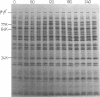
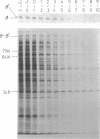
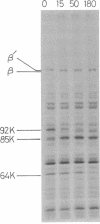
Abstract
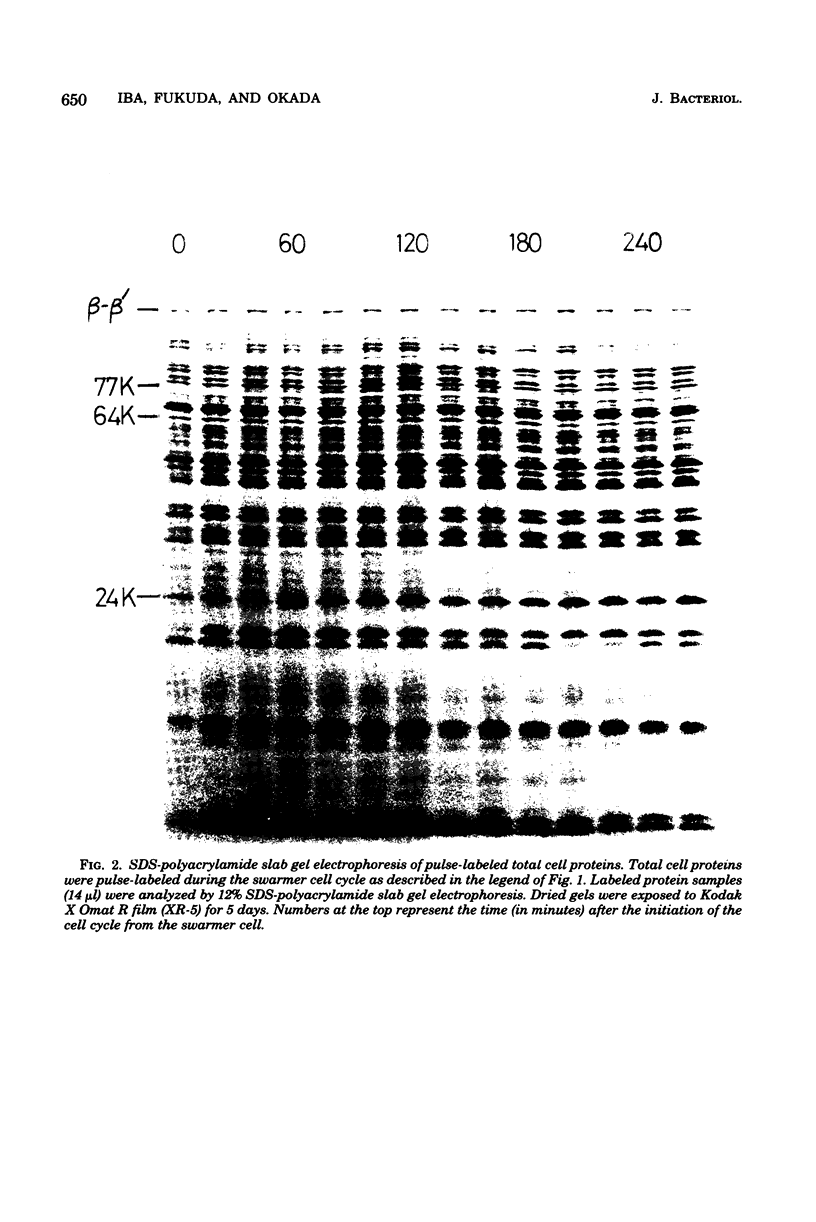
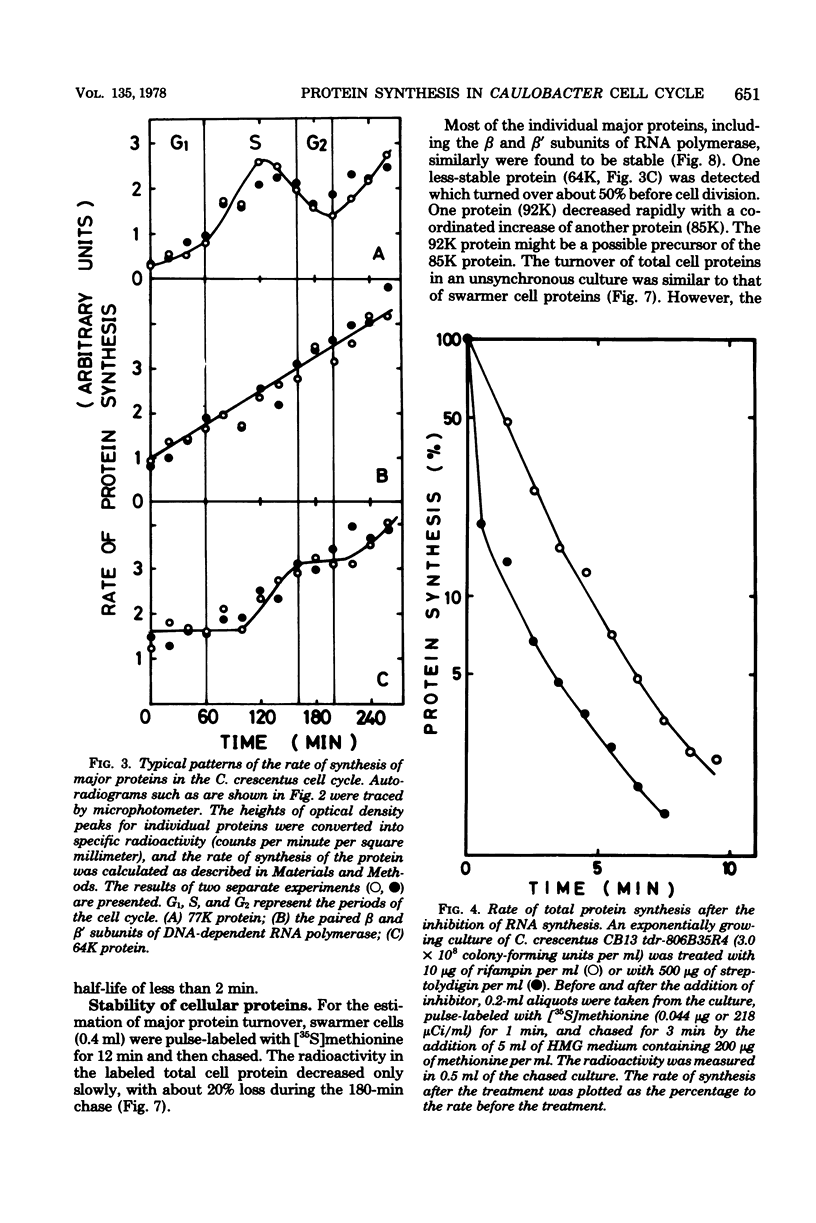
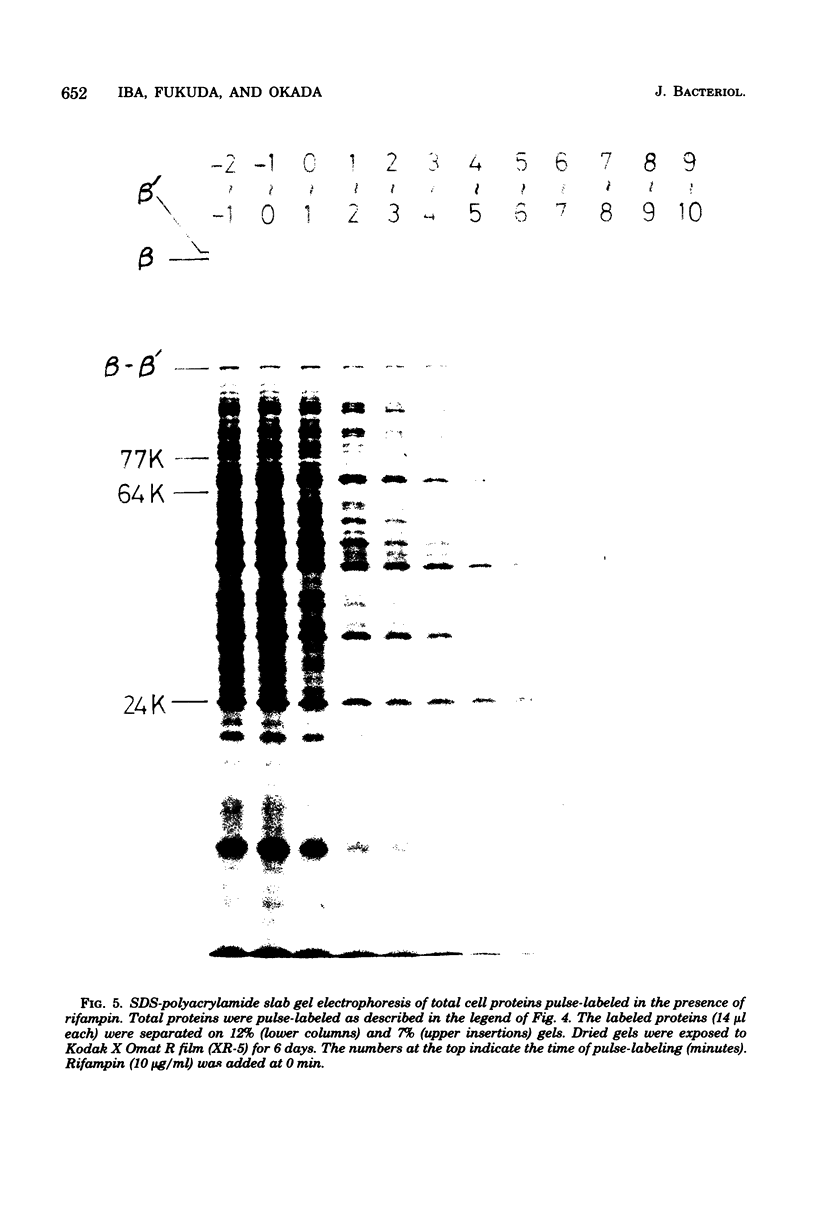
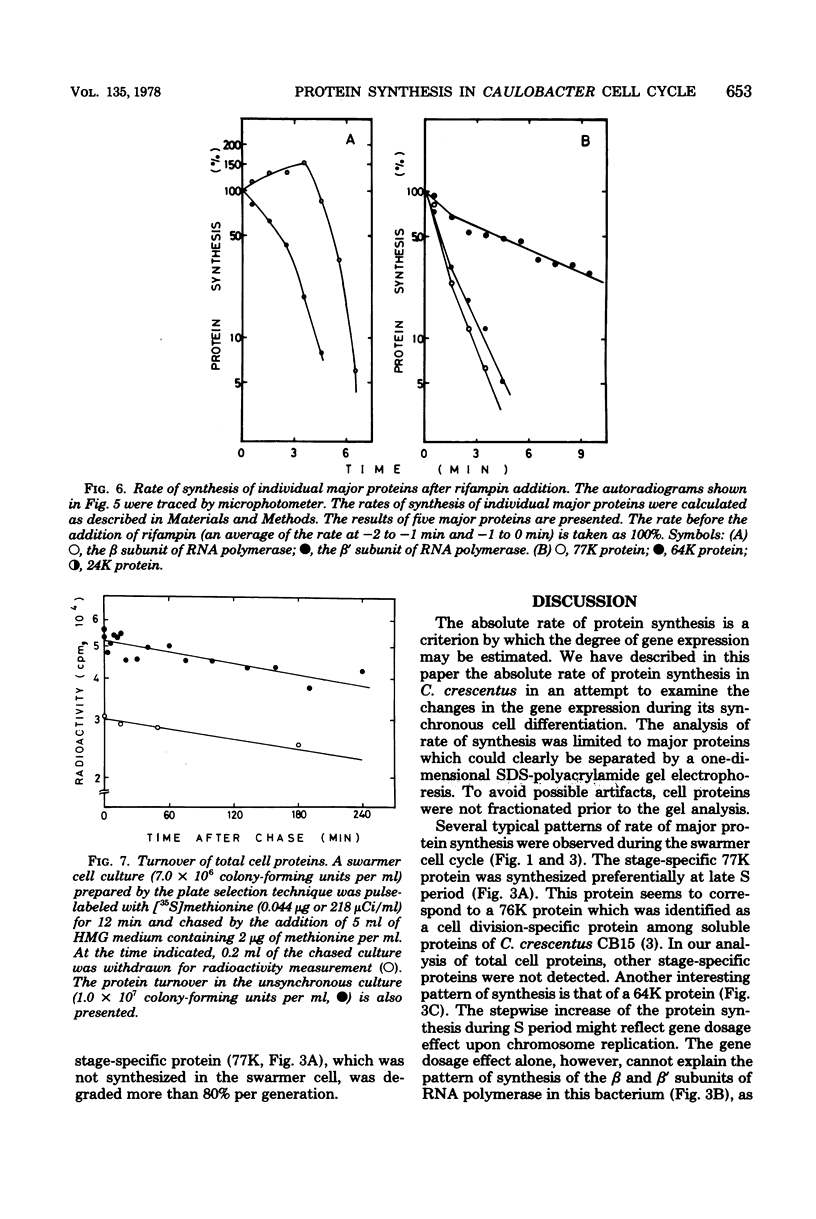
The rate of major protein synthesis was examined during the synchronous differentiation of Caulobacter crescentus. Total cell proteins were pulse-labeled with [35S]methionine at different times in the swarmer cell cycle and analyzed by sodium dodecyl sulfate- polyacrylamide gel electrophoresis. The rates of synthesis of total cell proteins and of about one-half of the individual major proteins examined increased through G1 and S periods but remained nearly constant during G2 period. The rates of synthesis of the other half of the individual major proteins either increased continuously throughout the swarmer cell cycle or doubled during S period. One stage-specific protein was also detected in late S period. For most of the major proteins examined, the rate of synthesis in the swarmer cell was less than that in the stalked cell. It seemed that, before the onset of G2 period, the Caulobacter