Abstract
Taeniasis due to Taenia solium is a disease with important public health consequences, since the larval stage is not exclusive to the animal intermediate, the pig, but also infects humans, causing neurocysticercosis. Early diagnosis and treatment of T. solium tapeworm carriers is important to prevent human cysticercosis. Current diagnosis based on microscopic observation of eggs lacks both sensitivity and specificity. In the present study, a nested-PCR assay targeting the Tso31 gene was developed for the specific diagnosis of taeniasis due to T. solium. Initial specificity and sensitivity testing was performed using stored known T. solium-positive and -negative samples. The assay was further analyzed under field conditions by conducting a case-control study of pretreatment stool samples collected from a population in an area of endemicity. Using the archived samples, the assay showed 97% (31/32) sensitivity and 100% (123/123) specificity. Under field conditions, the assay had 100% sensitivity and specificity using microscopy/enzyme-linked immunosorbent assay coproantigen testing as the gold standards. The Tso31 nested PCR described here might be a useful tool for the early diagnosis and prevention of taeniasis/cysticercosis.
Human taeniasis/cysticercosis is an important public health problem, particularly in developing countries. Taeniasis is a parasitic disease of the human intestinal tract caused by the adult stage of the closely related tapeworms Taenia solium, Taenia saginata, and Taenia saginata asiatica. The disease is a consequence of ingesting cysts present in raw or undercooked infected meat. After ingestion, the cyst reaches the human intestine, where it develops into an adult tapeworm, releasing proglottids filled with eggs that are passed in the stools (7). Ingestion of these eggs by the intermediate hosts results in the development of cysts in soft tissues, a disease known as cysticercosis. Cysticercosis affects swine (T. solium) and cattle (T. saginata), causing economic loss (1, 12). Unlike T. saginata eggs, T. solium eggs are capable not only of infecting the animal intermediate host, but also humans, causing human cysticercosis (10, 11). In humans, the larval stage often localizes in the central nervous system, causing a clinical disorder known as neurocysticercosis (8, 9), a major cause of seizures and epilepsy in most of the developing world. Human cysticercosis is highly endemic in Latin America, Asia, and Africa, especially in those countries where domestic pig husbandry is practiced. It is also increasing in industrialized countries due to immigration of tapeworm carriers (9, 30).
The human T. solium carrier is the sole source of cysticercosis infections in pigs and neurocysticercosis in humans. The early identification of taeniasis due to T. solium is of great importance due to its epidemiological implications. Currently, the diagnosis of taeniasis is based on the detection of eggs by microscopic observation of fecal samples. This technique lacks both sensitivity and specificity, since eggs of most members of the family Taenidae are morphologically indistinguishable and are shed intermittently. Detection of T. solium coproantigen by the enzyme-linked immunosorbent assay (ELISA) technique (2, 3, 5) is also used. The method is more sensitive than microscopy but cross-reacts with T. saginata. Differentiation of T. solium and T. saginata is based on the morphological characteristics of the scolex or gravid proglottids (21). Recovery of scolices after treatment is unusual for T. solium, and in many cases, both the scolex and proglottids can be recovered only after special treatment (18).
DNA-based differentiation of T. solium and T. saginata has been described and includes the use of probes (4, 6, 15), PCR with species-specific primers (13, 14, 31), and PCR followed by restriction enzyme analysis (PCR-REA) (21, 26, 32). However, these methods require pure parasite DNA, which means that the DNA has to be extracted from a single proglottid and adequately cleaned, because these primers may amplify any eukaryotic DNA, causing cross-amplification. A few reports have described the use of DNA-based techniques to differentiate T. solium from T. saginata from fecal samples (23, 24, 31), but they still lack sensitivity.
The goal of this study was to develop a rapid and sensitive PCR-based technique for the specific detection and diagnosis of taeniasis due to T. solium directly from fecal samples.
MATERIALS AND METHODS
Parasitic material from infected animals.
