Abstract
The global emergence of vancomycin-resistant Enterococcus faecium has been characterized as the clonal spread of clonal complex 17 (CC17) E. faecium. CC17 was defined upon multilocus sequence typing and is characterized by resistance to quinolones and ampicillin and the presence of the enterococcal surface protein (Esp) in the majority of isolates. The recently noticed increased incidence of vancomycin-susceptible CC17 E. faecium infections in our hospital initiated a nationwide study to determine ecological changes among enterococcal infections. The data and strain collections were obtained from 26 (38%) and 9 (14%) of 66 microbiology laboratories in The Netherlands. E. faecium and E. faecalis were distinguished by multiplex PCR; all E. faecium isolates were genotyped by multiple-locus variable-number tandem-repeat analysis (MLVA), and the presence of esp was identified by PCR. Average numbers of ampicillin-resistant enterococcal isolates from normally sterile body sites per hospital increased from 5 ± 1 in 1994 to 25 ± 21 in 2005. Among all enterococcal bloodstream infections, the proportions of ampicillin-resistant E. faecium (AREF) increased from 4% in 1994 to 20% in 2005 (P < 0.001). All E. faecalis isolates were susceptible to ampicillin, whereas 78% of the E. faecium isolates were resistant (49% of these contained esp). Genotyping revealed that 86% of AREF isolates belonged to CC17, including four dominant MLVA types found in ≥3 hospitals, accounting for 64% of the AREF isolates. Infections caused by CC17 E. faecium has increased nationwide, especially in university hospitals due to the clonal spread of four MLVA types, and seems associated with acquisition of the esp gene.
The emergence of vancomycin-resistant Enterococcus faecium (VREF) in the United States in the 1990s was preceded by the emergence of ampicillin-resistant Enterococcus faecium (AREF) in the 1980s (8, 11, 27, 28). Molecular epidemiological studies of human- and animal-derived E. faecium since then, revealed the existence of a genetic lineage, labeled clonal complex 17 (CC17), associated with nosocomial E. faecium outbreaks and infections in five continents. CC17 is characterized by ampicillin and quinolone resistance and the presence of a putative pathogenicity island, including the esp gene in the majority of isolates (2-4, 9, 12, 17-20, 31, 36). In retrospect, it seems likely that the acquisition of ampicillin resistance was an earlier step in hospital adaptation of E. faecium, facilitating the subsequent emergence of VREF (18, 36).
Since 2000, infection rates of VREF are rising in European hospitals (see EARSS Annual Report 2005 [www.rivm.nl/earss]), suggesting that the increase of VREF in Europe follows the American epidemiology with a 10-year delay. Little is known, though, about the molecular epidemiology of AREF.
In our hospital (the University Medical Center Utrecht [UMCU]), the proportion of invasive enterococcal infections caused by AREF increased from 2% in 1994 to 32% in 2005, with partial replacement of ampicillin-susceptible (Amps) E. faecalis by E. faecium (75% AREF) among enterococcal bloodstream infections (32). Based on these local findings, a nationwide study was initiated to determine the ecological changes among enterococcal infections from sterile body sites in hospitals in The Netherlands.
MATERIALS AND METHODS
Microbiology data.
All microbiology laboratories (n = 66) serving 9 university and 87 nonuniversity hospitals in The Netherlands were invited to submit data on annual numbers of ampicillin-resistant (Ampr) enterococci isolated from normally sterile body sites identified between 1994 and 2005. Normally sterile body sites included blood, abdominal and cerebrospinal fluid, intravascular catheter tips, and pus and wound specimens. These data did not differentiate enterococci to the species level.
Furthermore, the laboratories were invited to provide, for each year, the first 30 enterococcal bloodstream isolates, irrespective of antibiotic susceptibility (1 per patient). A species-specific multiplex PCR based on the ddl gene was performed to distinguish E. faecium and E. faecalis as previously described (6, 32). Susceptibilities to ampicillin were determined by inoculation of Mueller-Hinton agar containing ampicillin at 16 mg/liter according to Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards) guidelines.
Genotyping of E. faecium isolates.
