Abstract
Shiga toxin-producing Escherichia coli (STEC) strains from cattle and diarrheic children in a pastoralist community in Uganda were investigated. The STEC strains belonged to a variety of different serogroups, and 70% of the strains were positive for the intimin gene, eae. STEC strains from two of the children were closely related to bovine strains.
Cattle are the primary reservoir for human infections with Shiga toxin-producing Escherichia coli (STEC). These organisms may be transmitted from healthy, STEC-excreting cattle to humans via the food chain or by contact with animals or with their environment. The cattle corridor of Uganda is used by seminomadic pastoralist communities to graze and water their cattle. The incidence of STEC infections among cattle and humans living in these communities is unknown. The study area, Nyabushozi County, Uganda, is a remote district in the agroecological zone of the pastoral system within the cattle corridor. Health care is provided by small private clinics and three government outpatient clinics. The pastoralist communities live in close proximity to each other and to their cattle. Cattle keeping is characterized by the seminomadic, extensive grazing of large herds of freely mixing cattle on communal pastures, with access to water, all year round.
Stool samples or rectal swabs were obtained from 92 children (age range, 2 months to 12 years) reporting with diarrhea. With the intention of studying the relatedness of the STEC strains obtained from children to the corresponding isolates obtained from cattle, a guide assisted with the identification of cattle associated with the homestead of the child. Fresh fecal samples were collected from 10% of the cattle on these homesteads.
Of the 300 bovine fecal samples collected, E. coli was isolated from 196 adult animals, 10 heifers, and 10 calves. E. coli was isolated from the stools of 80 of the 92 children. Boiling was used to release the DNA from a sweep of E. coli colonies, which was used in multiplex PCR assays with primers (1) for the amplification of stx1 and stx2. PCR products of the expected sizes were obtained for 14 adult bovines and 1 calf (6.9%). With respect to the clinical specimens, amplicons were obtained from 7 of the 80 isolates (8.75%).
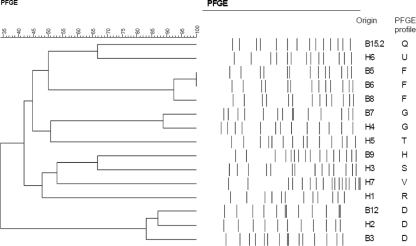
Ten single colonies from each of the 22 stx-positive sweeps (from 150 bovines and 70 humans) were analyzed for the presence of stx genes by PCR. Of the colonies tested, 126 (86 bovine and 40 human) were stx positive. Pulsed-field gel electrophoresis (PFGE) (5, 8) was carried out with these STEC isolates. Images were captured with a Kodak ID imaging program (Scientific Imaging System). The banding patterns were analyzed with the GelCompar II program; the similarity between the PFGE patterns was calculated by using the Dice coefficient similarity (tolerance, 1%). STEC isolates with profiles showing a greater than 69% coefficient of similarity were regarded as closely related, while strains with less than 65% similarity were considered unrelated. Seventeen banding patterns (patterns A to Q) were defined for the bovine isolates (Table 1; Fig. 1). While the majority of bovines (n = 12) were each infected with only one strain of STEC, three bovines were each infected with more than one strain of this organism (Table 1).
TABLE 1.
Virulence markers, serogroups, PFGE profiles, and phylogenetic backgrounds of STEC isolates from bovines and humans
| Origina | Presence of the following virulence marker:
|
Serogroup | PFGE profile | Phylogenetic background | |||
|---|---|---|---|---|---|---|---|
| stx1 | stx2 | stx1, stx2 | eae | ||||
| B1.1 | − | − | + | + | O111 | A | B1 |
| B1.2 | − | + | − | + | Poly9 | B | B1 |
| B2 | + | − | − | + | NTb | C | A |
| B3 | − | − | + | − | O76 | D | A |
| B4 | − | − | + | + | O28ac | E | B1 |
| B5 | − | − | + | + | O8 | F | A |
| B6 | − | − | + | + | O8 | F | B1 |
| B7 | − | − | + | + | O113 | G | A |
| B8 | − | − | + | − | O8 | F | B1 |
| B9 | + | − | − | + | O113 | H | B1 |
| B10 | + | − | − | − | NT | I | B1 |
| B11 | + | − | − | − | O142 | J | NT |
| B12 | + | − | − | + | O76 | D | B1 |
| B13 | + | − | − | − | NT | K | A |
| B14.1 | + | − | − | − | NT | L | A |
| B14.2 | + | − | − | + | O158 | M | A |
| B14.3 | − | − | + | + | O76 | N | D |
| B14.4 | + | − | − | + | O142 | O | B1 |
| B15.1 | + | − | − | + | O76 | P | A |
| B15.2 | + | − | − | + | O76 | Q | A |
| H1 | − | − | + | − | O74 | R | A |
| H2 | − | − | + | + | O76 | D | A |
| H3 | − | − | + | + | O113 | S | A |
| H4 | + | − | − | + | O113 | G | B1 |
| H5 | − | − | + | + | O78 | T | B1 |
| H6 | − | + | − | + | NT | U | A |
| H7 | − | + | − | − | O8 | V | A |
B, bovine; H, human. Two STEC isolates were collected from bovines B1 and B15, and four STEC isolates were collected from bovine B14.