T. solium cysticerci were dissected from naturally infected pigs, washed twice with 0.01 M Tris-HCl (pH 8.0), and stored at −70°C until they were needed. Immature tapeworms were obtained from hamsters that were infected under laboratory conditions with one to five T. solium cysticerci (20). Echinococcus granulosus scolices were obtained from hydatid cysts excised from naturally infected animals. DNA from Entamoeba hystolitica was kindly provided by W. A. Petri, Jr. (University of Virginia). DNA from Fasciola hepatica was provided by P. Herrera (Universidad Peruana Cayetano Heredia).
Tapeworms from infected individuals.
Taenia tapeworms were obtained from naturally infected patients after informed consent and treatment. After recovery, proglottids were identified as T. solium or T. saginata by PCR-REA as described previously (21). Recovered parasite material was identified by examination of the scolex (when recovered), uterine lateral-branch counting (histology), and PCR-REA. A total of 25 T. solium proglottids and 17 T. saginata proglottids obtained from different patients were used to test the specificity of the PCR assay.
Proglottids of Diphyllobothrium sp. (n = 2), Hymenolepis diminuta (n = 1), and Hymenolepis nana (n = 1) tapeworms were recovered from infected patients after informed consent and treatment. The specimens were identified by PCR-REA as described previously (21).
Positive stool samples.
All known positive samples were obtained from previous field studies (18). Taeniasis-positive patients identified upon microscopic examination were given standard medical treatment as indicated (18). Informed consent was approved by the ethical committee of the Johns Hopkins School of Public Health and the Universidad Peruana Cayetano Heredia. Stool samples were collected following treatment and preserved by diluting 1 volume of fecal sample with 2 volumes of 2.5% (wt/vol) potassium dichromate. Samples diluted in potassium dichromate were kept at room temperature or 4°C until used. A total of 35 fecal samples collected from patients with taeniasis were analyzed in the present study; 32 patients were identified as T. solium carriers and 3 as T. saginata carriers.
Stool samples positive for H. nana (n = 10), Diphyllobothrium sp. (n = 5), Strongyloides stercoralis (n = 3), and hookworm species (n = 2) were also tested by Tso31 nested PCR. All stool samples were positive for their respective parasites by microscopy and were preserved in potassium dichromate.
Testing of all previously identified positive samples was performed randomly in a blinded fashion, along with testing of 100 stool samples that were negative for taeniasis by microscopy. The negative samples were obtained in previous studies from a shantytown in Lima, Peru, where taeniasis is nonendemic. They included samples positive for parasites, such as Ascaris lumbricoides (n = 1), H. nana (n = 4), Giardia lamblia (n = 33), Cryptosporidium sp. (n = 1), Cyclospora cayetanensis (n = 2), Blastocystis hominis (n = 1), Chilomastix mesnili (n = 12), Iodamoeba butschlii (n = 2), Entamoeba coli (n = 29), and Endolimax nana (n = 22).
DNA extraction from stool samples.
One milliliter of potassium dichromate-preserved stool sample was washed three times with reagent grade water by centrifugation at 18,300 × g (14,000 rpm) for 10 min. The pellet was then extracted using the FastDNA spin kit for soil DNA extraction protocol (25).
Primers.
Primers were designed based on the recently published gene sequence that encodes the T. solium oncosphere-specific protein Tso31 (GenBank accession no. DQ861410). The outer PCR was performed using the primer pair F1 (5′ ATG ACG GCG GTG CGG AAT TCT G 3′) and R1 (5′ TCG TGT ATT TGT CGT GCG GGT CTA C 3′) and was predicted to amplify a 691-bp segment. The second PCR (nested PCR) was performed using primers F589 (5′ GGT GTC CAA CTC ATT ATA CGC TGT G 3′) and R294 (5′ GCA CTA ATG CTA GGC GTC CAG AG 3′) and was predicted to amplify a 234-bp DNA fragment.
PCR conditions.
The master mixture for the outer PCR was composed of 1× PCR buffer (Applied Biosystems, Foster City, CA), 3 mM of MgCl2 (Applied Biosystems), 200 μM of each deoxynucleoside triphosphate (Invitrogen, Carlsbad, CA), 0.2 μg/μl of bovine serum albumin, 0.8 μM of each primer, 0.125 U of Taq polymerase (Invitrogen), and 2.0 μl of extracted DNA in a total volume of 25 μl. The nested PCR was carried out in a PCR mixture similar to that used for the outer PCR, except that the final concentration of MgCl2 was 2.5 mM and 1 microliter of the first PCR amplification was used as a DNA template.