All E. faecium isolates, including 2006 isolates, were genotyped by using multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA), as described previously (31) with minor modifications (www.mlva.umcutrecht.nl). Identification of CC17-specific MLVA types (MTs) was performed by comparing each MLVA profile to the previously described seven different repeat combinations for VNTR-7, -8, and -10 with a positive predictive value of 87% and a specificity of 90% to belong to CC17 (31). The genetic relatedness of MTs was confirmed by multilocus sequence typing (MLST) on a subset of representative isolates (9). The obtained MLST profiles were clustered with 313 MLST profiles, representing 855 isolates from the database using the eBURST algorithm (7, 18). The presence of the putative pathogenicity island was determined by PCR using the esp gene as a marker (20).
Statistical analysis.
Statistical analysis of the data was performed with SPSS 12.0.1 for Windows (SPSS, Inc., Chicago, IL) using the chi-square test. The data from university hospitals were compared to those from nonuniversity hospitals.
RESULTS
Microbiology data invasive Ampr enterococci.
Of 66 microbiology laboratories serving 7 of 9 (78%) university hospitals (>500 beds) and 22 of 87 (25%) nonuniversity hospitals (250 to 500 beds [n = 6], >500 beds [n = 16]), 26 (39%) provided data on Ampr enterococci from normally sterile body sites. The data from our own hospital, already described previously (32), were included as well. The hospitals were dispersed throughout The Netherlands. Only one nonuniversity and three university hospitals could provide data going back as far as 1994.
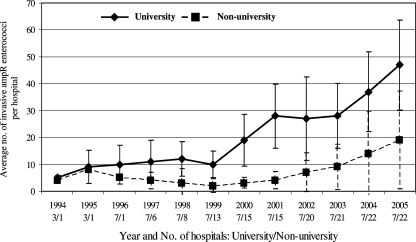
Average annual numbers of Ampr enterococci from normally sterile body sites per hospital increased from 5 ± 1 in 1994 to 25 ± 21 in 2005. The increase was most pronounced in university hospitals (from 5 ± 1 in 1994 to 47 ± 17 in 2005) (Fig. 1). The average annual numbers in nonuniversity hospitals increased from 4 ± 0 in 1994 to 19 ± 18 in 2005 (Fig. 1). Annual numbers per hospital varied between 1 and 14 for 250- to 500-bed hospitals and between 1 and 80 for larger hospitals (>500 beds).
FIG. 1.
Average annual numbers of invasive Ampr enterococci per hospital. Error bars denote standard deviations. University and nonuniversity hospitals were compared. For each year, the numbers of hospitals that provided data are indicated.
E. faecium/E. faecalis ratio among bloodstream isolates.
In all, 1,573 enterococcal bloodstream isolates were obtained from nine hospitals (five nonuniversity and four university). Three of the four university hospitals provided isolates from 1994 onward. The oldest isolates obtained from a nonuniversity hospital were from 1999.
Species identification revealed 1,121 E. faecalis, 303 E. faecium, and 149 non-E. faecalis and non-E. faecium isolates. The latter isolates were not further characterized. Discrepancies between the original identification, as provided by the submitting labs and identification based on the ddl gene, were found in 116 (7%) isolates. All E. faecalis isolates were Amps, whereas 237 of 303 (78%) E. faecium isolates were Ampr.
The proportions of AREF among all enterococcal bloodstream isolates increased from 4% (1994) to 20% (2005) (P = 0.01), while the proportions of Amps E. faecalis decreased from 89% (1994) to 77% (2005) (P = 0.5). Proportions of Amps E. faecium remained <9%, and no significant trend could be observed over time. In university hospitals the proportions of AREF increased from 4% in 1994 to 27% in 2005 (P < 0.001). For individual hospitals these proportions ranged from 0% in 1994 to 10% in 2005 (lowest) and from 27% in 1996 to 43% in 2005 (highest). In nonuniversity hospitals there was a slight, but nonsignificant increase in the proportions of AREF from 6% in 1999 to 12% in 2005.
Genotyping E. faecium isolates.
MLVA typing of 303 E. faecium isolates revealed 61 different MTs among 263 isolates, including 41 MTs that had not been previously detected. Incomplete MLVA profiles were obtained for 29 isolates due to repeatedly negative PCR results for ≥1 of the VNTR loci. In 11 isolates none of the VNTR loci were PCR positive. All 40 isolates that could not be assigned a MT appeared to be Amps. Twenty of the remaining 26 (77%) Amps E. faecium isolates yielded a unique MT.
