Abstract
Fimbriae are important virulence factors of pathogenic bacteria, facilitating their attachment to host and bacterial cells. In the periodontal pathogen Porphyromonas gingivalis, the fimA gene is classified into six types (genotypes I, Ib, II, III, IV, and V) on the basis of different nucleotide sequences, with fimA genotypes II and IV being prevalent in isolates from patients with periodontitis. The aims of this study were to examine the distribution of fimA genotypes in a collection of 82 P. gingivalis isolates from adult periodontitis patients of worldwide origin and to investigate the relationship between the fimA genotypes and the sequence types (STs), as determined by multilocus sequence typing (MLST), of the isolates. The fimA gene was amplified by PCR with primer sets specific for each genotype. The STs of all strains were assigned according to the MLST database for P. gingivalis (www.pubmlst.org/pgingivalis). The 82 strains showed extensive genetic diversity and were assigned to 69 STs. Only isolates with closely related STs harbored the same fimA genotype. Twenty-eight (34.1%) strains harbored fimA genotype II, while only the reference strain for fimA genotype V reacted with the primers specific for this genotype. Twenty-one isolates (25.6%) were positive by more than one of the fimA PCR assays; the most frequent combinations were genotypes I, Ib, and II (eight isolates) and genotypes I and II (four isolates). Sequencing of the fimA gene from selected isolates did not support the observed specific fimA genotype combinations, suggesting that the genotyping method used for the major fimbriae in P. gingivalis should be reevaluated.
Periodontal diseases are infections which result in various degrees of connective tissue attachment loss around the teeth, causing partial or total tooth loss in the affected individuals. In the United States, periodontal diseases affect about 75% of the adult population, with about 20% to 30% of adults having severe forms (1, 6). Worldwide, they represent a serious oral health problem, with an overall prevalence of about 10 to 20%. The proportion of affected people varies among geographically, ethnically, and racially different populations, as well as among different age groups (45).
Periodontal diseases are considered a heterogeneous disease entity caused by the complex actions and interactions of a number of pathogenic bacteria (18, 23, 50, 51) and are modified by various host factors (23, 51). There are also marked differences in the rates of progression and severities of these infectious disorders, as well as variations in the response to therapy.
Porphyromonas gingivalis is one of the main bacterial pathogens involved in the initiation and progression of periodontitis (4, 11, 23). It belongs to the red complex of periodontal bacteria (50) and is detected in patients with various forms of periodontitis (11). P. gingivalis harbors a number of virulence factors, e.g., fimbriae, lipopolysaccharide, collagenase, and cysteine proteinases (4, 18, 23, 25). It is capable of invading host tissue cells and can thereby be protected from the host's immune system and survive intracellularly in a nutritionally rich environment (4, 32). P. gingivalis can also be isolated from healthy individuals and is therefore recognized as a part of the commensal oral microflora (14, 19).
Cumulative evidence indicates that the ability of P. gingivalis to adhere to saliva components, host cells, solid surfaces, and bacterial cells is facilitated by its fimbriae (5, 21, 32, 43). These are curly and filamentous appendages arranged peritrichously on the cell surface of the organism (21) and have been classified into major and minor forms (5, 20). The major fimbriae (FimA) have been extensively studied (2, 3-5, 12, 35, 44, 47, 52). They are constructed from a fimbrillin monomer subunit protein of approximately 43 kDa and are directly responsible for many of the adhesive properties of the organism, binding specifically to and activating various host cells, such as human epithelial, endothelial, and spleen cells, and peripheral blood monocytes, resulting in the release of cytokines and several adhesion molecules (4, 5, 23, 32).
The fimA gene occurs as a single copy in the chromosome of P. gingivalis (12, 16, 43). It has been classified into six types (genotypes I, Ib, II, III, IV, and V) on the basis of the variation in its nucleotide sequence (2, 16, 37, 38). The amino acid sequences of recombinant fimbrillin from nine different P. gingivalis strains have also been investigated, and variations in the primary protein structures between the different isolates were described; these were separated into four types of proteins (16). The findings by Nakagawa et al. (39) demonstrated that recombinant FimA corresponding to fimA genotype II has a greater ability to adhere to and invade human epithelial cells than FimA corresponding to the proteins from other genotypes.
Several studies have evaluated the pathogenic difference in fimA genotypes in animals by the use of either a model for subcutaneous injection in rodents or an abscess model in mice (5, 17, 40, 42). Those studies showed that isolates harboring fimA genotypes II, Ib, and IV cause stronger infectious symptoms and inflammatory changes than strains harboring fimA genotypes I and III. The generation of P. gingivalis mutants in which the fimA type I gene was replaced by the type II gene also showed an enhanced bacterial adhesion/invasion of epithelial cells, whereas the replacement of type II with type I resulted in diminished efficiency (27). On the other hand, Umeda et al. (52) concluded, after studying the adhesion to and the invasion of different fimA genotypes with epithelial cells, that there were no significant differences between the genotypes.
Several clinical studies have indicated that the nucleotide variation in the gene is related to the virulence of the strains. In patients with chronic marginal periodontitis, P. gingivalis organisms with fimA genotype II are significantly more prevalent than isolates with other genotypes (2, 4, 7, 35, 37, 38). Studies concerning the pathogenic potentials of distinct fimA genotypes also indicate that genotype II organisms are more prevalent in patients with aggressive periodontitis (36). The relationship of periodontal health and specific fimA genotypes has also been investigated, and it was found that that fimA genotype I is the most prevalent among P. gingivalis-positive healthy adults, followed by genotype V (3).
Molecular typing methods have been used to clarify the genetic diversity of P. gingivalis (9, 13, 15, 29, 34). Currently, multilocus sequence typing (MLST) is the best method for the detection of the genetic diversity of bacterial pathogens and has a high discriminatory power (10). An MLST scheme was newly introduced for P. gingivalis and has facilitated the exact and worldwide classification of different isolates of the species (13). A publicly available MLST website for P. gingivalis isolates has been established at http://www.pubmlst.org/pgingivalis (26).
The aims of the present study were to examine the prevalence of different fimA genotypes of P. gingivalis strains obtained from adult periodontitis patients from worldwide sources and to examine the relationships between these fimA genotypes and the different sequence types (STs) of the species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Seventy-nine P. gingivalis isolates from three strain collections, all sampled from adult periodontitis patients, were analyzed. The first collection consisted of 38 isolates of worldwide origin and has been described earlier (13). Two of the original 40 isolates in the collection were left out because they were from monkeys. The second and third collections consisted of 28 isolates from Finland (provided by S. Asikainen, Oral Microbiology, Institute of Dentistry, Umeå University, Umeå, Sweden), and 13 isolates that were sent to us by A. J. van Winkelhoff, Department of Oral Microbiology, Academic Center for Dentistry, Amsterdam, Amsterdam, The Netherlands, respectively. In addition, three reference strains were included: the reference strains for fimA genotype Ib (strain HG1691; GenBank accession no. AB058848) (31, 38) and fimA genotype V (strain HNA99; GenBank accession no. AB027294) (37) were provided by T. Ooshima, Osaka University Graduate School of Dentistry, Osaka, Japan; and the reference strain for fimA genotype IV (strain W83 or ATCC BAA308; GenBank accession no. AAQ67087) (2, 43, 51) was purchased from LGC Promochem, Gothenburg, Sweden (Table 1). Reference strains for fimA types II (strain A7A1-28 or ATCC 53977) and III (strain BH 6/26; GenBank accession no. D17801) (2, 5) were found in the first collection, and the reference strain for fimA type I (strain ATCC 33277T; GenBank accession no. D17795) was present in the second collection (2) (Table 1).
TABLE 1.