PCR amplification was carried out in a PTC200-MJ research cycler (Bio-Rad, Hercules, CA). The outer PCR amplification consisted of an initial denaturation step at 95°C for 3 min, followed by 25 amplification cycles, each consisting of a denaturation step at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. The second PCR was carried out similarly to the outer PCR, except that 40 cycles were performed, with an annealing step at 60°C for 30 s. The amplification products were electrophoresed using 2% agarose gels. The gels were stained with ethidium bromide (10 μg/ml) for 10 min. The stained gels were then visualized under UV light and documented.
Analytical sensitivity of the Tso31 nested PCR.
A fecal sample, determined to be negative both by microscopy and a coproantigen test, was used to assess the detection limit of the Tso31 nested PCR. The sample was distributed in 250-mg aliquots, and each aliquot was contaminated in duplicate with 1, 2, 5, 10, 40, 50, or 100 T. solium eggs. The aliquots containing eggs were processed for DNA extraction as described above for stool samples.
To determine the pure-DNA detection limit of the Tso31 nested PCR, DNA extracted from T. solium cysts was quantitated using a spectrophotometer and diluted in PCR grade water in 10-fold dilutions. The diluted DNA was then amplified as described above.
Field testing.
To further test the Tso31 nested-PCR assay and to confirm the results of the PCR on archived specimens, we conducted a case-control study using samples obtained before treatment from a population in an area of endemicity in Puno, Peru. Out of 982 stool samples collected, 11 (1.1%) were positive by both ELISA-coproantigen testing (2) and microscopy. The tapeworm-infected patients were given standard treatment, and posttreatment stools were collected for confirmation. The 11 positive samples were considered cases. Four negative controls per case were randomly selected among the samples that tested negative for taeniasis both by ELISA-coproantigen testing and microscopy (44 out of 971). Specimens were coded so that the PCR was performed in a blinded fashion. All samples had DNA extracted and were tested by the Tso31 nested PCR as described above.
RESULTS
PCR from DNA extracted from parasites.
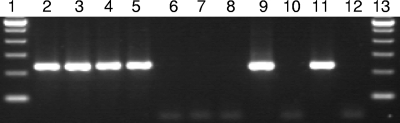
All DNA samples extracted from T. solium proglottids, cysts, and tapeworms from hamsters after Tso31 nested PCR gave a single amplification product of 234 bp (Fig. 1). All 25 DNA samples extracted from T. solium proglottids were positive by Tso31 nested PCR. None of the 17 DNA samples extracted from T. saginata proglottids amplified by Tso31 nested PCR. The Tso31 nested PCR produced no amplification when DNA obtained from E. granulosus scolices or proglottids of H. nana (n = 1), H. diminuta (n = 1), or Diphyllobothrium sp. (n = 2) was used as the target DNA. E. hystolitica and F. hepatica DNAs did not produce any amplification when assayed by the Tso31 nested PCR.
FIG. 1.
Tso31 nested-PCR amplification using DNA extracted from different sources. Electrophoresis was performed using 5 μl of amplification product. Lanes 1 and 13, 100-bp ladders; lanes 2 to 8, DNA from a contaminated sample with 100, 50, 20, 10, 5, 2, and 1 T. solium eggs, respectively; lane 9, DNA from a T. solium proglottid; lane 10, DNA from a T. saginata proglottid; lane 11, DNA from a T. solium-positive stool sample; lane 12, DNA from a T. saginata-positive stool sample.
Tso31 nested PCR with defined samples under laboratory conditions.
A total of 31 fecal samples out of the 32 known to be positive for T. solium were amplified using the Tso31 nested PCR described above. The only stool sample negative by the Tso31 nested PCR did not amplify even when dilutions were made to remove inhibitors or when DNA was extracted again from the original sample. However, the DNA extracted from the tapeworm recovered from this patient gave a positive amplification.