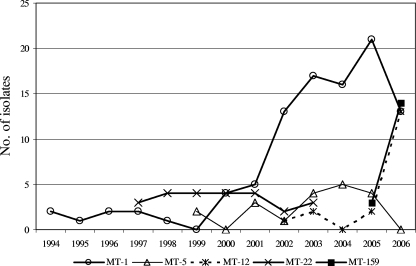
Sixty-seven percent (175 of 263) of typeable isolates belonged to five MTs, including four MTs detected in ≥3 hospitals: MT-1 (n = 97 in 9 hospitals) MT-5 (n = 19 in 5 hospitals), MT-12 (n = 18 in 6 hospitals), and MT-159 (n = 17 in 5 hospitals), together accounting for 64% (151 of 237) of Ampr isolates (Table 1). MT-22 (24 of 303 isolates) was detected in only one hospital, where it accounted for 29% (24 of 83) of all E. faecium isolates between 1999 and 2003 (Fig. 2).
TABLE 1.
Distribution of predominant MTs
| MT | MLVA profile (no. of repeats)
|
CC17-specific MTsa | Total no. of isolates | No. of Ampr isolates | No. of esp-positive isolates | No. of hospitals | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VNTR-1 | VNTR-2 | VNTR-7 | VNTR-8 | VNTR-9 | VNTR-10 | ||||||
| 1 | 5 | 7 | 3 | 3 | 2 | 3 | + | 97 | 97 | 19 | 9 |
| 5 | 5 | 7 | 3 | 2 | 2 | 3 | + | 19 | 19 | 17 | 5 |
| 12 | 5 | 7 | 3 | 3 | 1 | 3 | + | 18 | 18 | 16 | 6 |
| 22 | 5 | 7 | 4 | 2 | 2 | 1 | − | 24 | 24 | 24 | 1 |
| 159 | 5 | 7 | 3 | 3 | 1 | 2 | + | 17 | 17 | 17 | 5 |
+, MTs with the following repeat profiles for VNTR-7, -8, and -10: 3-3-3, 3-2-3, 3-3-2, 4-3-3, 3-4-3, 4-2-3, and 3-3-1.
FIG. 2.
Annual distribution of five predominant MTs.
Longitudinal analysis of the genotyping data revealed that MT-1 was already present in one hospital in 1994 and that its presence increased after 1999, with a documented presence in all nine hospitals (Fig. 2 and Table 1). MT-5 and MT-12 emerged from 1999 and 2002 onward (Fig. 2). The first MT-12 isolate was detected in one hospital in 2002, and it appeared in three other hospitals in 2006 (Table 1). Finally, MT-159 was found in two hospitals in 2005, with subsequent isolation in three additional hospitals in 2006.
The four most predominant MTs detected in ≥3 hospitals were closely related. MT-5 and MT-12 were single-locus variants from MT-1, while MT-159 was a double-locus variant from MT-1 and a single-locus variant from MT-12 (Table 1). Identification of CC17-specific MTs based on different repeat combinations for VNTR-7, -8, and -10 (31) revealed that 86% (204 of 237) of Ampr isolates belonged to CC17 (Table 1).
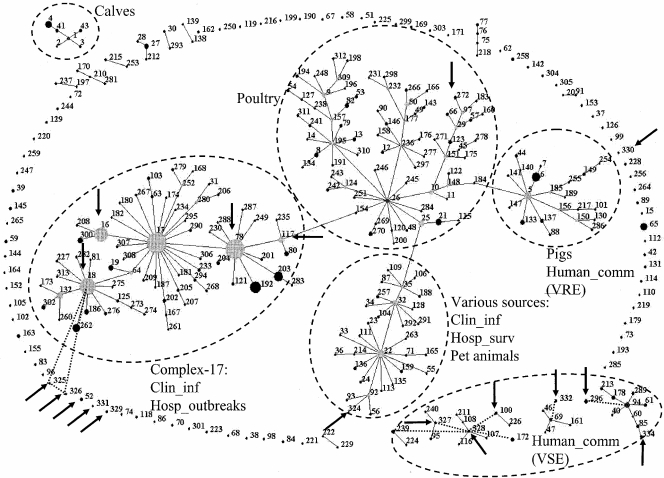
MLST was performed on 15 Ampr and 12 Amps isolates (Table 2). The seven MT-159 isolates of different hospitals revealed a single sequence type (ST), ST-78. In contrast, seven MT-12 isolates from different years represented five different STs. All STs representing Ampr isolates, except one (ST-324), grouped within or were linked to CC17 (Fig. 3). The Amps E. faecium isolates, all MLVA nontypeable, revealed different STs, including seven new STs, ST-326 to -332, ST-334, and ST-100, -52, -272, and -296. Six STs clustered with other ampicillin- and vancomycin-susceptible human community isolates, including MLVA nontypeable E. faecium isolates, four represented singletons, and one isolate grouped among poultry isolates, and one ST (ST-326) was linked to CC17 (Fig. 3).