Origins and STs of 82 P. gingivalis isolates examined by MLST
| Strain designation | STa | Type of infection | Isolation site | Geographic origin |
|---|---|---|---|---|
| A7A1-28 (ATCC 53977) | 1 | Periodontitis | Subgingival | Sacaton, AZ |
| CLN17-6-1 | 1 | Periodontitis | Subgingival | Sacaton, AZ |
| CLN16-6-4 | 1 | Periodontitis | Subgingival | Sacaton, AZ |
| ESO127 | 8 | Periodontitis | Subgingival | Okayama, Japan |
| SA28 | 17 | Periodontitis | Subgingival | Finland |
| OMZ481 | 20 | Periodontitis | Subgingival | Zurich, Switzerland |
| SP1-6 | 21 | Periodontitis | Subgingival | Oslo, Norway |
| 213PG3 | 22 | Periodontitis | Subgingival | Indonesia |
| 213PG1 | 22 | Periodontitis | Subgingival | Indonesia |
| 213PG4 | 22 | Periodontitis | Subgingival | Indonesia |
| 332-2 | 23 | Periodontitis | Subgingival | Umeå, Sweden |
| W83 (ATCC BAA-308) | 24 | Periodontitis | Abscess | Bonn, Germany |
| 13JC | 24 | Periodontitis | Subgingival | Rennes, France |
| W12 | 25 | Periodontitis | Subgingival | Birmingham, AL |
| Be-c | 25 | Root canal infection | Root canal | Umeå, Sweden |
| SA24 | 25 | Periodontitis | Subgingival | Finland |
| SA1 | 25 | Periodontitis | Subgingival | Finland |
| ESO75 | 26 | Periodontitis | Subgingival | Okayama, Japan |
| OMG406 | 27 | Periodontitis | Subgingival | Kenya |
| 196PG2 | 28 | Periodontitis | Subgingival | Indonesia |
| 196PG1 | 28 | Periodontitis | Subgingival | Indonesia |
| 195PG2 | 28 | Periodontitis | Subgingival | Indonesia |
| B42 | 29 | Odontogenic abscess | Incision | Hofu, Japan |
| 816B | 30 | Periodontitis | Subgingival | Malmö, Sweden |
| 34-4-4 | 31 | Periodontitis | Subgingival | Minneapolis, MN |
| 212PG3 | 34 | Periodontitis | Subgingival | Indonesia |
| 212PG2 | 35 | Periodontitis | Subgingival | Indonesia |
| 212PG1 | 36 | Periodontitis | Subgingival | Indonesia |
| 213PG2 | 37 | Periodontitis | Subgingival | Indonesia |
| 195PG3 | 38 | Periodontitis | Subgingival | Indonesia |
| 195PG1 | 39 | Periodontitis | Subgingival | Indonesia |
| OMZ479 | 40 | Periodontitis | Subgingival | Zurich, Switzerland |
| OMZ482 | 41 | Periodontitis | Subgingival | Zurich, Switzerland |
| 102 | 42 | Root canal infection | Root canal | Umeå, Sweden |
| 102822 | 43 | Periodontitis | Subgingival | Unknown |
| BH18/10 | 44 | Periodontitis | Subgingival | Winnipeg, Manitoba, Canada |
| AJW4 | 45 | Periodontitis | Subgingival | Buffalo, NY |
| OMG268 | 46 | Periodontitis | Subgingival | Gothenburg, Sweden |
| 295-1 | 47 | Periodontitis | Subgingival | Umeå, Sweden |
| B129 | 48 | Odontogenic abscess | Incision | Hofu, Japan |
| 212PG4 | 49 | Periodontitis | Subgingival | Indonesia |
| BH6/26 | 50 | Periodontitis | Subgingival | Winnipeg, Manitoba, Canada |
| SA5 | 51 | Periodontitis | Subgingival | Finland |
| SA6 | 52 | Periodontitis | Subgingival | Finland |
| SA11 | 53 | Gingivitis | Subgingival | Finland |
| SA9 | 53 | Gingivitis | Subgingival | Finland |
| SA12 | 54 | Periodontitis | Subgingival | Finland |
| SA21 | 55 | Periodontitis | Subgingival | Finland |
| SA32 | 55 | Periodontitis | Subgingival | Finland |
| SA22 | 56 | Periodontitis | Subgingival | Finland |
| SA25 | 57 | Periodontitis | Subgingival | Finland |
| SA29 | 58 | Gingivitis | Subgingival | Finland |
| SA31 | 58 | Gingivitis | Subgingival | Finland |
| SA30 | 59 | Periodontitis | Subgingival | Finland |
| SA36 | 60 | Periodontitis | Subgingival | Finland |
| SA37 | 61 | Periodontitis | Subgingival | Finland |
| 281 | 62 | Periodontitis | Subgingival | The Netherlands |
| 282 | 63 | Periodontitis | Subgingival | The Netherlands |
| 297 | 64 | Periodontitis | Subgingival | The Netherlands |
| 298 | 65 | Periodontitis | Subgingival | The Netherlands |
| 302 | 66 | Periodontitis | Subgingival | The Netherlands |
| 303 | 67 | Periodontitis | Subgingival | The Netherlands |
| 322 | 68 | Periodontitis | Subgingival | The Netherlands |
| 327 | 69 | Periodontitis | Subgingival | The Netherlands |
| SA2 | 70 | Periodontitis | Subgingival | Finland |
| SA8 | 71 | Periodontitis | Subgingival | Finland |
| SA18 | 72 | Periodontitis | Subgingival | Finland |
| SA23 | 73 | Periodontitis | Subgingival | Finland |
| SA45 | 74 | Periodontitis | Subgingival | Finland |
| SA46 | 75 | Periodontitis | Subgingival | Finland |
| 332 | 76 | Periodontitis | Subgingival | The Netherlands |
| 334 | 77 | Periodontitis | Subgingival | The Netherlands |
| SA10 | 90 | Periodontitis | Subgingival | Finland |
| SA16 | 91 | Periodontitis | Subgingival | Finland |
| SA40 | 92 | Periodontitis | Subgingival | Finland |
| 292 | 93 | Periodontitis | Subgingival | The Netherlands |
| 306 | 94 | Periodontitis | Subgingival | The Netherlands |
| 329 | 95 | Periodontitis | Subgingival | The Netherlands |
| ATCC 33277T | 96 | Gingivitis | Subgingival | United States |
| SA-17b | 97 | Periodontitis | Subgingival | Finland |
| HG1691 | 98 | Periodontitis | Subgingival | The Netherlands |
| HNA99 | 99 | Periodontitis | Subgingival | Japan |
New STs are in boldface.
Strain SA-17 was from the same patient from whom strain SA-18 was isolated.
The isolates in collections 1 and 3, as well as the reference strains from Japan, were received as pure cultures on prereduced anaerobically sterilized blood agar in sealed tubes, while the Finnish isolates had been stored as pure cultures in liquid nitrogen in Oslo since 1996. All isolates were grown on Colombia agar plates, incubated anaerobically (90% N2, 5% H2, 5% CO2) at 37°C (Anoxomat, WS9000; Mart; Lichtenvoorde, The Netherlands), and carefully inspected under a stereomicroscope (magnifications, ×8 to ×50; Zeiss Stemi SV 6) during and after the completion of the growth period for the detection of contaminants. After incubation for 7 to 14 days, a single colony from each culture of P. gingivalis was spread on a plate and further incubated anaerobically for 7 to 14 days before it was harvested for DNA extraction.
Preparation of DNA.
One loop of bacterial culture was suspended in 450 μl of distilled, sterile water, and the DNA was extracted by using the cetyltrimethylammonium bromide procedure (48). The DNA obtained from each sample was diluted 1:5 with distilled, sterile water.
fimA genotyping.
PCR amplification of the six fimA genotypes (genotypes I, Ib, II, III, IV, and V) was performed in MicroAmp reaction tubes (Applied Biosystems) in a GeneAMP system 9700 (ABI, Foster City, CA). The reaction volume of 50 μl consisted of 1 μl diluted DNA, 5 μl 10× PCR buffer, 1 μl (10 μM) of forward and reverse primers (MWG-Biotech Ag, Germany), 4 μl of a 10 μM deoxynucleoside triphosphate mix solution (Applied Biosystems), 0.25 μl of Ampli Taq DNA polymerase (Applied Biosystems), and 37.75 μl distilled, sterile water.
