroon. J. Acquir. Immune Defic. Syndr. 4315-22. [DOI] [PubMed] [Google Scholar]
- 15.Konings, F. A., and P. N. Nyambi. 2004. V118I substitution in the reverse transcriptase gene of HIV-1 type CRF02_AG strains infecting drug-naive individuals in Cameroon. AIDS Res. Hum. Retrovir. 20673-680. [DOI] [PubMed] [Google Scholar]
- 16.Konings, F. A., P. Zhong, M. Agwara, L. Agyingi, L. Zekeng, J. M. Achkar, L. Ewane, Saa, E. Afane Ze, T. Kinge, and P. N. Nyambi. 2004. Protease mutations in HIV-1 non-B strains infecting drug-naive villagers of Cameroon. AIDS Res. Hum. Retrovir. 20105-109. [DOI] [PubMed] [Google Scholar]
- 17.Laurent, C., C. Kouanfack, S. Koulla-Shiro, N. Nkoue, A. Bourgeois, A. Calmy, B. Lactuock, V. Nzeusseu, R. Mougnutou, G. Peytavin, F. Liegeois, E. Nerrienet, M. Tardy, M. Peeters, I. Andrieux-Meyer, L. Zekeng, M. Kazatchkine, E. Mpoudi-Ngole, and E. Delaporte. 2004. Effectiveness and safety of a generic fixed-dose combination of nevirapine, stavudine and lamivudine in HIV-1-infected adults in Cameroon: open-label multicentre-trial. Lancet 36429-34. [DOI] [PubMed] [Google Scholar]
- 18.Laurent, C., C. Kouanfack, L. Vergne, M. Tardy, L. Zekeng, N. Noumsi, C. Butel, A. Bourgeois, E. Mpoudi-Ngole, S. Koulla-Shiro, M. Peeters, and E. Delaporte. 2006. Antiretroviral drug resistance and routine therapy, Cameroon. Emerg. Infect. Dis. 121001-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luk, K. C., L. Kaptue, L. Zekeng, V. Soriano, L. Gurtler, S. G. Devare, G. Schochetman, and J. Hackett, Jr. 2001. Naturally occurring sequence polymorphisms within HIV type 1 group O protease. AIDS Res. Hum. Retrovir. 171555-1561. [DOI] [PubMed] [Google Scholar]
- 20.Mbanya, D., F. Assah, N. Ndembi, and L. Kaptue. 2007. Monitoring antiretroviral therapy in HIV/AIDS patients in resource-limited settings: CD4 counts or total lymphocyte counts? Int. J. Infect. Dis. 11157-160. [DOI] [PubMed] [Google Scholar]
- 21.Ndembi, N., J. Takehisa, L. Zekeng, E. Kobayashi, C. Ngansop, E. M. Songok, S. Kageyama, T. Takemura, E. Ido, M. Hayami, L. Kaptue, and H. Ichimura. 2004. Genetic diversity of HIV type 1 in rural eastern Cameroon. J. Acquir. Immune Defic. Syndr. 371641-1650. [DOI] [PubMed] [Google Scholar]
- 22.Ndongmo, C. B., D. Pieniazek, M. Holberg-Petersen, C. Holm-Hansen, L. Zekeng, S. L. Jeansson, L. Kaptue, and M. L. Kalish. 2006. HIV genetic diversity in Cameroon: possible public health importance. AIDS Res. Hum. Retrovir. 22812-816. [DOI] [PubMed] [Google Scholar]
- 23.Nkengasong, J. N., C. Adje-Toure, and P. J. Weidle. 2004. HIV antiretroviral drug resistance in Africa. AIDS Rev. 64-12. [PubMed] [Google Scholar]
- 24.Peeters, M., C. Toure-Kane, and J. N. Nkengasong. 2003. Genetic diversity of HIV in Africa: impact on diagnosis, treatment, vaccine development and trials. AIDS 172547-2560. [DOI] [PubMed] [Google Scholar]
- 25.Richard, N., M. Juntilla, A. Abraha, K. Demers, E. Paxinos, J. Galovich, C. Petropoulos, C. C. Whalen, F. Kyeyune, D. Atwine, C. Kityo, P. Mugyenyi, and E. J. Arts. 2004. High prevalence of antiretroviral resistance in treated Ugandans infected with non-subtype B human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 20355-364. [DOI] [PubMed] [Google Scholar]