TABLE 2.
MLST results for representative MTs
| Resistance | MT (no. of isolates typed) | No. of hospitals | esp gene | Yr | MLST |
|---|---|---|---|---|---|
| Ampr | MT-159 (7) | 5 | + | 2005/2006 | ST-78 |
| MT-12 (3) | 3 | − | 2002/2003 | ST-18, ST-324, ST-325 | |
| MT-12 (1) | 1 | + | 2005 | ST-78 | |
| MT-12 (3) | 3 | + | 2005/2006 | ST-117 | |
| MT-22 (1) | 1 | + | 2000 | ST-16 | |
| Amps | Nontypeable (12) | 8 | - | NAa | New: ST-326 to ST-332, ST-334; ST-100, ST-52, ST-272, ST-296 |
NA, not applicable.
FIG. 3.
eBURST clustering of 18 MLST profiles, indicated by an arrow, representing 27 isolates from present study, with 313 MLST profiles representing 855 E. faecium isolates from the database (www.mlst.net). Each ST is represented as a node; the relative size of each node is indicative of its prevalence among the isolates, and lines connect single-locus variants (STs that differ in only one of the seven housekeeping genes). Dashed lines indicate connections between double-locus variants. The sources of specific clusters of STs are indicated, including CC17 comprising hospital outbreaks and clinical isolates. Clin_inf, isolates from clinical sites (mainly blood) from hospitalized patients; Hosp_outbreak, hospital outbreak isolates; Hosp_surv, feces isolates from hospitalized patients without an enterococcal infection and not associated with an enterococcal hospital outbreak; Human_comm, feces isolates from human volunteers not connected to hospitals.
Determination of esp gene.
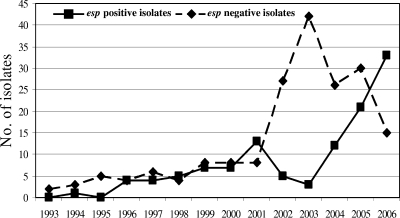
Forty-nine percent (115 of 237) of Ampr E. faecium isolates contained the esp gene, while none of the Amps isolates was esp positive. In longitudinal analysis a remarkable increase of esp-positive isolates occurred from 2004 onward (Fig. 4). The total numbers of esp-negative isolates peaked in 2003 (n = 40) and decreased subsequently. Interestingly, all MT-12 isolates from 2002 and 2003 were esp negative, whereas all MT-12 isolates from 2005 onward contained the esp gene. Similarly, the majority of esp-positive isolates among MT-1 isolates (15 of 19 [79%]) were found between 2004 and 2006. Before 2004, only 4 of 47 MT-1 isolates (9%) were esp positive. Finally, 17 of 19 MT-5 isolates (89%) and all MT-159 isolates were esp positive. These findings suggest that MT-1 and MT-12 isolates acquired the esp gene and that the presence of this gene was associated with nosocomial spread. On the other hand, esp-positive MT-22 isolates were only found in one hospital and apparently disappeared in 2003, and MT-5 esp-positive isolates were detected in low numbers in five hospitals, without evidence of increased nosocomial spread during the years (Table 1).
FIG. 4.
Comparison of the annual distribution of esp-positive and -negative isolates.
DISCUSSION
The present study demonstrates a nationwide increase of CC17 AREF isolates obtained from normally sterile body sites in The Netherlands. The molecular epidemiology is characterized by the emergence of several clones, with presumed intra- and interhospital spread. The presence of the esp gene, previously described as a marker of a putative pathogenicity island, seems strongly associated with the emergence of CC17 AREF. The partial replacement of Amps E. faecalis by CC17 AREF has consequences for antimicrobial treatment of enterococcal infections and, more importantly, may set the stage for the emergence of vancomycin-resistant E. faecium.
