Abstract
Three G3P[9] rotaviruses, detected in children hospitalized with gastroenteritis in Palermo, Italy, were found to be genetically related to strains of either human or feline origin in the VP7, VP4, and VP6 genes. In contrast, in the NSP4 gene the viruses resembled G2P[4] human strains, suggesting a reassortment between AU-1-like and Kun-like strains.
Group A rotaviruses are a major cause of acute gastroenteritis in humans and animals (14). The rotavirus genome is composed of 11 segments of double-stranded RNA (dsRNA) (4). The pattern of migration of the dsRNA by polyacrylamide gel electrophoresis (PAGE) allows the distinction of a long, short, or supershort electropherotype (e-type) (4). The viruses are classified as G and P types on the basis of the outer capsid proteins VP7 and VP4, respectively (4). Of the 15 G types and the 27 P types, 5 G types (types G1 to G4 and G9) and three P types (types P[4], P[6], and P[8]) appear to be common in human rotaviruses (21). On the basis of their reactivities to VP6-specific monoclonal antibodies, group A rotaviruses are classified into types SGI, SGII, SGI + SGII, and SG-non I-non II (4). The majority of SGI and SGII animal rotavirus strains and SGII human rotavirus strains display a long e-type of migration of the 11 dsRNA gene segments, while almost all SGI human rotaviruses possess a short e-type (14). The nonstructural protein NSP4 is able to induce diarrhea in experimental rodents through a Ca2+-dependent signaling pathway (18). Passively acquired antibodies to NSP4 appear to prevent watery diarrhea in mice (2), and this sets NSP4 as a potential vaccine target. Six NSP4 genotypes (genotypes A to F) have been established in group A rotaviruses. In humans, NSP4 genotypes A (Kun-like) and B (Wa-like) are common. Genotype C (AU-1-like) is common in feline and canine strains but is infrequent in human rotaviruses (3).
The G3 VP7 specificity has been identified in rotavirus strains from almost all susceptible animal species (4). In humans, G3 rotaviruses are usually associated with the P[8] VP4 genotype and, more rarely, with the P[9] genotype (10, 15, 21). Conversely, the G3P[9] combination is common in feline rotaviruses (11). Interspecies infections between animals and humans can be revealed by the detection of strains with unexpected antigenic and genetic features. The first human G3P[9] rotavirus strain, strain AU-1, was isolated in Israel in 1982. The strain displayed animal-like features (a long e-type in conjunction with SGI specificity) and was shown by RNA-RNA hybridization to be closely related to feline rotaviruses (19).
Uninterrupted surveillance activity for rotaviruses has been conducted in Palermo, Italy, since the mid-1980s. Between 1985 and 2005, 1,547 rotavirus-positive samples were collected and characterized either antigenically or genetically to predict the VP7, VP4, and VP6 specificities and the genome pattern. Three strains (strains PAF96/94, PAH136/96, and PAI58/96) displayed animal strain-like features, since they possessed a long e-type by PAGE analysis and SGI reactivity, and they were characterized as G3P[9]. The infections caused by the G3P[9] rotaviruses were diagnosed in children 18 to 42 months of age hospitalized with acute diarrhea at the G. Di Cristina Hospital of Palermo in January 1994 (strain PAF96/94), March 1996 (strain PAH136/96), and November 1996 (strain PAI58/96).
In order to characterize more precisely the three G3P[9] strains and to investigate their origins in a more in-depth manner, the nucleotide sequences of the VP7, VP4, VP6, and NSP4 genes were determined after amplification by reverse transcriptase PCR (5, 7, 12, 13, 17). The regions sequenced were from nucleotides (nt) 331 to 1009 for VP7, from nt 205 to 849 for VP8*, and from nt 873 to 1114 for VP6 (relative to the sequences of AU-1 strains D86271, D10970, and DQ490538, respectively) and from nt 126 to 452 for NSP4 (relative to the sequence of Kun strain D88829). Sequence alignment was performed by using the ClustalW program (22), and phylogenetic analysis was carried out with 226 amino acids (aa) of VP7, 214 aa of VP8*, 80 aa of VP6, and 109 aa of NSP4 by using MEGA software (version 3.0) (16).