There was no amplification using DNA samples extracted from the three T. saginata carriers. No amplification products were observed when 100 taeniasis-negative stool samples obtained from subjects living in Lima, a region of nonendemicity, were tested by Tso31 nested PCR. Similarly, none of the stool samples were positive by Tso31 nested PCR for T. solium when tested with specimens positive for Diphyllobothrium sp. (n = 10), H. nana (n = 5), S. stercolaris (n = 3), or hookworm (n = 2). Also, no amplification was observed when DNA extracted from stool samples positive for G. lamblia, Cryptosporidium sp., C. cayetanensis, B. hominis, C. mesnili, I. butschlii, E. coli, or E. nana was used. Using T. solium-positive samples that were previously identified after treatment (archived stool samples), the sensitivity and specificity of the Tso31 nested PCR were 97% and 100%, respectively (Table 1).
TABLE 1.
Results of the Tso31 nested-PCR assay on archived stool samples from T. solium-positive and -negative patients
| Tso31 nested PCR result |
T. solium taeniasisa
|
% Predictive value
|
||
|---|---|---|---|---|
| No. positive | No. negative | Positive | Negative | |
| Positive | 31 | 0 | 100.0 | |
| Negative | 1 | 123 | 99.2 | |
Negative samples included 100 from a population in an area of nonendemicity, 3 positive for T. saginata, 10 positive for H. nana, and 5 positive for Diphyllobothrium. Sensitivity and specificity were 96.9% for and 100.0%, respectively.
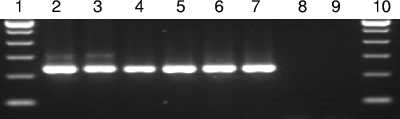
The lower detection limit of the Tso31 nested PCR in a fecal sample was 10 eggs per 250 mg of stool sample (40 eggs/g stool sample) using the Fast DNA spin kit for soil DNA extraction (Fig. 1). The detection limit using purified DNA was 100 femtograms (Fig. 2).
FIG. 2.
Analytical sensitivity of the Tso31 nested PCR. Lanes 1 and 10, 100-bp ladders. DNA extracted from T. solium cysts was spectrophotometrically quantitated and diluted. PCR was carried out using different DNA concentrations as follows; lanes 2 to 9, 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg, respectively.
Tso31 nested PCR with samples from a field epidemiological study.
Out of the 55 samples (11 cases and 44 controls) analyzed in the case-control study, 10 of the initial 11 cases gave a positive amplification when tested by the Tso31 nested-PCR assay. Tapeworms recovered after treatment of the 11 cases were identified at the species level by PCR-REA. One tapeworm recovered from a taeniasis-positive patient by microscopy and ELISA-coproantigen testing (2, 3) was further identified as T. saginata by PCR-REA. The stool sample obtained from this patient consistently did not give any amplification product in the Tso31 nested PCR. Ten of the recovered tapeworms were identified as T. solium, and all of these samples were positive when tested by the Tso31 nested-PCR assay. Thus, under field conditions, both the specificity and the sensitivity of the Tso31 nested PCR were 100%. Although samples tested in the case-control study were positive by microscopy for other parasites and multiple infections were common (29 were infected with more than two parasites), the Tso31 nested PCR had no cross-reactions with any of these parasites. Among the control samples, four were infected with A. lumbricoides, two with Enterobius vermicularis, and four with H. nana.
DISCUSSION
Specific identification of T. solium is important because of the clinical and epidemiological consequences for public health. Patients with T. solium tapeworms have a risk of developing cysticercosis, while those with T. saginata do not. Several DNA-based assays have recently been developed to differentiate T. solium from T. saginata; however, most of them rely on the use of pure parasite DNA. Parasite DNA, however, can only be obtained from proglottids recovered after treatment of tapeworm carriers, and elimination of proglottids after treatment is often erratic.