Our study was based on the voluntary collaboration of microbiological laboratories in The Netherlands and therefore has some potential limitations. In all, 39% of all laboratories provided information on annual numbers of Ampr enterococci obtained from normally sterile body sites. Eight laboratories did not have computerized data information, and thirty hospitals never responded to our (once-repeated) request. Since we failed to obtain information from all laboratories, some selection bias cannot be fully excluded. However, only one of the participating hospitals (large nonuniversity hospital) had identified a nosocomial outbreak with AREF before our study request. On the other hand, three hospitals that did not participate reported the emergence of AREF infections (all MT-159) in 2006 (unpublished data).
Furthermore, one of the participating nonuniversity laboratories, representing 4.4% of the total nonuniversity isolates and 1.9% of the total number of isolates, could not provide information on isolation sites and, therefore, some isolates might not reflect invasive infections. However, this would account only for urine isolates, since surveillance for asymptomatic carriage with Ampr enterococci had not been performed in any hospital. Few laboratories had stored enterococcal isolates, and nine could provide enterococcal bloodstream isolates. It is highly unlikely that hospitals preferably stored either E. faecalis or E. faecium isolates and, therefore, the reported proportions of AREF probably reflect an unbiased estimate. For all of these reasons, we consider the present study to be a reliable reflection of the enterococcal epidemiology in The Netherlands.
The increase and replacement of AREF was most pronounced in university hospitals and large nonuniversity hospitals (>500 beds), a finding that probably reflects differences in patient populations, compared to smaller hospitals. Hematology and transplant patients are generally considered at highest risk for enterococcal bacteremia (5, 32). In our hospital, the increase in AREF bloodstream infections was associated with increased fecal carriage of AREF among hospitalized patients (32). Point-prevalence studies revealed intestinal colonization with AREF in up to 35% of hospitalized patients, especially among high-risk patients on hematology and nephrology wards. Although colonization data are absent for other centers, the endemicity of intestinal colonization with AREF has probably been established in multiple hospitals in The Netherlands.
MLVA typing revealed four highly related types to be responsible for the nationwide emergence of AREF. MT-159 E. faecium isolates first appeared in one hospital in 2005, with documented presence in five hospitals in 2006. However, outbreaks of AREF documented in three other hospitals and not included in the present study were also caused by MT-159 isolates (data not shown). MLST of representative MT-159 isolates (also from the three hospitals not included in the study) revealed ST-78. Nosocomial outbreaks of ST-78 have been described in Korea and Europe, including Germany and Italy (1, 14, 16, 24).
Interestingly, the majority (57%) of esp-positive isolates were found from 2004 onward, and this gene was contained in MT-1 and its genetically related variants MT-12 and -159. We consider the esp gene to be a marker of a putative pathogenicity island (17). This sudden increase of esp-positive isolates suggests that MT-1 and MT-12 acquired the putative pathogenicity island via conjugative transfer, as has been shown in vitro (23), which might contribute to an increased ability to spread and cause infections. In a recent study, Esp expression on the surface of E. faecium varied substantially between isolates and was correlated with initial adherence to polystyrene and biofilm formation (35). Therefore, a role for Esp in the early stages of colonization and subsequent infection has been hypothesized (35).
Previously, MLST of several MT-1 isolates indicated that MT-1 is comprised of multiple STs, including ST-17, the presumed founder of CC17, thus representing a polyclonal population (31, 36). The observation that particular MTs, such as the MT-12 isolates from the present study, are represented by different types determined by MLST and vice versa has been reported before (31), and probably results from differences in the frequency in occurrence of changes in repeat numbers compared to DNA polymorphisms, mutation, and recombination in housekeeping genes.
MLVA typing of 40 Amps E. faecium isolates revealed incomplete MLVA profiles. Southern blot hybridization of three representative isolates confirmed the absence of at least one of the VNTR regions (data not shown). MLST of 12 MLVA nontypeable isolates confirmed that the Amps E. faecium isolates are not linked to CC17 but clustered with other MLVA nontypeable Amps E. faecium isolates that were not involved in hospital outbreaks.