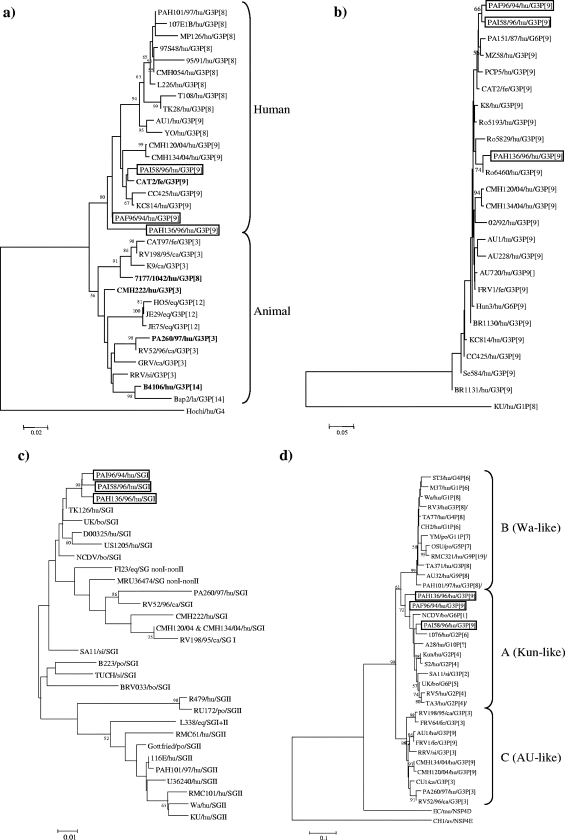
By sequence analysis of VP7, strains PAF96/94, PAH136/96, and PAI58/96 displayed 95.5 to 98.2% amino acid identity to each other. The highest amino acid identity (94.7 to 98.7%) was to G3P[9] human rotaviruses CC425, KC814, CMH120, and CMH134 and to feline strain CAT2, which is G3P[9] (Fig. 1a). The Italian G3P[9] viruses were more distantly related to the reference G3P[9] strain, strain AU-1 (92.8 to 96.9% amino acid identity), and to G3P[8] human rotaviruses, which included G3P[8] human strain PAH101/97, isolated in Palermo in 1997 (92.4 to 96.4% amino acid identity). Three distinctive amino acid changes were found in VP7 of the G3P[9] rotaviruses, i.e., 43Val→Ile within variable region 3 (VR3), 122Ala→Thr within VR6, and 213Ser→Asn in VR8. These three amino acid residues were also conserved in feline strain CAT2.
FIG. 1.
Phylogenetic analysis of deduced amino acid sequence of the VP7 (a), VP4 (b), VP6 (c), and NSP4 (d) genes. The phylogeny was inferred by the neighbor-joining method and by use of the Kimura two-parameter model as the method of substitution. The statistical significance was estimated by bootstrap analysis with 1,000 pseudoreplicate data sets. The origins of the rotavirus strains are indicated as follows: hu, human; fe, feline; ca, canine; eq, equine; si, simian; la, lapine; bo, bovine; po, porcine; mu, murine; and av, avian. In panel a, strains with atypical positions in the VP7 tree are highlighted in boldface.
Sequence analysis of the VP8* portion of VP4 revealed 93 to 100% amino acid identity to human and animal P[9] rotaviruses. Partial sequence analysis of the VP6 gene revealed 91 to 98.9% amino acid identity with SGI sequences (Fig. 1b and c). The only available VP6 sequences of human G3P[9] rotaviruses (strains CMH120 and CMH134) are also SGI and showed an amino acid identity of 92.3% with Italian G3P[9] strains. The NSP4 genes of strains PAF96/94, PAH136/96, and PAI58/96 were characterized as NSP4 genotype A, since the viruses displayed 94 to 95.3% amino acid identity to G2P[4] strain Kun and only 83 to 88.3% amino acid identity to NSP4 genotype B Wa-like strains and NSP4 genotype C AU-1-like strains (Fig. 1d).