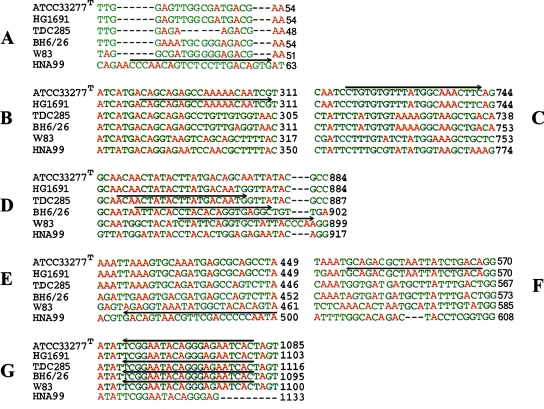
The fimA genotyping was performed with primer sets specific for the different genotypes of fimA (3, 7, 35, 36, 38). The primer sets were synthesized by Operon Biotechnologies GmbH (Cologne, Germany). The specific fimA primer sets (primer sets fimAIf and fimAr, fimAIIf and fimAr, fimAIIIf and fimAr, and fimAIVf and fimAr) were designed by Amano et al. (2), based on earlier findings for the gene sequence; the fimAIbf and fimAIbr primer set was designed by Nakagawa et al. (38); and the fimAVf and fimAVr primer set was also designed by Nakagawa et al. (37). A multiple alignment of these sequences was made by using the ClustalW program (http://www.ebi.ac.uk./clustalw). The sequences of the different specific forward primers in the alignment were located by identifying the corresponding identical sequence in the multiple alignment of the reference sequences of fimA representing fimA genotypes I (ATCC 33277T; GenBank accession no. D17795), Ib (strain HG1691; GenBank accession no. AB058848), II (strain TDC285; GenBank accession no. AB195793), III (GenBank accession no. D17801), IV (strain W83; GenBank accession no. AAQ67087), and V (strain HNA99; GenBank accession no. AB027294), which were obtained from GenBank (NCBI Entrez Gene) (Fig. 1). All isolates were amplified with all primer sets (Fig. 1), and a positive control and a negative control for each set of reactions were always included. The PCR amplification conditions for fimA genotyping were identical to those of Missailidis et al. (35), except that a slightly lower annealing temperature (56°C instead of 58°C) was used.
FIG. 1.
Sequences of forward and reverse primers specific for fimA types I to V superimposed on the multiple-sequence alignment of the reference sequences obtained with the ClustalW program. (A) Forward primer specific for fimA type V; (B) forward primer specific for fimA type Ib; (C) forward primer specific for fimA type I; (D) forward primers specific for fimA types II, III, and IV; (E) reverse primer specific for fimA type V; (F) reverse primer specific for fimA type Ib; (G) reverse primer identical to the sequences of fimA types I, II, III, and IV.
PCR products were examined by agarose gel electrophoresis in Tris-borate-EDTA buffer. The gels were stained for 10 min in a solution of ethidium bromide, washed for 10 min in deionized, distilled water, and photographed under UV light. The results of the PCRs for specific fimA genotyping were recorded as positive or negative after visual examination of photographs of the gels.
fimA sequencing.
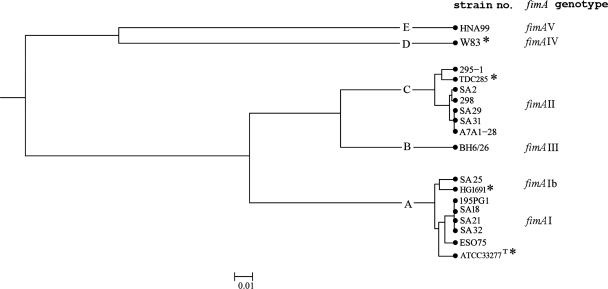
To further explore the sequence variation in the fimA gene, 14 isolates (Fig. 2) in our population were selected for sequencing of the whole gene. The selection was concentrated mostly on isolates that were specifically of fimA genotypes I, Ib, and II or isolates that showed combined positive PCRs with several of these specific primer sets. Isolates BH6/26 (fimA genotype III) and HNA99 (fimA genotype V) were also included (Fig. 2).
FIG. 2.
Distance matrix of selected sequenced isolates obtained by UPGMA after multiple-sequence alignment with the ClustalW program (http://www.ebi.ac.uk./clustalw/) of different fimA genotypes of P. gingivalis. Clusters at a genetic distance of 0.02 were designated A to E. The corresponding fimA genotypes are shown in the right column. The fimA nucleotide sequences from GenBank are marked with an asterisk.
A universal fimA primer set (primers unifim2f and fimAr; 5′-AGTTCTTGCCTGCCTTCAAA-3′ and 5′-AACCCCGCTCCCTGTATTCCGA-3′, respectively) was designed for PCR amplification of the whole fimA gene by using the conserved region at the beginning of the fimA reference sequence alignment for the forward primer (primer unifim2f) (http://www.ebi.ac.uk./clustalw), while fimAr was used as the reverse primer, as its sequence was located in a conserved region downstream for the stop codon of the reference sequences in the gene (Fig. 1). The primers were synthesized by MWG-Biotech Ag, and amplifying conditions were identical to those described above for the reactions performed with the fimA-specific primer sets.
The PCR products obtained from the reactions with the universal primer set (primers unifi2f and fimAr) (14 isolates) were purified by using an exonuclease-alkaline phosphate kit (ExoSAP-IT; USB Corporation, Cleveland, OH), as described by the manufacturer. The primers for PCR amplification were diluted to 1.6 μM and were used to sequence the PCR products from both strands.
In the sequencing reaction with the fimA genes of the 14 isolates, the unifim2f and fimAr primer set and two additional designed sequencing primer sets (forward primer unifim3f1 for fimA types I, Ib, and II [5′-GCTCGCATGGCTTTCACC-3′], forward primer unifim3f2 for fimA type III [5′-GCCCGTATTGCGTTCACC-3′], and universal reverse primer unifimr [5′-TAGACAAACTATGAAAGT-3′]) were used together with the products obtained from the PCR amplification with the universal fimA primer set (primers unifim2f and fimAr). This was done so that both strands of the whole fimA gene (1,044 bp) could be sequenced.
The sequencing reaction was performed with an ABI Prism BigDye Terminator (version 1.1) cycle sequencing ready reaction kit (version 2.0), according to the manufacturer's recommendation. The results were analyzed with an ABI 3730 DNA analyzer (Applied Biosystems).
fimA sequence analysis.
The sequence data obtained by sequencing the 14 isolates were assembled and edited by using the Sequencher (version 4.6) program (Gene Codes Corporation), with a nucleotide sequence fragment of each isolate of about 600 to 700 bp being obtained. By coupling the sequences together, we obtained a nucleotide sequence result for each isolate, and each sequence was linked together in a fasta file. The sequence with GenBank accession no. D17795 (strain ATCC 33277T) refers to the total nucleotide sequence of fimA genotype I, which consists of 1,290 nucleotides. In this sequence the exact number of nucleotides in the fimA gene starts at nucleotide 206 (5′-GTG-3′), ends at nucleotide 1249 (5′-TAA-3′), and consists of 1,044 bp. The 5′ end of each sequence obtained was located at the starting point of the gene (5′-GTG-3′), and all nucleotides upstream were excluded. The sequences obtained were all thoroughly checked against the raw data for misinterpretations, and the end of the gene at the 3′ end of the alignment was identified (5′-TAA-3′; nucleotide 1044). The sequences downstream were excluded before the fasta file was created.
The relationship among the fimA genes of the selected isolates was analyzed by creating a dendrogram of the nucleotide sequences obtained in the fasta file by the unweighted pair group method with arithmetic averages (UPGMA) with S.T.A.R.T.2 software (version 05.13 beta; Sequence Type Analysis and Recombinational Tests; keith.jolley@ceid.ox.ac.uk) (Fig. 2). In addition, the sequences of reference strains ATCC 33277T, HG1691, TDC285, and W83 (GenBank) were included in the dendrogram (Fig. 2).