Other researchers have developed and used PCR-based assays for the detection of T. solium. The PCR-restriction fragment length polymorphism described by Nunes et al. (23) has a detection limit of 17 eggs/g of stool sample, but it is not highly sensitive (66.6%). The multiplex PCR based on the HDP2 sequence (13, 24) has been reported to have a DNA detection level equivalent to 2 eggs, but it has not been tested with fecal specimens; furthermore, when fecal samples were artificially inoculated with known numbers of eggs, the detection limit was of 4,375 T. saginata eggs (35,000 pg of DNA) (23). The multiplex PCR targeting the mitochondrial cox1 gene provides reliable results with more than 50 eggs/g feces, but it was only 48% sensitive when tested with 23 positive samples (31). Although nested PCR is a two-step procedure, it seems to be the best way to overcome the challenge of amplifying DNA extracted from stool samples. We tested a battery of positive samples using the primers of the Tso31 nested PCR in a simple PCR format. The results of these tests were unreliable, and weak amplifications and poor sensitivity were observed.
Reports on the specific detection of T. solium or differentiation of T. solium from T. saginata directly from stool samples are scarce, probably because of the difficulties encountered in extracting DNA from parasite eggs and the presence of PCR inhibitors, which affects the sensitivity of PCR assays. Releasing oncospheres from eggs is a difficult process that requires the use of extreme and often detrimental procedures, such as treatment with bleach. The use of such procedures usually results in damage to the DNA and absence of PCR amplification. The Fast DNA spin kit has been successfully used for DNA extraction from cryptosporidium cysts and bacteria present in human fecal samples (19, 22). Extraction of DNA from cryptosporidium cysts presents challenges similar to those encountered with DNA extraction from taenia eggs. The high sensitivity achieved by this study suggests that the Fast DNA spin kit for soil is a method that overcomes both the difficulty of obtaining DNA from taenia eggs and the presence of inhibitors in stool samples. It is also probable that the Fast DNA extraction extracts DNA not only from eggs, but also from the tapeworm tegument cells, since we were able to amplify DNA extracted from stool samples from hamsters that had infertile T. solium tapeworms (data not shown).
We tested a large collection of samples from both tapeworm carriers and noncarriers that were stored with no special storage conditions other than the suspension of the specimens in 2% potassium dichromate. Some samples had been stored for up to 3 years under room temperature conditions without apparent adverse effects on the performance of the Tso31 nested PCR. Potassium dichromate is widely used for preservation and viability studies of different parasites present in fecal specimens (17, 27, 28, 29); it has also been reported to be used for DNA recovery from cryptosporidium parasites (16).
The Tso31 nested-PCR test amplifies only DNA from T. solium, and it does not cross-react with DNA extracted from other parasites or from stool samples positive for other parasites. However, the assay has not been performed using DNA obtained from T. saginata asiatica or samples positive for that parasite. It is probable that no cross-reaction will occur, since the parasite is a T. saginata subspecies.
The Tso31 nested PCR developed here for the detection of T. solium in stool samples is highly sensitive and specific. The test is reliable under conditions of endemicity for taeniasis, and it constitutes a powerful tool for the early diagnosis of taeniasis and is a potential tool for the control of human taeniasis/cysticercosis.
Acknowledgments
This study was supported by Bill and Melinda Gates Foundation grant no. 23981, NIH grant no. P01 AI51976, Global Research Training grant no. D43 TW006581, and NIH BRAVO/MIRT grant no. 5 T37 MD001427.
We express our gratitude to Cesar Jeri for his contribution to the present study. We also appreciate the technical assistance of M.-C. Camila, J. B. Phu, and D. Sara.
Footnotes
Published ahead of print on 7 November 2007.