During a 20-year surveillance for rotavirus infections in children admitted with diarrhea to a hospital in Palermo, marked fluctuations in the rotavirus G types have been observed. G3 rotaviruses were identified at yearly rates of 0 to 7% until the end of the 1990s, while they reached peaks of 16.9% in the subsequent years. Strain PAF96/94 was the only G3 rotavirus detected in 1994, while strains PAH136/96 and PAI58/96 were detected in 1996, along with a few G3P[8] viruses. The P[9] genotype is responsible for less than 2.5% of the rotavirus infections among humans worldwide, while it is common in feline rotaviruses (15). After their initial description in 1982 in Israel, G3P[9] strains have been detected in humans on more occasions, but only in a sporadic fashion. Recently, G3P[9] rotaviruses have been documented in Japan, Thailand, and Spain (6, 8-10, 15, 20).
The molecular characterization of strains PAF96/94, PAH136/96, and PAI58/96 revealed that they were genetically related in the VP7, VP4, and VP6 genes to reference/established G3P[9] strains, including reference strain AU-1. Unlike AU-1-like strains, however, the Italian G3P[9] strains possessed an NSP4 of genotype A (Kun-like). To our knowledge, NSP4 genotype A has never been identified in G3P[9] rotaviruses, and this may suggest that such viruses acquired the 10th genome segment by genetic reassortment with human G2P[4] rotaviruses that were epidemic, almost in the same years, in Palermo (1). However, the Italian G3P[9] strains were not of clonal origin, and this is more consistent with the spillover introduction from an unidentified animal source. Although multiple reassortment events and the emergence of novel strains have also been documented in the past (i.e., G9 strains in humans), there is a concern that the introduction of human rotavirus vaccines might alter the forces and balances that drive rotavirus evolution and determine the spread of novel strains that are antigenically different from those included in the vaccine formulations. The G3P[9] strains completely fulfill these requirements, since they match poorly or do not match at all the main antigenic determinants included in the monovalent vaccine (G1P[8], SGII VP6, NSP4 genotype B) or in the polyvalent vaccine (G1 to G4, P[8], SGI, NSP4 genotype A). This may warrant efforts to evaluate the efficacy of the vaccines against G3P[9] viruses and to investigate the ecology of such viruses in animals.
Nucleotide sequence accession numbers.
The sequences of human strains PAF96/94, PAH136/96, and PAI58/96 have been registered in GenBank. The accession numbers are as follows: EU076911 (VP6), EU076905 (VP7), EU076908 (VP8*), and EU076914 (NSP4) for PAF96/94; EU076912 (VP6), EU076906 (VP7), EU076909 (VP8*), and EU076915 (NSP4) for PAH136/96; and EU076913 (VP6), EU076907 (VP7), EU076910 (VP8*), and EU076916 (NSP4) for PAI58/96.
Acknowledgments
This study was supported by the Ministero dell'Istruzione, dell'Università e della Ricerca (Italian Ministry of Education, University and Reserch) (Fondi di Ateneo ex 60%).
Footnotes
Published ahead of print on 6 December 2007.