The S.T.A.R.T.2 software (keith.jolley@ceid.ox.ac.uk) (26) was also used to calculate the pairwise ratios of nonsynonymous substitutions to synonymous substitutions (the dN/dS ratio) for the fimA sequences (41). The average dN/dS value was calculated for the variation in the 14 sequenced isolates and for the variation in the six fimA reference sequences based on the other fasta file containing the corresponding reference sequences (for fimA types I, Ib, II, III, IV, and V) (NCBI Entrez Gene).
Translation to protein sequences.
The prepared fasta file was imported into the Mega (version 3.1) program (30), and an alignment was created in the program before translation to primary protein sequences. These sequences were then exported, and a new fasta file was created from the protein multiple-sequence alignment in the program. In the search for a predicted secondary structure, the translated protein sequences were analyzed by using the PSIPRED protein prediction server (http://www.bioinf.cs.ucl.ac.uk/psipred), while the prediction of the tertiary structure of fimbrillin was done by using the protein search engine Phyre (http://www.sbg.bio.ic.ac.uk/).
MLST.
The PCR primers and amplification conditions for the MLST method have been described earlier (13, 29) (www.pubmlst.org/pgingivalis). Fragments of 380 to 420 bp from seven ubiquitous, chromosomal P. gingivalis genes (ftsQ, cell division protein; gdpxJ, pyridoxal phosphate synthetase; hagB, hemagglutinin; mcmA, methylmalonyl coenzyme A mutase; pepO, endothelin-converting enzyme; pga, transposase; recA, DNA recombination, repair) were amplified. The purification of the PCR products and the sequencing reaction for MLST were performed as described above for the fimA sequencing section.
The allelic profiles and sequencing data from the first strain collection (13) were directly imported into this project. Complementary strands of the sequencing data acquired from the other strain collections and reference strains were assembled, and the sequences were edited by using the Sequencher (version 4.6) program. For each gene, the sequenced fragments of the 44 isolates were compared by use of the single-locus query at the MLST website (http://www.pubmlst.org/pgingivalis). New alleles were given consecutive and higher numbers. On the basis of the combination of the alleles at the seven loci, each strain was assigned an allelic profile, and the distinct profiles were designated STs, according to the database.
The standard MLST statistics (the number of polymorphic sites, the average G+C content, and the allele frequencies) were computed, and a phylogenetic tree was constructed by use of the S.T.A.R.T.2 software (keith.jolley@ceid.ox.ac.uk) (26). The genetic diversity for each locus (h) among the isolates was calculated as 1 − ∑ xi2 [n/(n − 1)], where xi is the frequency of the ith allele at the locus and n is the number of isolates (46).
To assess the genetic relationships among the isolates on the basis of their allelic profiles, another UPGMA dendrogram was also constructed (Fig. 3). In addition, the S.T.A.R.T.2 program was used to calculate the index of association (IA), based on 69 STs (49). The standardized index of association (IAS) was calculated as (1/l − 1)IA, where l is the number of loci (22). Furthermore, the program was used to calculate the dN/dS ratios for the MLST data (41).
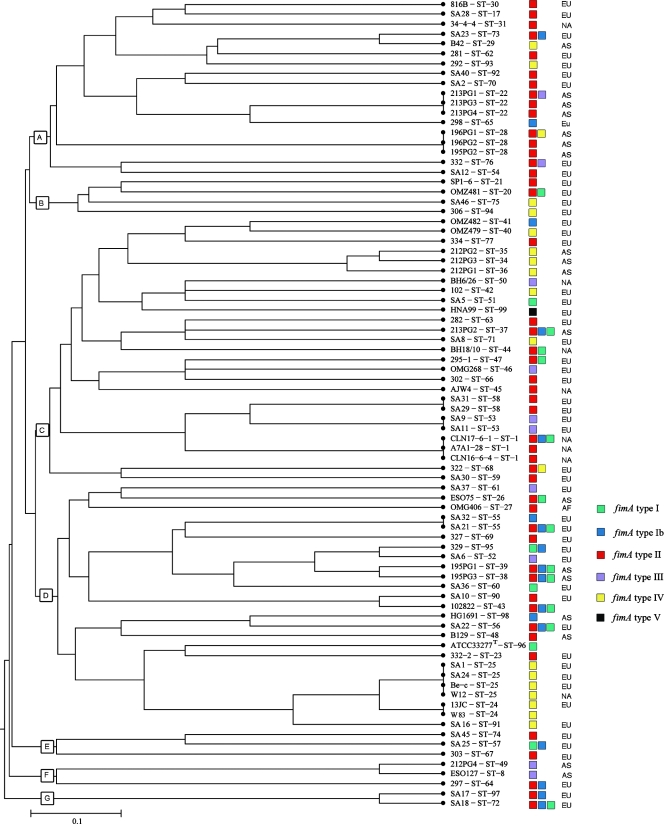
FIG. 3.
Genetic relationships among 82 P. gingivalis isolates. A distance matrix was calculated by UPGMA on the basis of the differences in the allelic profiles. Clusters at a genetic distance of 0.5 were designated A to G. Columns to the right of the tree give the ST, the strain designation, the fimA genotype (see below), and the continents where the strains originated (EU, Europe; NA, North America; AF, Africa; AS, Asia).
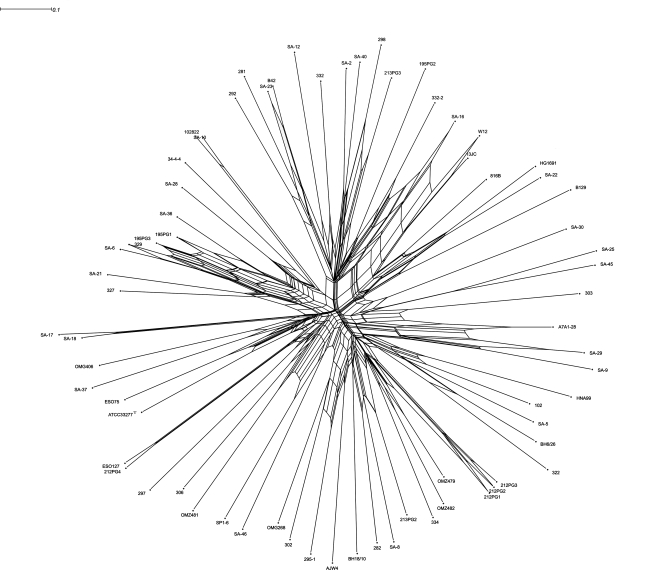
Split decomposition analysis can be used as a tool to illustrate graphically whether recombination plays a role in the evolutionary history of the separate genes investigated by MLST. The analysis was performed with SplitsTree (version 4.6) software, downloaded from http://www.splitstree.org. By pasting the allelic profiles from the MLST data set for the 82 isolates included in the study into the web tool provided at http://www.pubmlst.org, a nexus file was created, and that file was further imported into the SplitsTree (version 4.6) software. When the allelic profiles of the seven gene fragments were used, the SplitsTree graph obtained visualizes the isolates according to the degree of recombination between the isolates in the bacterial population (13, 24, 29).
RESULTS
fimA genotyping.
The distribution of positive results obtained by PCR with the specific primer sets among the 82 P. gingivalis isolates from patients with periodontitis is shown in Table 2. Sixty-one isolates (74.4%) showed a positive reaction with a single set of specific fimA primers. In this group we found that fimA genotype II dominated and was detected in 34.1% of the isolates, while fimA genotype IV was detected in 20.7%. fimA genotypes III, Ib, and I were found in 9.8%, 4.9%, and 3.7% of the isolates, respectively; and fimA type V was detected only in isolate HNA99, a positive control for fimA genotype V.
TABLE 2.