In the United States, the emergence of AREF preceded the nationwide nosocomial epidemic of vancomycin-resistant enterococci. A changing E. faecalis/E. faecium ratio in hospital infections was reported in three longitudinal microbiology-based studies (10, 22, 34). In Europe, several reports on the increase in invasive AREF have been published (4, 29, 33), but to our knowledge ours is the first nationwide study in Europe on the molecular epidemiology of AREF. The emergence of CC17 AREF, resulting in changing E. faecalis/E. faecium ratios among bloodstream isolates, and with 78% of E. faecium isolated determined to be Ampr will impact the treatment of enterococcal infections. The preferred antibiotic for invasive enterococcal infections, amoxicillin, must now be replaced by vancomycin, linezolid, or daptomycin. Increased use of these agents may create selective antibiotic pressure, facilitating the emergence of VREF due to the horizontal transfer of vancomycin resistance genes (13, 30, 32), to mutations leading to resistance to linezolid (15, 26, 37), or to an as-yet-undescribed resistance to daptomycin (21, 25).
Acknowledgments
We thank the following individuals and laboratories in The Netherlands for providing data and/or isolates: J. H. Sloos, Laboratory for Medical Microbiology, Medical Center, Alkmaar; L. Spanjaard, Medical Microbiology, Academic Medical Center, Amsterdam; E. R. Heddema, Medical Microbiology, VU Medical Center, Amsterdam; Medical Microbiology and Infection Prevention, Gelre Hospitals, Apeldoorn; E. Mascini, Medical Microbiology, Rijnstate Arnhem; P. Willemse, Laboratory for Medical Microbiology and Infection Prevention, Amphia Hospital, Breda; R. Brimicombe, Haga Hospitals, The Hague; F. W. Sebens, Laboratory for Medical Microbiology and Infectious Diseases, Deventer; M. A. Schouten, Medical Microbiology, Gelderse Vallei Hospital, Ede; E. Roelofsen, Laboratory for Microbiology, Enschede; A. Demeulemeester, Laboratory for Medical Microbiology and Immunology, Goes; E. Mooi-Kokenberg, Medical Microbiology Groene Hart Hospital, Gouda; K. E. Veldkamp, Laboratory for Infectious Diseases, Groningen; J. P. Arends, Medical Microbiology, University Medical Centre, Groningen; C. G. van Mameren, Medical Microbiology, St. Jansdal Harderwijk; J. H. T. Wagenvoort, Atrium Medical Centre, Heerlen; M. A. Muijsken, Medical Microbiology Westfriesgasthuis, Hoorn; J. H. van Zeijl, Public Health Laboratory Friesland, Leeuwarden; L. Dijkshoorn, Department of Infectious Disease, Leiden University Medical Center, Leiden; B. M. de Jongh, Medical Microbiology and Immunology St. Antonius Hospital, Nieuwegein; P. Sturm, Medical Microbiology, University Medical Center, Nijmegen; H. Dahmen, Medical Microbiology, Laurentius Hospital, Roermond; A. van Belkum, Medical Microbiology, Erasmus Medical Center, Rotterdam; A. Buiting, Medical Microbiology, St. Elisabeth Hospital Tilburg; Medical Microbiology, Veldhoven; and J. Berkhout, Medical Microbiology, VieCuri Medical Center, Venlo.
Footnotes
Published ahead of print on 31 October 2007.