- 26.Rodes, B., D. C. Mendoza, M. Rodgers, A. Newell, V. Jimenez, R. M. Lopez-Brugada, and V. Soriano. 2005. Treatment response and drug resistance in patients infected with HIV type 1 group O viruses. AIDS Res. Hum. Retrovir. 21602-607. [DOI] [PubMed] [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 28.Vergne, L., A. Bourgeois, E. Mpoudi-Ngole, R. Mougnutou, J. Mbuagbaw, F. Liegeois, C. Laurent, C. Butel, L. Zekeng, E. Delaporte, and M. Peeters. 2003. Biological and genetic characteristics of HIV infections in Cameroon reveal dual group M and O infections in correlation between SI-inducing phenotype of the predominant CRF02_AG variant and disease stage. Virology 310254-266. [DOI] [PubMed] [Google Scholar]
- 29.Vergne, L., G. Malonga-Mouellet, I. Mistoul, R. Mavoungou, H. Mansaray, M. Peeters, and E. Delaporte. 2002. Resistance to antiretroviral treatment in Gabon: need for implementation of guidelines on therapy use and HIV-1 drug resistance in developing countries. J. Acquir. Immune Defic. Syndr. 29165-168. [DOI] [PubMed] [Google Scholar]
- 30.Vergne, L., C. T. Kane, C. Laurent, N. Diakhate, N. F. Gueye, P. M. Gueye, P. S. Sow, M. A. Faye, F. Liegeois, A. Ndir, I. Laniece, M. Peeters, I. Ndoye, S. Mboup, and E. Delaporte. 2003. Low rate of genotypic HIV-1 drug resistant strains in the Senegalese government initiative of access to antiretroviral therapy. AIDS 17(Suppl. 3)S31-S38. [DOI] [PubMed] [Google Scholar]
- 31.Vergne, L., M. Peeters, E. Mpoudi-Ngole, A. Bourgeois, F. Liegeois, C. Toure-Kane, S. Mboup, C. Mulanga-Kabeya, E. Saman, J. Jourdan, J. Reynes, and E. Delaporte. 2000. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: evidence of many minor drug resistance mutations in treatment-naïve patients. J. Clin. Microbiol. 383919-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vergne, L., S. Diagbouga, C. Kouanfack, A. Aghokeng, C. Butel, C. Laurent, N. Noumssi, M. Tardy, A. Sawadogo, J. Drabo, H. Hien, L. Zekeng, E. Delaporte, and M. Peeters. 2006. HIV-1 drug-resistance mutations among newly diagnosed patients before scaling-up programmes in Burkina Faso and Cameroon. Antivir. Ther. 11575-579. [PubMed] [Google Scholar]
- 33.Wensing, A. M., D. A. van de Vijver, G. Angarano, B. Asjo, C. Balotta, E. Boeri, R. Camacho, M. L. Chaix, D. Costagliola, A. De Luca, I. Derdelinckx, Z. Grossman, O. Hamouda, A. Hatzakis, R. Hemmer, A. Hoepelman, A. Horban, K. Korn, C. Kucherer, T. Leitner, C. Loveday, E. MacRae, I. Maljkovic, C. de Mendoza, L. Meyer, C. Nielsen, E. L. Op de Coul, V. Ormaasen, D. Paraskevis, L. Perrin, E. Puchhammer-Stockl, L. Ruiz, M. Salminen, J. C. Schmit, F. Schneider, R. Schuurman, V. Soriano, G. Stanczak, M. Stanojevic, A. M. Vandamme, K. Van Laethem, M. Violin, K. Wilbe, S. Yerly, M. Zazzi, C. A. Boucher, and SPREAD Programme. 2005. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J. Infect. Dis. 192958-966. [DOI] [PubMed] [Google Scholar]
- 33a.WHO/UNAIDS. June 2005. 3 by 5 progress report. WHO/UNAIDS, Geneva, Switzerland.
- 34.Yamaguchi, J., P. Bodelle, L. Kaptue, L. Zekeng, L. G. Gurtler, S. G. Devare, and C. A. Brennan. 2003. Near full-length genomes of 15 HIV type 1 group O isolates. AIDS Res. Hum. Retrovir. 19979-988. [DOI] [PubMed] [Google Scholar]
- 35.Yamaguchi, J., R. Coffey, A. Vallari, C. Ngansop, D. Mbanya, N. Ndembi, L. Kaptue, L. G. Gurtler, P. Bodelle, G. Schochetman, S. G. Devare, and C. A. Brennan. 2006. Identification of HIV type 1 group N infections in a husband and wife in Cameroon: viral genome sequences provide evidence for horizontal transmission. AIDS Res. Hum. Retrovir. 2283-92. [DOI] [PubMed] [Google Scholar]
- 36.Zekeng, L., R. Salla, L. Kaptue, P. Pinay, P. Barth, B. Schmidt-Ehry, and T. Rehle. 1992. HIV-2 infection in Cameroon: no evidence of indigenous cases. J. Acquir. Immune Defic. Syndr. 5319-320. [PubMed] [Google Scholar]