Allelic variation of the fimA genes of 82 P. gingivalis isolates from patients with periodontitis
| Allelic variant | No. of isolates | % of all isolates tested (% total combination prevalencea) |
|---|---|---|
| fimA I | 3 | 3.7 |
| fimA Ib | 4 | 4.9 |
| fimA II | 28 | 34.1 |
| fimA III | 8 | 9.8 |
| fimA IV | 17 | 20.7 |
| fimA V | 1 | 1.2 |
| fimA I/Ib/II | 8 | 9.8 (38.1) |
| fimA I/II | 4 | 4.9 (19.0) |
| fimA Ib/II | 3 | 3.7 (14.3) |
| fimA II/III | 2 | 2.4 (9.5) |
| fimA II/IV | 2 | 2.4 (9.5) |
| fimA I/Ib | 2 | 2.4 (9.5) |
Prevalence of different combinations of positive PCR results from the total prevalence of combinations of 25.6%.
Twenty-one isolates (25.6%) had positive PCR results with more than one specific primer set. The most prevalent combination was found in eight isolates (38.1%), with a positive reaction for genotypes I, Ib, and II. In four isolates (19.0%) combinations of fimA genotypes I and II were observed, and in three isolates (14.3%) combinations of types Ib and II were observed. Combinations of types II and III (9.5%), types II and IV (9.5%), and types I and Ib (9.5%) were found in two isolates each (Table 2).
fimA sequencing.
The whole coding sequence of the fimA gene (1,044 bp) of 14 isolates was sequenced. Six isolates from among those with positive PCR results for multiple genotypes were selected (strains SA18, SA21, and 195PG1, which were positive for fimA types I, Ib, and II; strains 295-1 and ESO75, which were positive for fimA types I and II; and strain SA25, which was positive for fimA types I and Ib). Five isolates represented those with positive PCR results for a single type by the fimA genotyping method (strains SA2, SA29, and SA31, which were positive for fimA type II, and strains SA32 and 298, which were positive for fimA type Ib), together with isolates A7A1-28, BH6/26, and HNA99 (reference strains for fimA genotypes II, III, and V, respectively) (Fig. 2). Multiple-sequence alignment of all the fimA sequences obtained for the 14 isolates with the ClustalW program (www.ebi.ac.uk/clustalw/) showed a high degree of conservation within the isolates, with some isolates of each genotype differing from the reference sequences included by separate, single nucleotides. In some regions, point mutations could be located at the same nucleotide or at neighboring nucleotides across several sequences. In other regions, the mutations were restricted to only one part of the alignment.
An UPGMA dendrogram was created from the sequence data obtained for these isolates by using the S.T.A.R.T.2 program (Fig. 2). At a linkage distance of 0.02, the 14 isolates were divided into five clusters (clusters A to E), with clusters B, D, and E represented by one isolate each (isolates BH6/26, W83, and HNA99, respectively).
The sequences of eight isolates were found in cluster A. Isolates SA18, SA21, SA32, and 195PG1 showed identical fimA sequences that clustered in one branch with isolate ESO75 and the reference sequence of strain ATCC 33277T (GenBank) (Fig. 2). For the isolates in this branch, the ClustalW alignment showed 11 separate regions with point mutations. In isolate ESO75, a single repeat (5′-ACC-3′) was located at nucleotide positions 594 to 596; this repeat was unique to ESO75. In the other branch of cluster A, isolate SA25 appeared together with the sequence of isolate HG1691 (GenBank accession no. AB058848). The two sequences differed at 14 separate nucleotides throughout the whole gene; in addition, nucleotides 872 to 875 were changed from 5′-ATGG-3′ in HG1691 to 5′-GCAA-3′ in our isolate.
Cluster C contained the reference sequence of TDC285 (GenBank accession no. AB195793) together with that of isolate 295-1 in the first branch and the sequences of isolates 298, SA2, SA29, SA31, and A7A1-28 in the second branch. In the last branch, isolates SA2 and 298 had fimA sequences with few variations between them; and their fimA sequences also differed from those of isolates SA29, SA31, and A7A1-28, which were identical. Isolates in cluster C presented 14 separate regions of point mutations. The sequence of isolate 295-1 was unique in that group, in that it showed identity with the sequence of BH6/26 at two triplets (6 bp) at nucleotide positions 42 to 48, which may suggest a recombinational event.
Except for one point mutation in the first part of the gene of isolate BH6/26, the sequences that we determined for isolates BH6/26 and HNA99 were identical to the respective fimA reference sequences corresponding to fimA genotypes III and V (GenBank, accession nos. D17801 and AB027294), respectively. The clustering of the isolates versus the corresponding specific fimA genotypes is shown in Fig. 2. Comparison of regions of point mutations between fimA genotypes I, Ib, and II indicated no pattern of recombination.
Specific fimA genotyping versus fimA sequencing.
Comparison of the specific fimA genotyping results and the sequencing results for selected isolates concentrated on those assigned to fimA genotypes I, Ib, and II and isolates with positive results by PCR with several of the specific primer sets. Isolates SA18, SA21, and 195PG1 showed positive PCR results with all of the primer sets specific for fimA genotypes I, Ib, and II. These isolates were assigned to cluster A, corresponding to fimA genotype I.
Isolate SA32 showed a positive result by PCR with fimA genotype Ib-specific primers; but the sequencing results assigned the isolate to cluster A, corresponding to fimA genotype I, and the isolate had a nucleotide sequence identical to the sequences of isolates SA18, SA21, and 195PG1. The specific genotyping of isolates ESO75 and 295-1 gave positive PCR results for fimA genotypes I and II. After it was sequenced, isolate ESO75 showed a sequence similar to the sequences of isolates assigned to cluster A, corresponding to fimA genotype I, while isolate 295-1 was assigned to cluster C, corresponding to fimA genotype II. For isolate SA25, the specific genotyping resulted in a positive PCR result for fimA genotypes I and Ib; and it was identified in one branch in cluster A, corresponding to fimA genotype Ib, after it was sequenced.
For isolates A7A1-28, SA2, SA29, and SA31, sequencing placed the isolates in cluster C and confirmed the results of the specific genotyping, while isolate 298 was positive by PCR for fimA genotype Ib and was placed in cluster C, which also corresponds to fimA genotype II, after sequencing.
dN/dS ratio for fimA.
The average dN/dS ratio, based on sequence variation, for the 14 sequenced isolates was 0.5392. This indicated that the frequency of synonymous substitutions was almost twice as high as the frequency of changes leading to alterations in the primary structure of the amino acid sequence.
Protein sequences.
Translation of the DNA sequences of the fimA gene to the protein sequences of the selected isolates showed variations in the primary structure of fimbrillin. For each of the isolates, the same number of primary protein structures was produced, as there were different nucleotide fimA sequences (for cluster A, three; for cluster B, one; for cluster C, four, and for clusters D and E, one each) (Fig. 2).
The amino acid compositions of the FimA proteins from isolates SA18, SA21, SA32, 195PG1, and ESO75 (cluster A, corresponding to fimA genotype I) varied at 7 single, separate amino acids. Those of isolates SA25 (cluster A, corresponding to fimA genotype Ib) and ESO75 or SA18, SA21, SA32, and 195PG1 (cluster A, corresponding to fimA genotype I) varied at 16 amino acids; and the variations were located at different positions in the two comparisons. In cluster C, variations in only two amino acids between the fimA sequences of isolates SA2 and 298 and the identical sequences of isolates A7A1-28, SA29, and SA31 were observed. The variation between these sequences and the protein sequence of isolate 295-1 was 19 amino acids. Comparison of the primary structures of the translated reference sequences of fimA genotypes I and II showed variations in 80 amino acids.
Comparison of the proteins after a search with the PSIPRED protein prediction server (http://bioinf.cs.ucl.ac.uk/psipred) (8), a highly accurate method for prediction of the secondary protein structure, showed that the fimbrillin proteins contain both α helices and β sheets; but significant differences in the predicted secondary structure elements of the translated primary protein structures could not be identified. Attempts to predict the protein fold by using the Phyre protein homology/analogy recognition engine (http://www.sbg.bio.ic.ac.uk/) were not successful due to the lack of an experimental structure of a protein with a high degree of homology with FimA.