REFERENCES
- 1.Bonora, M. G., M. Ligozzi, M. De Fatima, L. Bragagnolo, A. Goglio, G. C. Guazzotti, and R. Fontana. 2004. Vancomycin-resistant Enterococcus faecium isolates causing hospital outbreaks in northern Italy belong to the multilocus sequence typing C1 lineage. Microb. Drug Resist. 10114-123. [DOI] [PubMed] [Google Scholar]
- 2.Bonten, M. J., R. Willems, and R. A. Weinstein. 2001. Vancomycin-resistant enterococci: why are they here, and where do they come from? Lancet Infect. Dis. 1314-325. [DOI] [PubMed] [Google Scholar]
- 3.Coque, T. M., R. Willems, R. Canton, R. Del Campo, and F. Baquero. 2002. High occurrence of esp among ampicillin-resistant and vancomycin-susceptible Enterococcus faecium clones from hospitalized patients. J. Antimicrob. Chemother. 501035-1038. [DOI] [PubMed] [Google Scholar]
- 4.Coque, T. M., R. J. Willems, J. Fortun, J. Top, S. Diz, E. Loza, R. Canton, and F. Baquero. 2005. Population structure of Enterococcus faecium causing bacteremia in a Spanish university hospital: setting the scene for a future increase in vancomycin resistance? Antimicrob. Agents Chemother. 492693-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubberke, E. R., J. M. Hollands, P. Georgantopoulos, K. Augustin, J. F. DiPersio, L. M. Mundy, and H. J. Khoury. 2006. Vancomycin-resistant enterococcal bloodstream infections on a hematopoietic stem cell transplant unit: are the sick getting sicker? Bone Marrow Transplant. 38813-819. [DOI] [PubMed] [Google Scholar]
- 6.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 3324-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grayson, M. L., G. M. Eliopoulos, C. B. Wennersten, K. L. Ruoff, P. C. De Girolami, M. J. Ferraro, and R. C. Moellering, Jr. 1991. Increasing resistance to beta-lactam antibiotics among clinical isolates of Enterococcus faecium: a 22-year review at one institution. Antimicrob. Agents Chemother. 352180-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. Van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 401963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwen, P. C., D. M. Kelly, J. Linder, S. H. Hinrichs, E. A. Dominguez, M. E. Rupp, and K. D. Patil. 1997. Change in prevalence and antibiotic resistance of Enterococcus species isolated from blood cultures over an 8-year period. Antimicrob. Agents Chemother. 41494-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, R. N., H. S. Sader, M. E. Erwin, S. C. Anderson, et al. 1995. Emerging multiply resistant enterococci among clinical isolates. I. Prevalence data from 97 medical center surveillance study in the United States. Diagn. Microbiol. Infect. Dis. 2185-93. [DOI] [PubMed] [Google Scholar]
- 12.Jureen, R., J. Top, S. C. Mohn, S. Harthug, N. Langeland, and R. J. Willems. 2003. Molecular characterization of ampicillin-resistant Enterococcus faecium isolates from hospitalized patients in Norway. J. Clin. Microbiol. 412330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawalec, M., M. Gniadkowski, M. Zaleska, T. Ozorowski, L. Konopka, and W. Hryniewicz. 2001. Outbreak of vancomycin-resistant Enterococcus faecium of the phenotype VanB in a hospital in Warsaw, Poland: probable transmission of the resistance determinants into an endemic vancomycin-susceptible strain. J. Clin. Microbiol. 391781-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klare, I., C. Konstabel, S. Mueller-Bertling, G. Werner, B. Strommenger, C. Kettlitz, S. Borgmann, B. Schulte, D. Jonas, A. Serr, A. M. Fahr, U. Eigner, and W. Witte. 2005. Spread of ampicillin/vancomycin-resistant Enterococcus faecium of the epidemic-virulent clonal complex-17 carrying the genes esp and hyl in German hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 24815-825. [DOI] [PubMed] [Google Scholar]
- 15.Kloss, P., L. Xiong, D. L. Shinabarger, and A. S. Mankin. 1999. Resistance mutations in 23 S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 29493-101. [DOI] [PubMed] [Google Scholar]
- 16.Ko, K. S., J. Y. Baek, J. Y. Lee, W. S. Oh, K. R. Peck, N. Lee, W. G. Lee, K. Lee, and J. H. Song. 2005. Molecular characterization of vancomycin-resistant Enterococcus faecium isolates from Korea. J. Clin. Microbiol. 432303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leavis, H., J. Top, N. Shankar, K. Borgen, M. Bonten, J. van Embden, and R. J. Willems. 2004. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J. Bacteriol. 186672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leavis, H. L., M. J. Bonten, and R. J. Willems. 2006. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 9454-460. [DOI] [PubMed] [Google Scholar]
