Abstract
Here we report the development of a more-sensitive immunoassay for severe acute respiratory syndrome (SARS) based on an enzyme-linked immunosorbent assay using chemiluminescence (CLEIA) to detect the viral nucleocapsid (N) antigen in nasopharyngeal aspirate (NPA) from patients infected with SARS coronavirus (CoV). The CLEIA was established with an optical combination of monoclonal antibodies (MAbs) against SARS CoV N protein prepared from mice immunized with recombinant N protein without cultivating the virus. The capture and detecting MAbs of the CLEIA reacted to the carboxyl-terminal and amino-terminal peptides of the N protein, respectively. The CLEIA was capable of detecting recombinant N protein at 1.56 pg/ml and viral N protein in SARS CoV cell culture lysates at 0.087 of 50% tissue culture infective doses/ml. The CLEIA showed no cross-reactivities to recombinant N proteins of common human CoV (229E, OC43, and NL63) or lysates of cells infected with 229E and OC43. In addition, an evaluation with 18 SARS-positive NPA samples, all confirmed SARS positive by quantitative PCR and antibodies to SARS CoV, revealed that all (18/18) were found positive by the CLEIA; thus, the sensitivity of detection was 100%. When we tested 20 SARS-negative NPA samples, the CLEIA was shown to have high specificity (100%). The sensitivity of our novel SARS CLEIA was significantly higher than the previous EIA and comparable to the other methods using reverse transcription-PCR.
Since the identification of a novel type of human coronavirus, severe acute respiratory syndrome-associated coronavirus (SARS CoV) (19, 26, 28, 33), sensitive diagnostic tests that can detect viral RNA genomes and antigens are in demand for the effective and immediate containment of the patients. Firstly, reverse transcription-PCR (RT-PCR) was found to be useful to detect the SARS CoV during the early phase of infection due to its high sensitivity (13, 32, 44). Hourfar et al. (13) demonstrated that RT-PCR could detect SARS CoV in clinical samples of nasopharyngeal aspirates (NPA) at a sensitivity of about 80% during the first few days after the onset of clinical symptoms. However, the RT-PCR test requires specific laboratories with expertise in molecular diagnostics. Secondly, although the real-time loop-mediated amplification assay is a simpler procedure with a single-step reaction (36), it showed lower sensitivity than the RT-PCR method (31). Thirdly, whereas serological tests gave good sensitivity in diagnosing SARS infection using sera collected 10 to 40 days after the onset of symptoms (40, 41), they showed only 65.4% sensitivity using sera obtained 6 to 10 days after the onset (35). Fourthly, an immunochromatography test for detecting SARS CoV nucleocapsid (N) protein was developed (17). The major merit of this test system is that it can be performed without any particular instrument or extensive training and thus can be applied immediately in any clinical setting. However, the minimum detection limit was 199 50% tissue culture infective doses (TCID50)/ml (17) and was not sensitive enough to detect the SARS CoV N protein in clinical samples such as NPA (20). Finally, the enzyme-linked immunosorbent assay (ELISA) for detecting SARS virus antigens, especially the N protein, was developed (2, 4, 10, 20, 42). Interestingly, although the concentration of SARS CoV is very low in clinical specimens such as NPA, urine, and feces obtained from SARS patients 6 to 10 days after the onset of illness (20), antibodies against the SARS CoV N protein can be detected in the early phase of viral infection (21, 25). Here, we introduce a highly sensitive chemiluminescence enzyme immunoassay (CLEIA) system that can readily and reproducibly detect the SARS CoV N protein antigen and thus can be used for early detection of human SARS CoV infection. The results with detection performance in NPA are also demonstrated.
On the other hand, some reports suggest that the sensitivity of direct N protein detection in serum decreased gradually in the late stage in infection (after 11 days from the onset of symptoms) (2, 23). The viral roads in serum/plasma of SARS patients were also lower in the late stage in infection (5, 8, 15). From those previous reports, a novel direct antigen detection assay with higher sensitivity, for example CLEIA, may be required for diagnostics in the late stage in infection.
MATERIALS AND METHODS
Viral strains.
SARS CoV (HKU-39849) (6) isolated from a patient with SARS CoV pneumonia in Hong Kong was propagated in FRhk4 cells in Dulbecco's modified Eagle's medium (Gibco BRL) with 10% fetal calf serum. Common human CoV 229E (ATCC number VR-740) (9) and OC43 (ATCC number VR-759) (27) were obtained from ATCC.
Clinical specimens.