MLST analysis, STs, and allelic profiles of P. gingivalis isolates.
Altogether, 69 STs were identified among the 82 P. gingivalis isolates included in this study, and 37 new STs were identified among the isolates from the second and third strain collections, including isolates HG1691, HNA99, and W83 (these new STs were detected in a total of 44 isolates not characterized earlier by MLST) (Table 2) (http://www.pubmlst.org/pgingivalis). Sixty-one STs were represented by a single isolate. Three STs (ST1, ST22, and ST28) were represented by three isolates each (3.7%), while ST25 was represented by four isolates (isolates SA1, SA24, W12, and Be-c) (4.9%). Furthermore, ST24, ST53, ST55, and ST58 were represented by two isolates each (2.4%) (Table 1). Only three isolates from the Finnish strain collection (isolates SA1, SA24, and SA28) and W83 (ATCC BAA-308) had STs (ST17, ST24, and ST25) that were identified previously (13, 29) (Table 1).
The average number of alleles per locus for the seven genes was 23.7 (range, 11 to 29) (http://www.pubmlst.org/pgingivalis). The average G+C contents for each gene were estimated to range from 48.4% for the pepO gene to 58.0% for the hagB gene (Table 3). No insertions or deletions were observed in any of the sequences, and for all seven loci, the 82 sequences were aligned without gaps and numbered (www.pubmlst.org/pgingivalis). The sequences revealed from 2.3% (recA) to 7.1% (ftsQ) variations in the nucleotides, and the number of polymorphic sites ranged from 9 to 30 for the different genes. The variation between alleles was always located at one or a few nonadjacent nucleotide sites. The average dN/dS values shown in Table 3 were all <1, indicating that most of the sequence variability was selectively neutral for the genes included in the MLST analysis. For each of the seven loci, the number of synonymous substitutions was at least three times larger than the number of substitutions that led to amino acid changes when the sequence was translated (Table 3).
TABLE 3.
Genetic diversity at seven loci in 69 STs of 82 isolates of P. gingivalis
| Locus | Fragment length (bp) | No. of alleles | No. of polymorphic sites | % Variable nucleotide sites | % Avg G+C content | Avg dN/dS ratio | Genetic diversity |
|---|---|---|---|---|---|---|---|
| ftsQ | 420 | 29 | 30 | 7.1 | 49.3 | 0.2431 | 0.929 |
| gdpxJ | 380 | 28 | 22 | 5.8 | 55.5 | 0.2164 | 0.919 |
| hagB | 380 | 25 | 18 | 4.7 | 58.0 | 0.3214 | 0.813 |
| mcmA | 420 | 22 | 15 | 3.6 | 51.4 | 0.0202 | 0.861 |
| pepO | 400 | 29 | 20 | 5.0 | 48.4 | 0.2175 | 0.936 |
| pga | 400 | 22 | 23 | 5.8 | 53.4 | 0.0635 | 0.840 |
| recA | 400 | 11 | 9 | 2.3 | 53.0 | 0.0252 | 0.789 |
The maximum frequencies of the individual alleles ranged from 34.8% (24/69) for the pga and recA genes to 11.6% (8/69) for the pepO gene (data not shown). The genetic diversity at individual loci ranged from 0.789 for the recA gene to 0.936 for the pepO gene, with a mean of 0.870 (Table 3).
Among the 44 isolates with new STs, new alleles were detected at all seven MLST genes (http://www.pubmlst.org/pgingivalis). For the pga gene we detected only two new alleles, each of which was recorded with a prevalence of 1.4%; and for the other genes we detected from 4 (recA) to 10 (gdpxJ, pepO) new alleles, with the prevalence never being greater than 4.3% for the whole strain collection.
Cluster analysis.
The evolutionary relationship among the STs in this study was illustrated by UPGMA, with the phylogenetic tree shown in Fig. 3. The 69 STs clustered at a genetic distance of 0.5. Seven clusters (clusters A to G) were observed at a linkage distance at 0.45 (Fig. 3). Except for clusters E and G, which consisted of only three and two isolates, respectively, all from Europe, the other clusters contained isolates from different continents.
Congruence and recombination analysis.
The SplitsTree graph was created for all isolates with each of the gene fragments individually (data not shown) and for all the seven gene fragments together by using the allelic profiles. This showed an overall clonal population structure with some degree of recombination along some of the branches (Fig. 4).
FIG. 4.
SplitsTree graph illustrating the degree of recombination between 82 isolates of P. gingivalis for the seven ubiquitous chromosomal gene fragments used for MLST.
The calculation of IA, based on the allelic profile data with one representative of each ST detected in the study, gives a good estimate of the degree of association between the alleles at different loci. IA was calculated to be 0.324, indicating a significant linkage disequilibrium in the population and a weak clonal population structure. IAS has been claimed (22) to give a better estimate of the linkage disequilibrium than the simpler IA value because it takes the number of loci included in the study into account. It was calculated to be 0.054.
fimA and MLST.
In Fig. 3, the results of specific fimA genotyping (Table 2) were superimposed on the dendrogram constructed from the MLST data by UPGMA to illustrate the relationship between the results of MLST, the geographic origins of the isolates, and the distributions of the fimA genotypes of the P. gingivalis isolates. Forty-eight of the different STs included in the study (69.6%) showed a single specific fimA genotype. The dendrogram showed an even distribution of different fimA genotypes in relation to the ST and the geographic origin. Furthermore, the dendrogram indicated some tendency for clustering of the fimA genotypes in relation to the STs of the strains obtained by MLST, with isolates of the same or closely related STs harboring the same fimA genotype.
DISCUSSION
In this study we investigated the prevalence of different fimA genotypes in a collection of P. gingivalis isolates sampled and cultured from periodontitis patients from four continents. The specific fimA genotypic distribution of the isolates examined was related to their STs determined by MLST in order to find a possible association between variations in the fimA gene and the genotypic framework provided by MLST.
The results of the MLST analysis in this investigation supported our earlier results and conclusion about the weak clonal population structure of P. gingivalis (13). The 44 isolates added to the earlier population investigated were assigned to 37 new STs, providing further evidence of the great genotypic diversity of the species (Fig. 3). The estimates of the IA and IAS values in this study were similar to our previous findings, which indicated a significant linkage disequilibrium (13). Together with the congruence and recombinational analysis performed in the split decomposition analysis, these results further supported the conclusion that P. gingivalis has a weak clonal population structure.
The fimA genotypes of the 82 isolates included in this study were determined by using the same specific fimA primer sets and similar experimental conditions used in a number of studies that have used the direct PCR technique with clinical material for investigation of the prevalence of the fimA genotypes of P. gingivalis from sites in patients with various periodontal conditions (2, 3, 7, 35, 36, 38). The results that we obtained with cultured isolates confirmed the results of clinical investigations that fimA genotypes II and fimA IV are predominant in chronic periodontitis-affected sites (2, 7). Other investigators (35, 36, 38) have confirmed that genotype II is the most prevalent genotype at diseased sites, with genotype Ib being the second most prevalent in patients with aggressive or chronic periodontitis. These observations and the results from a similar study with healthy subjects with a predominance of genotypes I, III, and V (3) could indicate a difference in the fimbrillin monomer and, therefore, a difference in the composition of the fimbriae that could be responsible for the variation in the pathogenic potential of the major fimbriae of P. gingivalis. The recombinant FimA protein from genotype II strains demonstrated a greater ability to adhere to and invade human epithelial cells than the recombinant FimA protein from strains of other genotypes (39). Any variation in the secondary protein structure between the different proteins produced in this study was not detectable due to a nonexistent overall structure of the protein itself or a protein with a high degree of homology with FimA.
A study of the adhesion and invasion abilities of different fimA genotypes of P. gingivalis in KB cells (originally derived from an epidermal carcinoma of the mouth), however, did not indicate any significant difference between the fimA genotypes, suggesting that other characteristics besides the fimA gene variation could be responsible for the observations (5, 28, 52). The absence of genotype V in our material was in accordance with the results of Missailidis et al. (35), who directly genotyped isolates from clinical samples from a Brazilian population.