- 19.Leavis, H. L., R. J. Willems, J. Top, and M. J. Bonten. 2006. High-level ciprofloxacin resistance from point mutations in gyrA and parC confined to global hospital-adapted clonal lineage CC17 of Enterococcus faecium. J. Clin. Microbiol. 441059-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leavis, H. L., R. J. Willems, J. Top, E. Spalburg, E. M. Mascini, A. C. Fluit, A. Hoepelman, A. J. de Neeling, and M. J. Bonten. 2003. Epidemic and nonepidemic multidrug-resistant Enterococcus faecium. Emerg. Infect. Dis. 91108-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, J. S., A. Owens, J. Cadena, K. Sabol, J. E. Patterson, and J. H. Jorgensen. 2005. Emergence of daptomycin resistance in Enterococcus faecium during daptomycin therapy. Antimicrob. Agents Chemother. 491664-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murdoch, D. R., S. Mirrett, L. J. Harrell, J. S. Monahan, and L. B. Reller. 2002. Sequential emergence of antibiotic resistance in enterococcal bloodstream isolates over 25 years. Antimicrob. Agents Chemother. 463676-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oancea, C., I. Klare, W. Witte, and G. Werner. 2004. Conjugative transfer of the virulence gene, esp, among isolates of Enterococcus faecium and Enterococcus faecalis. J. Antimicrob. Chemother. 54232-235. [DOI] [PubMed] [Google Scholar]
- 24.Peta, M., E. Carretto, D. Barbarini, A. Zamperoni, L. Carnevale, L. Perversi, M. Pagani, M. G. Bonora, R. Fontana, P. Marone, and M. Langer. 2006. Outbreak of vancomycin-resistant Enterococcus spp. in an Italian general intensive care unit. Clin. Microbiol. Infect. 12163-169. [DOI] [PubMed] [Google Scholar]
- 25.Poutsiaka, D. D., S. Skiffington, K. B. Miller, S. Hadley, and D. R. Snydman. 2007. Daptomycin in the treatment of vancomycin-resistant Enterococcus faecium bacteremia in neutropenic patients. J. Infect. 54567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 452154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice, L. B. 2001. Emergence of vancomycin-resistant enterococci. Emerg. Infect. Dis. 7183-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shepard, B. D., and M. S. Gilmore. 2002. Antibiotic-resistant enterococci: the mechanisms and dynamics of drug introduction and resistance. Microbes. Infect. 4215-224. [DOI] [PubMed] [Google Scholar]
- 29.Simonsen, G. S., L. Smabrekke, D. L. Monnet, T. L. Sorensen, J. K. Moller, K. G. Kristinsson, A. Lagerqvist-Widh, E. Torell, A. Digranes, S. Harthug, and A. Sundsfjord. 2003. Prevalence of resistance to ampicillin, gentamicin and vancomycin in Enterococcus faecalis and Enterococcus faecium isolates from clinical specimens and use of antimicrobials in five Nordic hospitals. J. Antimicrob. Chemother. 51323-331. [DOI] [PubMed] [Google Scholar]
- 30.Suppola, J. P., E. Kolho, S. Salmenlinna, E. Tarkka, J. Vuopio-Varkila, and M. Vaara. 1999. vanA and vanB incorporate into an endemic ampicillin-resistant vancomycin-sensitive Enterococcus faecium strain: effect on interpretation of clonality. J. Clin. Microbiol. 373934-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Top, J., L. M. Schouls, M. J. Bonten, and R. J. Willems. 2004. Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J. Clin. Microbiol. 424503-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Top, J., R. Willems, H. Blok, M. de Regt, K. Jalink, A. Troelstra, B. Goorhuis, and M. Bonten. 2007. Ecological replacement of Enterococcus faecalis by multiresistant clonal complex 17 Enterococcus faecium. Clin. Microbiol. Infect. 13316-319. [DOI] [PubMed] [Google Scholar]
- 33.Torell, E., O. Cars, B. Olsson-Liljequist, B. M. Hoffman, J. Lindback, and L. G. Burman. 1999. Near absence of vancomycin-resistant enterococci but high carriage rates of quinolone-resistant ampicillin-resistant enterococci among hospitalized patients and nonhospitalized individuals in Sweden. J. Clin. Microbiol. 373509-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treitman, A. N., P. R. Yarnold, J. Warren, and G. A. Noskin. 2005. Emerging incidence of Enterococcus faecium among hospital isolates (1993 to 2002). J. Clin. Microbiol. 43462-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Wamel, W. J., A. P. Hendrickx, M. J. Bonten, J. Top, G. Posthuma, and R. J. Willems. 2007. Growth condition-dependent Esp expression by Enterococcus faecium affects initial adherence and biofilm formation. Infect. Immun. 75924-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willems, R. J., J. Top, M. van Santen, D. A. Robinson, T. M. Coque, F. Baquero, H. Grundmann, and M. J. Bonten. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woodford, N., L. Tysall, C. Auckland, M. W. Stockdale, A. J. Lawson, R. A. Walker, and D. M. Livermore. 2002. Detection of oxazolidinone-resistant Enterococcus faecalis and Enterococcus faecium strains by real-time PCR and PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 404298-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]