Human NPA samples were obtained from 18 individual patients with SARS CoV infection whose infection was confirmed by both RT-PCR and serology during the course of their illness. Sex, age, and period after the onset of symptoms of the 18 individual SARS patients are shown in Table 1. The mean age of the 18 SARS patients (10 males and 8 females) from whom positive NPA samples were collected was 40.4 years old. The mean period after the onset of symptoms in the 18 SARS patients was 9.7 days (Table 1). All donors of SARS-positive NPA were Asian people. SARS-negative NPA (n = 20) and NPA obtained from hospitalized patients because of unrelated illnesses were used as negative controls. All SARS-negative NPA were analyzed by immunofluorescence assay (for SARS, influenza A and B, parainfluenza 1, 2, and 3, respiratory syncytial virus, and adenovirus) and were culture negative. These NPA samples were stored frozen until use.
TABLE 1.
SARS patients from whom NPA specimens were collected
| Patient | Sexa | Age (yr) | Days from onset of symptoms |
|---|---|---|---|
| P1 | M | 44 | 9 |
| P2 | F | 39 | 10 |
| P3 | F | 39 | 8 |
| P4 | M | 49 | 8 |
| P5 | M | 35 | 12 |
| P6 | M | 26 | 11 |
| P7 | M | 24 | 12 |
| P8 | F | 71 | 18 |
| P9 | F | 57 | 4 |
| P10 | M | 58 | 12 |
| P11 | F | 34 | 17 |
| P14 | M | 43 | 15 |
| P15 | M | 35 | 7 |
| P16 | M | 34 | 4 |
| P17 | M | 32 | 6 |
| P18 | F | 55 | 5 |
| P19 | F | 34 | 8 |
| P20 | M | 18 | 8 |
| Mean | 40.4 | 9.7 |
M, male; F, female.
Expression and purification of recombinant SARS CoV N protein and common human CoV (229E, OC43, and NL63).
The genome RNA sequences of SARS CoV and other human coronaviruses, 229E, OC43, and NL63, encoding the N protein (GenBank accession number AY274119) were utilized to synthesize the respective cDNA sequences in accordance with the codon usage for Escherichia coli. The cDNA sequences encoding these CoV N proteins were synthesized by PCR as previously described (38). Two halves of the cDNA fragments for N-terminal and C-terminal peptides of SARS CoV N protein, respectively, were synthesized by PCR (twice with first PCR and second PCR primers, as shown in Table 2). These two cDNA fragments were linked at the SmaI restriction enzyme site located at the central part of the cDNA encoding N protein, allowing the generation of full-length cDNA. The cDNA fragments of the truncated SARS CoV N protein, having deletions in amino acid regions 283 to 422 (NΔ283-422) or amino acid regions 1 to 141 (NΔ1-141), were prepared from cloned full-length cDNA using two single-stranded DNA primers shown in Table 2. These synthesized cDNA fragments encoding various N proteins were cloned into the pQE30 expression vector (Qiagen) at the BamHI sites in frame and downstream of a series of six-histidine residues. These clones were sequenced to confirm the proper PCR amplification and construction at least twice for each clone. The His6-tagged recombinant nucleocapsid protein was expressed and purified by using a Ni2-loaded HiTrap chelating system (Amersham Pharmacia) according to the manufacturer's instructions and as previously reported (37). The synthesis of cDNAs of other human coronaviruses (229E, OC43, and NL63 [12]) encoding the N protein (GenBank accession numbers NC_002645, NC_005147, and AY567487) and production of recombinant N proteins were carried out similarly.
TABLE 2.