NT, nontypeable.
FIG. 1.
Dendrogram showing the relatedness of human STEC strains and selected bovine STEC strains determined by macrorestriction of genomic DNA with XbaI.
All of the children (n = 7) were each infected with one strain of STEC (Table 1). The profile of the isolate from child H2 was 80% and 86% similar to profile D of the isolates from bovines B3 and B12, respectively, suggesting that infection may have been acquired from the animals or their environment. It is notable that individuals H2 and B3 resided on the same homestead. The profile of the STEC isolate from child H4 showed 88% similarity with the corresponding profile (profile G) of the isolate from bovine B7 (Table 1; Fig. 1). The profiles (profiles S and U) of the STEC isolates from children H3 and H6 showed 66% similarity to the profiles (profiles H and Q) of the STEC isolates from bovines B9 and B15.2, suggesting a possible relatedness between the human strains and the corresponding bovine isolate. The profiles of the STEC isolates from the three remaining children were unique.
PCR assays with primers specific for eae (14) detected this gene in 14 of 20 bovine STEC isolates and 5 of 7 human STEC isolates (Table 1). Although the proportion of intimin-positive strains from cattle varies with the time of sampling (10), the overall high frequency of eae-positive isolates (14 of 20 bovines) was unexpected, as previous studies have shown a much lower prevalence of eae-positive STEC isolates in healthy cattle (2, 4, 10, 12).
Phylogenetic analyses (6) showed that the bovine STEC strains separated into phylogenetic groups A (nine strains), B1 (nine strains), and D (one strain); one strain could not be classified (Table 1). A review of the phylogenetic distributions of the stx and eae genes showed that six of the nine group A strains (66.6%) possessed stx1 alone, and four of these strains were eae positive (Table 1). The three remaining group A strains possessed stx1 and stx2, and two of three strains were eae positive. Seven of the nine group B1 isolates (77.7%) were eae positive, and three of these strains possessed stx1 and one strain contained only stx2 (Table 1).
Similar analyses segregated the human STEC strains into group A (five strains) and group B1 (two strains). Three of the group A strains were eae positive; two of these strains contained both stx genes, and one strain was positive only for stx2 (Table 1). Both of the group B1 STEC strains were eae positive, and one of these strains possessed stx1 alone.
The presence of eae in STEC strains belonging to phylogenetic group A (six of nine strains), particularly in strains possessing only stx1 (four of six strains), was unexpected, as eae was characteristically absent in group A strains in the study of Girardeau et al. (9), suggesting that group A strains may be “nonvirulent.”
The STEC strains (Table 1) from the cattle and humans were serotyped at the Enteric Diseases Reference Unit, National Institute of Communicable Diseases, National Health Laboratory Service, Sandringham, Johannesburg, South Africa, into 10 serogroups. Five strains expressed nontypeable O antigens. Of the 22 typeable STEC strains, six were O76, while serogroups O113 and O8 accounted for four strains each. Four of the five bovine O76 STEC strains were eae positive, and PFGE indicated the heterogeneity of the strains within this serogroup (Table 1). The O76 STEC strains from bovines B3 and B12 were closely related to the STEC strain from one of the children (child H2). Surprisingly, the two bovine STEC O113 strains were eae positive, and one of these strains was closely related to the isolate from child H4. Strains belonging to this serogroup are prevalent in cattle (3, 12), and O113 strains have been implicated in sporadic cases of hemolytic-uremic syndrome (3, 7, 11, 13, 15); however, these STEC O113 strains have typically been eae negative (3, 7, 12, 13, 15). O158 (eae positive), a previously unreported STEC serogroup, was isolated from one of the bovines infected with multiple strains of STEC.