REFERENCES
- 1.Arista, S., G. M. Giammanco, S. De Grazia, C. Colomba, V. Martella, A. Cascio, and M. Iturriza-Gomara. 2005. G2 rotavirus infections in an infantile population of the south of Italy: variability of viral strains over time. J. Med. Virol. 77587-594. [DOI] [PubMed] [Google Scholar]
- 2.Ball, J. M., P. Tian, C. Q. Zeng, A. P. Morris, and M. K. Estes. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272101-104. [DOI] [PubMed] [Google Scholar]
- 3.Ciarlet, M., F. Liprandi, M. E. Conner, and M. K. Estes. 2000. Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analyses of animal rotaviruses. Arch. Virol. 145371-383. [DOI] [PubMed] [Google Scholar]
- 4.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 301365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gollop, R., O. Nakagomi, I. Silberstein, L. M. Shulman, H. B. Greenberg, E. Mendelson, and I. Shif. 1998. Three forms of AU-1 like human rotaviruses differentiated by their overall genomic constellation and by the sequence of their VP8*. Arch. Virol. 143263-277. [DOI] [PubMed] [Google Scholar]
- 7.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin, D. D., T. Nakagomi, Y. Hoshino, O. Nakagomi, C. D. Kirkwood, U. D. Parashar, R. I. Glass, and J. R. Gentsch. 2002. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant (P2A[6],G12) and rare P3[9] strains related to bovine rotaviruses. Virology 294256-269. [DOI] [PubMed] [Google Scholar]
- 9.Iizuka, M., M. Chiba, O. Masamune, E. Kaga, T. Nakagomi, and O. Nakagomi. 1994. A highly conserved genomic RNA constellation of Japanese isolates of human rotaviruses carrying G serotype 3 and P serotype 9. Res. Virol. 14521-24. [DOI] [PubMed] [Google Scholar]
- 10.Inoue, Y., and Y. Kitahori. 2006. Rare group a rotavirus P[9]G3 isolated in Nara Prefecture, Japan. Jpn. J. Infect. Dis. 59139-140. [PubMed] [Google Scholar]
- 11.Isegawa, Y., O. Nakagomi, T. Nakagomi, and S. Ueda. 1992. A VP4 sequence highly conserved in human rotavirus strain AU-1 and feline rotavirus strain FRV-1. J. Gen. Virol. 73(Pt 8)1939-1946. [DOI] [PubMed] [Google Scholar]
- 12.Iturriza-Gomara, M., G. Kang, and J. Gray. 2004. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J. Clin. Virol. 31259-265. [DOI] [PubMed] [Google Scholar]
- 13.Iturriza Gomara, M., C. Wong, S. Blome, U. Desselberger, and J. Gray. 2002. Molecular characterization of VP6 genes of human rotavirus isolates: correlation of genogroups with subgroups and evidence of independent segregation. J. Virol. 766596-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 15.Khamrin, P., N. Maneekarn, S. Peerakome, S. Tonusin, T. G. Phan, S. Okitsu, and H. Ushijima. 2007. Molecular characterization of rare G3P[9] rotavirus strains isolated from children hospitalized with acute gastroenteritis. J. Med. Virol. 79843-851. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 17.Lee, C. N., Y. L. Wang, C. L. Kao, C. L. Zao, C. Y. Lee, and H. N. Chen. 2000. NSP4 gene analysis of rotaviruses recovered from infected children with and without diarrhea. J. Clin. Microbiol. 384471-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris, A. P., J. K. Scott, J. M. Ball, C. Q. Zeng, W. K. O'Neal, and M. K. Estes. 1999. NSP4 elicits age-dependent diarrhea and Ca2+-mediated I− influx into intestinal crypts of CF mice. Am. J. Physiol. 277G431-G444. [DOI] [PubMed] [Google Scholar]
- 19.Nakagomi, T., and O. Nakagomi. 1989. RNA-RNA hybridization identifies a human rotavirus that is genetically related to feline rotavirus. J. Virol. 631431-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez-Fauquier, A., V. Montero, S. Moreno, M. Sole, J. Colomina, M. Iturriza-Gomara, A. Revilla, I. Wilhelmi, and J. Gray. 2006. Human rotavirus G9 and G3 as major cause of diarrhea in hospitalized children, Spain. Emerg. Infect. Dis. 121536-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 1529-56. [DOI] [PubMed] [Google Scholar]
- 22.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]