In our study, a relatively large number of isolates positive by PCR for multiple fimA genotypes was seen, similar to the prevalence reported in clinical studies (2, 3, 7, 35, 37), without the cultivation of P. gingivalis. Since the fimA gene occurs as a single copy in the chromosome of the species (12), the cross-reactions to different specific primer sets in these investigations have been explained by the possible existence of new unidentified genotypes (35, 37, 38). The nontypeable strains reported in the literature have also been explained by the presence of new genotypes (2, 7, 37, 38). In our study, however, all isolates were typeable.
Another explanation for the multiple positive PCR observations in the clinical studies could be that several different genotypes colonize the same site (2, 35). Although there is a general acceptance that only one clone is present in a periodontitis patient, some investigators have demonstrated that individual patients may harbor several clones (13, 33, 35). Whether these clones were sampled from the same or different sites was not reported, however.
Since we have dealt with cultured isolates in our material and have made the same observations as those made in clinical studies concerning positive PCR results for multiple genotypes (2, 4, 7, 35, 37, 38), we anticipated that the answer to the positive PCR results for multiple genotypes could be explained by a previously unidentified variation within the fimA genes of our isolates. We therefore designed a set of universal primers with the purpose of sequencing a selected number of isolates in an attempt to explain our findings. The isolates were chosen on the basis of the results of the specific genotyping (Fig. 3), and we mainly focused on those isolates with multiple genotypes that were positive by PCR with primers specific for fimA genotypes I, Ib, and II because of the high prevalence of these combinations. Two reference strains of fimA genotypes III and V were also selected for sequencing.
Although we found minor variations, mainly point mutations, in the sequences of most of the selected isolates, the results for the 14 different isolates in our material indicated that the fimA gene is conserved, with few signs of recombination within the gene, except in isolate 295-1. This may present a recombination of a fimA type III fragment within the fimA type II gene. Our multiple-sequence alignments of the fimA genes of several isolates also resembled the alignments published by Fujiwara et al. (16).
A search of the GenBank database for reference strains of fimA genotypes revealed several fimA genotype II sequences, and a multiple-sequence alignment of these alleles with the ClustalW program demonstrated the same small variations in the fimA type II gene observed with the sequenced isolates in our material.
The specific fimA primers superimposed on the multiple-sequence alignments are shown in Fig. 1. An important observation was that the sequence of the forward primer specific for fimA type Ib (primer fimAIbf) was identical to the reference sequences for both fimA type I and type Ib and was located in a conserved region. In the region of the forward primer specific for fimA type I (primer fimAIf), the sequences of both genotypes were conserved and identical as well. The sequences of the reverse primers specific for types I and Ib were also selected from two separate but conserved and identical regions of the fimA gene. On the basis of these observations, i.e., the variation between sequences in fimA genotypes I and Ib and the reported 97.1% homology (38) between the reference sequences of these genotypes, the relevance of distinguishing fimA genotype Ib may be questioned. Theoretically, all isolates belonging to fimA genotype I should also be positive by the genotype Ib-specific PCR. However, this was not what we observed. A possible explanation could be the existence of a minor variation in the primer binding site in some isolates.
Since the sequences of the forward primers specific for types I, II, III, and IV (Fig. 1) were selected from conserved regions or from almost the same region of the gene with small variation and partly overlap and because the sequence of the reverse primer was identical for the four genotypes and is located in a conserved region downstream of the end of the gene, one might obtain positive PCR results for multiple genotypes for isolates belonging to these genotypes, as supported by Missailidis et al. (35). Our sequencing results therefore indicated that the specific method for the genotyping of the major fimbriae of P. gingivalis needs to be improved. This fact may have previously been concealed, because genotyping of the major fimbriae of P. gingivalis has been done directly with clinical material.
Whether the variation in the sequences of the fimA gene may result in changes in the amino acid composition or not can be calculated by using the average dN/dS ratio of a number of DNA sequences (41). Since the average dN/dS value for the 14 isolates sequenced was 0.5392 (i.e., <1), the estimation of the unchanged amino acid composition is almost doubled compared to the likelihood of a resulting change in the primary protein structure after translation. The average dN/dS value for the fimA gene compared to the dN/dS value for the MLST gene with the highest value (hagB, for which the dN/dS value was 0.3214) indicates a greater likelihood of a change in the amino acid composition for fimA than for the MLST genes.
Translation of the various nucleotide sequences of the fimA genes of the 14 isolates selected resulted in various primary protein structures, which resulted in the same number of proteins as the number of sequence variants of the fimA gene. Although our findings indicate that the fimA gene is conserved, we have shown that there is a small variation between isolates belonging to the same genotype, in addition to the variation between different genotypes, resulting in a corresponding variation in the primary protein sequence of the FimA monomer after translation. This was also shown by Fujiwara et al. (16). Whether these mutations result in a FimA monomer with a changed structure that influences the pathogenicity of the isolate may partly be dependent on how the secondary structure of the molecule folds into a tertiary structure. Through bioinformatic resources on the Internet, we were not able to predict either the secondary structure of the protein or how it folds into a tertiary structure due to the lack of an experimental structure resembling a protein with a high degree of homology with FimA.
The relationship between the distribution of fimA genotypes and MLST data for P. gingivalis has not been investigated before. The tendency of the fimA genotypes in relation to the multilocus STs to cluster with isolates of the same or closely related STs harboring the same fimA genotype strengthens our conclusion that P. gingivalis has a weak clonal population structure, but no strong relationship between fimA genotypes and the STs of the species was evidenced. While our findings support the relationship between periodontitis and the prevalence of fimA genotypes II and IV, they also indicate that the method for the genotyping of the major fimbriae of P. gingivalis should be reevaluated.
Acknowledgments
We are indebted to the Faculty of Dentistry, University of Oslo, Oslo, Norway, for financial support.
We thank S. Asikainen, T. Ooshima, and A. J. van Winkelhoff for providing strains of P. gingivalis; K. Jolley, P. H. Backe, B. Dalhus, and T. Rognes for scientific advice; and T. Alvestad, A. M. Klem, and J. Oksnes for technical assistance.
Footnotes
Published ahead of print on 31 October 2007.