Primers for synthesizing cDNA of SARS CoV N protein
| Primera | Sequence |
|---|---|
| S1 | 5′-ATGAGCGATAACGGCCCGCAGAGCAACCAGCGTAGCGCGCCGCGTATTACCTTTGGCGGCCCGACCGATAGCACCGATAACAACCAGAACGGCGGCCG-3′ |
| S2 | 5′-GTTTACCGCGCTGACCCAGCATGGCAAAGAAGAACTGCGTTTTCCGCGTGGCCAGGGCGTGCCGATTAACACCAACAGCGGCCCGGATGATCAGATTGGC-3′ |
| S3 | 5′-CGCGTTGGTATTTTTATTATCTGGGCACCGGCCCGGAAGCGAGCCTGCCGTATGGCGCGAACAAAGAAGGCATTGTGTGGGTGGCGACCGAAGGCGCGCTG-3′ |
| S4 | 5′-CAGCTGCCGCAGGGCACCACCCTGCCGAAAGGCTTTTATGCGGAAGGCAGCCGTGGCGGCAGCCAGGCGAGCAGCCGTAGCAGCAGCCGTAGCCGTGGC-3′ |
| A1 | 5′-GCTGGGTCAGCGCGGTAAACCAGCTCGCGGTGTTGTTCGGCAGGCCCTGCGGACGACGCTGTTTCGGACGCGCGCCGTTACGGCCGCCGTTCTGGTTGTTATC-3′ |
| A2 | 5′-GATAATAAAAATACCAACGCGGGCTCAGTTCTTTCATTTTGCCATCGCCGCCACGCACACGACGGGTCGCACGACGATAATAGCCAATCTGATCATCCGGGC-3′ |
| A3 | 5′-GTGGTGCCCTGCGGCAGCTGCAGCACGGTCGCCGCGTTGTTGTTCGGGTTACGGGTGCCAATATGATCTTTCGGGGTGTTCAGCGCGCCTTCGGTCGCCAC-3′ |
| A4 | 5′-CAGCGCGGTTTCGCCGCCGCCGCTCGCCATACGCGCCGGGCTGTTGCCACGGCTGCTGCCCGGGGTGCTGTTACGGCTGTTGCCACGGCTACGGCTGCTGC-3′ |
| S5 | 5′-ACAGCCGTAACAGCACCCCGGGCAGCAGCCGTGGCAACAGCCCGGCGCGTATGGCGAGCGGCGGCGGCGAAACCGCGCTGGCGCTGCTGCTGCTGGATCGTCTG-3′ |
| S6 | 5′-AAAGCGCGGCGGAAGCGAGCAAAAAACCGCGTCAGAAACGTACCGCGACCAAACAGTATAACGTGACCCAGGCGTTTGGCCGTCGTGGCCCGGAACAGACCCAGGG-3′ |
| S7 | 5′-GCGCAGTTTGCGCCGAGCGCGAGCGCGTTTTTTGGCATGAGCCGTATTGGCATGGAAGTGACCCCGAGCGGCACCTGGCTGACCTATCATGGCGCGATTAAAC-3′ |
| S8 | 5′-ACCTTTCCGCCGACCGAACCGAAAAAAGATAAAAAAAAAAAAACCGATGAAGCGCAGCCGCTGCCGCAGCGTCAGAAAAAACAGCCGACCGTGACCCTGC-3′ |
| A5 | 5′-GCTCGCTTCCGCCGCGCTTTTTTTGGTCACGGTCTGGCCCTGCTGCTGCTGGCCTTTGCCGCTCACTTTGCTTTCCAGCTGGTTCAGACGATCTAGCAGCAGCA-3′ |
| A6 | 5′-CGCGCTCGGCGCAAACTGCGCAATCTGCGGCCAATGTTTATAATCGGTGCCCTGACGAATCAGATCCTGATCGCCAAAGTTGCCCTGGGTCTGTTCCGGGCC-3′ |
| A7 | 5′-GGTTCGGTCGGCGGAAAGGTTTTATACGCATCAATATGTTTGTTCAGCAGAATCACGTTATCTTTAAACTGCGGATCTTTATCATCCAGTTTAATCGCGCCATGATAG-3′ |
| A8 | 5′-TTACGCCTGGGTGCTATCCGCGCTCGCGCCGCTCATGCTGTTCTGCAGCTGACGGCTAAAATCATCCATATCCGCCGCCGGCAGCAGGGTCACGGTCGGCTG-3′ |
| NS | 5′-CCCGGATCCATGAGCGATAACGGCCCGCA-3′ |
| NA | 5′-CCCACAGCCGTAACAGCACCCCG-3′ |
| CS | 5′-CCCGGATCCCAGCGCGGTTTCGCCGCCGC-3′ |
| CA | 5′-CCCTTACGCCTGGGTGCTATCCG-3′ |
| N2/3 | 5′-GGATCCCTGTTCCGGGCCACGACGGC-3′ |
| C2/3 | 5′-GGATCCACCCCGAAAGATCATATTGG-3′ |
S1, S2, S3, S4, A1, A2, A3, and A4: primer set of the 1st PCR for the N-terminal cDNA fragment of SARS CoV N protein from the 1st to 220th amino acid residues. S5, S6, S7, S8, A5, A6, A7, and A8: primer set of the 1st PCR for the C-terminal cDNA fragment of SARS CoV N protein from the 215th to 422nd amino acid residues. NS and NA: primer set of the 2nd PCR for the N-terminal cDNA fragment of SARS CoV N protein from 1st to 220th amino acid residues. CS and CA: primer set of the 2nd PCR for the C-terminal cDNA fragment of SARS CoV N protein from the 215th to 422nd amino acid residues. NS and CA: primer set of PCR for the full-length cDNA fragment of SARS CoV N protein from the 1st to 422nd amino acid residues. NS and N2/3: primer set of PCR for the N-terminal cDNA fragment of SARS CoV N protein from the 1st to 282nd amino acid residues. N2/3 and CA: primer set of PCR for the C-terminal cDNA fragment of SARS CoV N protein from the 143rd to 422nd amino acid residues.