REFERENCES
- 1.Abuseir, S., C. Epe, T. Schnieder, G. Klein, and M. Kuhne. 2006. Visual diagnosis of Taenia saginata cysticercosis during meat inspection: is it unequivocal? Parasitol. Res. 99405-409. [DOI] [PubMed] [Google Scholar]
- 2.Allan, J. C., G. Avila, J. Garcia Noval, A. Flisser, and P. S. Craig. 1990. Immunodiagnosis of taeniasis by coproantigen detection. Parasitology 101473-477. [DOI] [PubMed] [Google Scholar]
- 3.Allan, J. C., P. P. Wilkins, V. C. Tsang, and P. S. Craig. 2003. Immunodiagnostic tools for taeniasis. Acta Trop. 8787-93. [DOI] [PubMed] [Google Scholar]
- 4.Chapman, A., V. Vallejo, K. G. Mossie, D. Ortiz, N. Agabian, and A. Flisser. 1995. Isolation and characterization of species-specific DNA probes from Taenia solium and Taenia saginata and their use in an egg detection assay. J. Clin. Microbiol. 331283-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deplazes, P., J. Eckert, Z. S. Pawlowski, L. Machowska, and B. Gottstein. 1991. An enzyme-linked immunosorbent assay for diagnostic detection of Taenia saginata copro-antigens in humans. Trans. R. Soc. Trop. Med. Hyg. 85391-396. [DOI] [PubMed] [Google Scholar]
- 6.Flisser, A. 1994. Taeniasis and cysticercosis due to Taenia solium. Prog. Clin. Parasitol. 477-116. [PubMed] [Google Scholar]
- 7.Flisser, A., A. Reid, E. Garcia-Zepeda, and D. P. McManus. 1988. Specific detection of Taenia saginata eggs by DNA hybridisation. Lancet ii1429-1430. [DOI] [PubMed] [Google Scholar]
- 8.Garcia, H. H., O. H. Del Brutto, T. E. Nash, A. C. White, Jr., V. C. Tsang, and R. H. Gilman. 2005. New concepts in the diagnosis and management of neurocysticercosis (Taenia solium). Am. J. Trop. Med. Hyg. 723-9. [PubMed] [Google Scholar]
- 9.Garcia, H. H., O. H. Del Brutto, and the Cysticercosis Working Group in Peru. 2005. Neurocysticercosis: updated concepts about an old disease. Lancet Neurol. 4653-661. [DOI] [PubMed] [Google Scholar]
- 10.Garcia, H. H., A. E. Gonzalez, C. A. Evans, R. H. Gilman, and the Cysticercosis Working Group in Peru. 2003. Taenia solium cysticercosis. Lancet 362547-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia, H. H., A. E. Gonzalez, R. H. Gilman, and the Cysticercosis Working Group in Peru. 2003. Diagnosis, treatment and control of Taenia solium cysticercosis. Curr. Opin. Infect. Dis. 16411-419. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, A. E., H. H. Garcia, R. H. Gilman, V. C. Tsang, and the Cysticercosis Working Group in Peru. 2003. Control of Taenia solium. Acta Trop. 87103-109. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez, L. M., E. Montero, L. J. Harrison, R. M. Parkhouse, and T. Garate. 2000. Differential diagnosis of Taenia saginata and Taenia solium infection by PCR. J. Clin. Microbiol. 38737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez, L. M., E. Montero, E. Sciutto, L. J. Harrison, R. M. Parkhouse, and T. Garate. 2002. Differential diagnosis of Taenia saginata and Taenia solium infections: from DNA probes to polymerase chain reaction. Trans. R. Soc. Trop. Med. Hyg. 96S243-S250. [DOI] [PubMed] [Google Scholar]
- 15.Harrison, L. J., J. Delgado, and R. M. Parkhouse. 1990. Differential diagnosis of Taenia saginata and Taenia solium with DNA probes. Parasitology 100459-461. [DOI] [PubMed] [Google Scholar]
- 16.Higgins, J. A., R. Fayer, J. M. Trout, L. Xiao, A. A. Lal, S. Kerby, and M. C. Jenkins. 2001. Real-time PCR for the detection of Cryptosporidium parvum. J. Microbiol. Methods 47323-337. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, M., S. Uga, T. Oda, S. K. Rai, G. Vesey, and H. Hotta. 2006. Changes of physical and biochemical properties of Cryptosporidium oocysts with various storage conditions. Water Res. 40881-886. [DOI] [PubMed] [Google Scholar]