cell was already able to synthesize each major protein at the additive rate of the two progeny cells. Compared to the stability of cellular proteins, the functional degradation rate of mRNA coding for individual major proteins was rapid, with half-lives of 0.4 to 5.8 min. It thus seems that the rate of major protein synthesis mainly reflects the transcriptional control of gene expression.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnsley P. G., Sells B. H. Functional inactivation rates of the messenger RNA molecules coding for the individual ribosomal proteins in Escherichia coli. Mol Gen Genet. 1977 Jun 8;153(2):121–127. doi: 10.1007/BF00264726. [DOI] [PubMed] [Google Scholar]
- Bendis I. K., Shapiro L. Deoxyribonucleic acid-dependent ribonucleic acid polymerase of Caulobacter crescentus. J Bacteriol. 1973 Sep;115(3):848–857. doi: 10.1128/jb.115.3.848-857.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K. K., Newton A. Patterns of protein synthesis during development in Caulobacter crescentus. Dev Biol. 1977 Apr;56(2):417–425. doi: 10.1016/0012-1606(77)90281-0. [DOI] [PubMed] [Google Scholar]
- Degnen S. T., Newton A. Chromosome replication during development in Caulobacter crescentus. J Mol Biol. 1972 Mar 14;64(3):671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- Heil A., Zillig W. Reconstitution of bacterial DNA-dependent RNA-polymerase from isolated subunits as a tool for the elucidation of the role of the subunits in transcription. FEBS Lett. 1970 Dec;11(3):165–168. doi: 10.1016/0014-5793(70)80519-1. [DOI] [PubMed] [Google Scholar]
- Iba H., Fukuda A., Okada Y. Chromosome replication in Caulobacter crescentus growing in a nutrient broth. J Bacteriol. 1977 Mar;129(3):1192–1197. doi: 10.1128/jb.129.3.1192-1197.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba H., Fukuda A., Okada Y. Gamma-ray sensitivity during synchronous cell differentiation in Caulobacter crescentus. J Bacteriol. 1977 Jul;131(1):369–371. doi: 10.1128/jb.131.1.369-371.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakura Y., Ito K., Ishihama A. Biosynthesis of RNA polymerase in Escherichia coli. I. Control of RNA polymerase content at various growth rates. Mol Gen Genet. 1974;133(1):1–23. doi: 10.1007/BF00268673. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matzura H., Hansen B. S., Zeuthen J. Biosynthesis of the beta and beta' subunits of RNA polymerase in Escherichia coli. J Mol Biol. 1973 Feb 15;74(1):9–20. doi: 10.1016/0022-2836(73)90350-1. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Yura T. Effects of rifampicin on synthesis and functional activity of DNA-dependent RNA polymerase in Escherichia coli. Mol Gen Genet. 1976 Jun 15;145(3):227–237. doi: 10.1007/BF00325817. [DOI] [PubMed] [Google Scholar]
- Newton A. Role of transcription in the temporal control of development in Caulobacter crescentus (stalk-rifampin-RNA synthesis-DNA synthesis-motility). Proc Natl Acad Sci U S A. 1972 Feb;69(2):447–451. doi: 10.1073/pnas.69.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L. Differentiation in the Caulobacter cell cycle. Annu Rev Microbiol. 1976;30:377–407. doi: 10.1146/annurev.mi.30.100176.002113. [DOI] [PubMed] [Google Scholar]
- Siddhikol C., Erbstoeszer J. W., Weisblum B. Mode of action of streptolydigin. J Bacteriol. 1969 Jul;99(1):151–155. doi: 10.1128/jb.99.1.151-155.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Wehrli W., Staehelin M. Actions of the rifamycins. Bacteriol Rev. 1971 Sep;35(3):290–309. doi: 10.1128/br.35.3.290-309.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]