To the best of our knowledge, this is the first study on the occurrence of STEC in bovines in a seminomadic pastoralist community in Uganda. Furthermore, on the basis of the findings of this study, we conclude that there is probable transmission of STEC strains from cattle or their environment to children living in the community.
Acknowledgments
Samuel Majalija and this study were funded by the Rockefeller and Carnegie Foundations through the University Science, Humanities, and Engineering Partnerships in Africa (USHEPiA) program. S.M. is grateful for the financial assistance provided by the University of Makerere during the course of his studies.
Footnotes
Published ahead of print on 7 November 2007.
REFERENCES
- 1.Blanco, M., J. E. Blanco, A. Mora, J. Rey, J. M. Alonso, M. Hermoso, J. Hormoso, M. P. Alfonso, G. Dhabi, E. A. González, M. I. Bernárdez, and J. Blanco. 2003. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J. Clin. Microbiol. 411351-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco, M., J. E. Blanco, J. Blanco, E. A. Gonzalez, A. Mora, C. Prado, L. Fernandez, M. Rio, J. Ramos, and M. P. Alfonso. 1996. Prevalence and characteristics of Escherichia coli serotype O157:H7 and other verotoxin-producing E. coli in healthy cattle. Epidemiol. Infect. 117251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett, K. N., M. A. Hornitzky, K. A. Bettelheim, M. J. Walker, and S. P. Djordjevic. 2003. Bovine non-O157 Shiga 2-containing Escherichia coli isolates commonly posses stx2-EDL933 and/or stx2vhb subtypes. J. Clin. Microbiol. 412716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caprioli, A., S. Morabito, H. Brugère, and E. Oswald. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36289-311. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1998. One-day (24-28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, non-typhoidal Salmonella serotypes, and Shigella sonnei by pulsed field gel electrophoresis (PFGE). PulseNet document (http://www.cdc.gov/pulsenet/protocols/ecoli_salmonella_shigella_protocols.pdf). Centers for Disease Control and Prevention, Atlanta, GA.
- 6.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 664555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott, E. J., R. M. Robins-Browne, E. V. O'Loughlin, V. Bennett-Wood, J. Bourke, P. Henning, G. G. Hogg, J. Knight, H. Powell, and D. Redmond. 2001. Nationwide study of haemolytic uraemic syndrome: clinical, microbiological, and epidemiological features. Arch. Dis. Child. 85125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 352977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girardeau, J. P., A. Dalmasso, Y. Bertin, C. Ducrot, S. Bord, V. Livrelli, C. Vernozy-Rozand, and C. Martin. 2005. Association of virulence genotype with phylogenetic background in comparison to different seropathotypes of Shiga toxin-producing Escherichia coli isolates. J. Clin. Microbiol. 436098-6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenkins, C., M. C. Pearce, H. Chart, T. Cheasty, G. A. Willshaw, G. J. Gunn, G. Dougan, H. R. Smith, B. A. Synge, and G. Frankel. 2002. An eight-month study of a population of verocytotoxigenic Escherichia coli (VTEC) in a Scottish cattle herd. J. Appl. Microbiol. 93944-953. [DOI] [PubMed] [Google Scholar]
- 11.Karch, H., P. I. Tarr, and M. Bielaszewska. 2005. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295405-418. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi, H., A. Miura, H. Hayashi, T. Ogawa, T. Endô, E. Hata, M. Eguchi, and K. Yamamoto. 2003. Prevalence and characteristics of eae-positive Escherichia coli from healthy cattle in Japan. Appl. Environ. Microbiol. 695690-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mellmann, A., M. Bielaszewska, L. B. Zimmerhackl, R. Prager, D. Harmsen, H. Tschäpe, and H. Karch. 2005. Enterohemorrhagic Escherichia coli in human infection: in vivo evolution of a bacterial pathogen. Clin. Infect. Dis. 41785-792. [DOI] [PubMed] [Google Scholar]
- 14.Mora, A., M. Blanco, J. E. Blanco, G. Dahbi, C. López, P. Justel, M. P. Alonso, A. Echeita, M. I. Bernárdez, E. A. González, and J. Blanco. 2007. Serotypes, virulence genes and intimin types of Shiga toxin (verocytotoxin)-producing Escherichia coli isolates from minced beef in Lugo (Spain) from 1995 through 2003. BMC Microbiol. 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 373357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]