- 18.Jeri, C., R. H. Gilman, A. G. Lescano, H. Mayta, M. E. Ramirez, A. E. Gonzalez, R. Nazerali, and H. H. Garcia. 2004. Species identification after treatment for human taeniasis. Lancet 363949-950. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, J., K. A. Alderisio, A. Singh, and L. Xiao. 2005. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl. Environ Microbiol. 711135-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letonja, T. 1975. The hamster (Mesocricetus auratus) as an experimental definitive host of Taenia solium. Bol. Chil. Parasitol. 3032-33. [PubMed] [Google Scholar]
- 21.Mayta, H., A. Talley, R. H. Gilman, J. Jiménez, M. Verastegui, M. Ruiz, H. H. Garcia, and A. E. Gonzalez. 2000. Differentiating Taenia solium and Taenia saginata infections by simple hematoxylin-eosin staining and PCR-restriction enzyme analysis. J. Clin. Microbiol. 38133-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McOrist, A. L., M. Jackson, and A. R. Bird. 2002. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J. Microbiol. Methods 50131-139. [DOI] [PubMed] [Google Scholar]
- 23.Nunes, C. M., A. K. Dias, F. E. Dias, S. M. Aoki, H. B. de Paula, L. G. Lima, and J. F. Garcia. 2005. Taenia saginata: differential diagnosis of human taeniasis by polymerase chain reaction-restriction fragment length polymorphism assay. Exp. Parasitol. 110412-415. [DOI] [PubMed] [Google Scholar]
- 24.Nunes, C. M., L. G. Lima, C. S. Manoel, R. N. Pereira, M. M. Nakano, and J. F. Garcia. 2003. Taenia saginata: polymerase chain reaction for taeniasis diagnosis in human fecal samples. Exp. Parasitol. 10467-69. [DOI] [PubMed] [Google Scholar]
- 25.Qbiogene. 2006. FastDNA SPIN kit for soil: application manual. Qbiogene, Irvine, CA.
- 26.Rodriguez-Hidalgo, R., D. Geysen, W. Benitez-Ortiz, S. Geerts, and J. Brandt. 2002. Comparison of conventional techniques to differentiate between Taenia solium and Taenia saginata and an improved polymerase chain reaction-restriction fragment length polymorphism assay using a mitochondrial 12S rDNA fragment. J. Parasitol. 881007-1011. [DOI] [PubMed] [Google Scholar]
- 27.Sathyanarayanan, L., and Y. Ortega. 2006. Effects of temperature and different food matrices on Cyclospora cayetanensis oocyst sporulation. J. Parasitol. 92218-222. [DOI] [PubMed] [Google Scholar]
- 28.Verastegui, M., R. H. Gilman, H. H. Garcia, A. E. Gonzalez, Y. Arana, C. Jeri, I. Tuero, C. M. Gavidia, M. Levine, V. C. Tsang, and the Cysticercosis Working Group in Peru. 2003. Prevalence of antibodies to unique Taenia solium oncosphere antigens in taeniasis and human and porcine cysticercosis. Am. J. Trop. Med. Hyg. 69438-444. [PubMed] [Google Scholar]
- 29.Waldenstedt, L., K. Elwinger, A. Lunden, P. Thebo, and A. Uggla. 2001. Sporulation of Eimeria maxima oocysts in litter with different moisture contents. Poult. Sci. 801412-1415. [DOI] [PubMed] [Google Scholar]
- 30.Willingham, A. L., III, and D. Engels. 2006. Control of Taenia solium cysticercosis/taeniosis. Adv. Parasitol. 61509-566. [DOI] [PubMed] [Google Scholar]
- 31.Yamasaki, H., J. C. Allan, M. O. Sato, M. Nakao, Y. Sako, K. Nakaya, D. Qiu, W. Mamuti, P. S. Craig, and A. Ito. 2004. DNA differential diagnosis of taeniasis and cysticercosis by multiplex PCR. J. Clin. Microbiol. 42548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamasaki, H., M. Nakao, Y. Sako, K. Nakaya, M. O. Sato, and A. Ito. 2006. Mitochondrial DNA diagnosis for taeniasis and cysticercosis. Parasitol. Int. 55S81-S85. [DOI] [PubMed] [Google Scholar]