REFERENCES
- 1.Al-Zahrani, M. S., N. Bissada, and E. A. Borawski. 2003. Obesity and periodontal disease in young, middle-aged and older adults. J. Periodontol. 74610-615. [DOI] [PubMed] [Google Scholar]
- 2.Amano, A., I. Nakagawa, K. Kataoka, I. Morisaki, and S. Hamada. 1999. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J. Clin. Microbiol. 371426-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano, A., M. Kubinowa, I. Nakagawa, S. Akiyama, I. Morisaki, and S. Hamada. 2000. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 791664-1668. [DOI] [PubMed] [Google Scholar]
- 4.Amano, A. 2003. Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J. Periodontol. 7490-96. [DOI] [PubMed] [Google Scholar]
- 5.Amano, A., I. Nakagawa, N. Okahashi, and N. Hamada. 2004. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J. Periodontal Res. 39136-142. [DOI] [PubMed] [Google Scholar]
- 6.American Academy of Periodontology. 1996. Position paper: epidemiology of periodontal diseases. J. Periodontol. 67935-945. [PubMed] [Google Scholar]
- 7.Beikler, T., U. Peters, S. Prajaneh, K. Prior, B. Ehmke, and T. F. Fleming. 2003. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur. J. Oral Sci. 111390-394. [DOI] [PubMed] [Google Scholar]
- 8.Bryson, K., L. J. McGuffin, R. L. Marsden, J. J. Ward, J. S. Sodhi, and D. T. Jones. 2005. Protein structure prediction servers at University College London. Nucleic Acids Res. 33(web server issue)W36-W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Califano, J. V., T. Arimoto, and T. Kitten. 2003. The genetic relatedness of Porphyromonas gingivalis clinical and laboratory strains assessed by analysis of insertion sequence (IS) element distribution. J. Periodontal Res. 38411-416. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, J. E., and E. J. Feil. 2004. Multilocus sequence typing—what is resolved? Trends Microbiol. 12373-377. [DOI] [PubMed] [Google Scholar]
- 11.Darveau, R. P., A. Tanner, and R. C. Page. 1997. The microbial challenge in periodontitis. Periodontol. 2000 1412-32. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson, D. P., M. A. Kubiniec, F. Yoshimura, and R. J. Genco. 1988. Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis. J. Bacteriol. 1701658-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enersen, M., I. Olsen, A. J. van Winkelhoff, and D. A. Caugant. 2006. Multilocus sequence typing of Porphyromonas gingivalis strains from different geographic origins. J. Clin. Microbiol. 4435-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, Z., and A. Weinberg. 2006. Role of bacteria in health and disease of periodontal tissues. Periodontol. 2000 4050-76. [DOI] [PubMed] [Google Scholar]
- 15.Frandsen, E. V., K. Poulsen, M. A. Curtis, and M. Kilian. 2001. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect. Immun. 694479-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujiwara, T., S. Morishima, I. Takahashi, and S. Hamada. 1993. Molecular cloning and sequencing of the fimbrillin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem. Biophys. Res. Commun. 197241-247. [DOI] [PubMed] [Google Scholar]
- 17.Genco, C. A., T. Van Dyke, and S. Amar. 1998. Animal models for Porphyromonas gingivalis-mediated periodontal disease. Trends Microbiol. 6444-449. [DOI] [PubMed] [Google Scholar]
- 18.Haffajee, A. D., and S. S. Socransky. 1994. Microbial etiological agents of destructive periodontal diseases. Periodontol. 2000 578-111. [DOI] [PubMed] [Google Scholar]
- 19.Haffajee, A. D., M. A. Cugini, A. Tanner, R. P. Pollack, C. Smith, R. L. Kent, Jr., and S. S. Socransky. 1998. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J. Clin. Periodontol. 25346-353. [DOI] [PubMed] [Google Scholar]
- 20.Hamada, N., H. T. Sojar, M. Cho, and R. J. Genco. 1996. Isolation and characterization of minor fimbria from Porphyromonas gingivalis. Infect. Immun. 594788-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamada, S., A. Amano, S. Kimura, I. Nakagawa, S. Kawabata, and I. Morisaki. 1998. The importance of fimbriae in the virulence and ecology of some oral bacteria. Oral Microbiol. Immunol. 13129-138. [DOI] [PubMed] [Google Scholar]
- 22.Haubold, B., and R. R. Hudson. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16847-848. [DOI] [PubMed] [Google Scholar]
- 23.Holt, S., and J. Ebersole. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 3872-122. [DOI] [PubMed] [Google Scholar]
- 24.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 1468-73. [DOI] [PubMed] [Google Scholar]
- 25.Imamura, T. 2003. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 74111-118. [DOI] [PubMed] [Google Scholar]
- 26.Jolley, K., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato, T., S. Kawai, K. Nakano, H. Inaba, M. Kubinowa, I. Nakagawa, K. Tsuda, H. Omori, T. Ooshima, T. Yoshimon, and A. Amano. 2007. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell. Microbiol. 9753-765. [DOI] [PubMed] [Google Scholar]
- 28.Kilian, M., E. V. G. Frandsen, D. Haubek, and K. Poulsen. 2006. The etiology of periodontal disease revisited by population genetic analysis. Periodontol. 2000 42158-179. [DOI] [PubMed] [Google Scholar]
- 29.Koehler, A., H. Karch, T. Beikler, T. F. Flemmig, S. Suerbaum, and H. Schmidt. 2003. Multilocus sequence analysis of Porphyromonas gingivalis indicates frequent recombination. Microbiology 1492407-2415. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 31.Laine, M. L., B. J. Appelmelk, and A. J. van Winkelhoff. 1996. Novel polysaccharide capsular serotypes in Porphyromonas gingivalis. J. Periodontal Res. 31278-284. [DOI] [PubMed] [Google Scholar]
- 32.Lamont, R. J., and H. F. Jenkinson. 2000. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol. Immunol. 15341-349. [DOI] [PubMed] [Google Scholar]
- 33.Loos, B. G., A. J. van Winkelhoff, R. G. Dunford, R. J. Genco, J. De Graaff, D. P. Dickinson, and D. W. Dyer. 1992. A statistical approach to the ecology of Porphyromonas gingivalis. J. Dent. Res. 71353-358. [DOI] [PubMed] [Google Scholar]
- 34.Loos, B. G., D. W. Dyer, T. S. Whittam, and R. K. Selander. 1993. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect. Immun. 61204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Missailidis, C. G., J. E. Emeda, C. Ota-Tsuzuki, D. Anzai, and M. P. A. Mayer. 2004. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol. Immunol. 19224-229. [DOI] [PubMed] [Google Scholar]
- 36.Miura, M., T. Hamachi, O. Fujise, and K. Maeda. 2005. The prevalence and pathogenic differences of Porphyromonas gingivalis fimA genotypes in patients with aggressive periodontitis. J. Periodontal Res. 40147-152. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa, I., A. Amano, R. K. Kimura, T. Nakamura, S. Kawabata, and S. Hamada. 2000. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of fimA gene. J. Clin. Microbiol. 381909-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakagawa, I., A. Amano, Y. Ohara-Nemoto, N. Endoh, I. Morisaki, S. Kimura, S. Kawabata, and S. Hamada. 2002. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J. Periodontal Res. 37425-432. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa, I., A. Amano, M. Kubinowa, T. Nakamura, S. Kawabata, and S. Hamada. 2002. Functional differences among fimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epithelial cells. Infect. Immun. 70277-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakano, K., M. Kubinowa, I. Nakagawa, T. Yamamura, R. Nomura, N. Okahashi, T. Ooshima, and A. Amano. 2004. Comparison of inflammatory changes caused by Porphyromonas gingivalis with distinct fimA genotypes in a mouse abscess model. Oral Microbiol. Immunol. 19205-209. [DOI] [PubMed] [Google Scholar]
- 41.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3418-426. [DOI] [PubMed] [Google Scholar]
- 42.Neiders, M. E., P. B. Chen, H. Suido, H. S. Reynolds, J. J. Zambon, M. Shlossman, and R. J. Genco. 1989. Heterogeneity of virulence among strains of Bacteroides gingivalis. J. Periodontal Res. 24192-198. [DOI] [PubMed] [Google Scholar]
- 43.Nelson, K., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. Duncan, F. L. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 1855591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuda, K., J. Slots, and R. J. Genco. 1981. Bacteroides gingivalis, Bacteroides asaccharolyticus and Bacteroides melaninogenicus subspecies: cell surface morphology and adherence to erythrocytes and human buccal epithelial cells. Curr. Microbiol. 67-12. [Google Scholar]
- 45.Papapanou, P. N. 1999. Epidemiology of periodontal diseases. J. Int. Acad. Periodontol. 4110-116. [PubMed] [Google Scholar]
- 46.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slots, J., and R. J. Gibbons. 1978. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect. Immun. 19254-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, G. L., S. S. Socransky, and C. M. Smith. 1989. Rapid method for the purification of DNA from subgingival microorganisms. Oral Microbiol. Immunol. 447-51. [DOI] [PubMed] [Google Scholar]
- 49.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 904384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25134-144. [DOI] [PubMed] [Google Scholar]
- 51.Socransky, S. S., and A. D. Haffajee. 2005. Periodontal microbial ecology. Periodontol. 2000 38135-187. [DOI] [PubMed] [Google Scholar]
- 52.Umeda, J. E., C. Missailidis, P. L. Longo, D. Anzai, M. Wikstrom, and M. P. A. Mayer. 2006. Adhesion and invasion to epithelial cells by fimA genotypes of Porphyromonas gingivalis. Oral Microbiol. Immunol. 21415-419. [DOI] [PubMed] [Google Scholar]