Preparation of N protein-specific MAbs.
Seven-week-old BALB/c mice were immunized with purified recombinant nucleocapsid protein mixed with an equal volume of complete Freund adjuvant and then with an equal volume of monophosphoryl lipid A and trehalose dicorynomycolate adjuvant five times every three weeks. Splenocytes from the immunized mice were fused with χ63 myeloma cells. The fused hybridoma cells were selected in RPMI 1640 medium supplemented with 10% fetal calf serum, hypoxanthine, aminopterin, and thymidine (18). The hybridoma clones producing antibodies against the SARS CoV N protein were screened by an ELISA with the recombinant SARS CoV N protein as the coated antigen and immunoprecipitated with recombinant nucleocapsid proteins, respectively. The hybridoma clones thus identified were further cloned by limiting dilution. Monoclonal antibodies (MAbs) were purified from mouse ascites with a protein A affinity column.
Western blot analysis.
One microgram of purified His6-tagged recombinant N protein of SARS CoV was loaded to each well of 5 to 20% gradient sodium dodecyl sulfate (SDS)-polyacrylamide gel, electrophoresed, and subsequently electroblotted to a polyvinylidene difluoride membrane (Millipore). The blot was cut into strips and incubated separately with 10 μg/ml of each MAb against SARS CoV N protein. Strips were washed three times and incubated with 1:1,000 dilution of peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) polyclonal antibody. Antigen and antibody interactions were detected with 4-chloro-1-naphthol (Bio-Rad).
Conventional EIA by color detection for SARS CoV N protein.
Immunoplates (Nunc International) were coated with 100 μl of 1.0 μg/ml anti-nucleocapsid monoclonal antibody (capture MAb) for 15 h and blocked with 10 mM phosphate-buffered saline (pH 7.4) containing 2.5 mM EDTA and 1% bovine serum albumin. Fixed concentrations of purified SARS CoV N protein in 100 μl sample buffer were added to each well and incubated at room temperature for 1 h with shaking. Wells were washed three times and 100 μl of 1.0 μg/ml biotinylated anti-SARS CoV nucleocapsid MAbs (detecting MAb) was added to the well, and the plates were further incubated for 30 min at room temperature with shaking. Wells were washed three times, and 1:12,000-diluted peroxidase-conjugated streptavidin was added to each well. After adding 100 μl of substrate solution (5% o-phenylenediamine) per well and incubating for 5 min, the spectrophotometric absorbance at 492 to 690 nm was measured by a microplate reader.
CLEIA for SARS CoV N protein.
Immunoplates (Nunc International) were coated with 100 μl of 1 μg/ml anti-nucleocapsid MAb (capture MAb) for 15 h and blocked with phosphate-buffered saline containing 2.5 mM EDTA, 1% bovine serum albumin, and 0.5% casein (CLEIA blocking buffer). Fifty microliters of sample buffer {6 mM ACES [N-(2-acetamido)-2-aminoethanesulfonic acid], 0.3% NP-40, 220 mM sodium chloride, 15 mM EDTA 2Na, 60 mM NaOH, 15 mM NaN3} containing various concentrations of purified nucleocapsid protein was added to each well. The plates were then incubated at room temperature for 1 h with shaking. After the wells were washed three times, 100 μl of 1 μg/ml biotinylated MAb against SARS CoV N protein (detecting MAb) was added to each well, and the plates were incubated at room temperature for 30 min with shaking. Wells were then washed three times, and 1:12,000-diluted peroxidase-conjugated streptavidin were added to each well. A hundred microliters per well of substrate solution (Femtogrow; Michigan Diagnostics) was added, and after incubation for 5 min at room temperature, chemiluminescence signals were measured by luminometer.
Measurement of clinical specimens.
We also examined the efficacy of CLEIA in detecting SARS CoV N protein in the clinical specimen. Fifty microliters of NPA and four hundred and fifty microliters of sample buffer containing final 2 mM phenylmethylsulfonyl fluoride were mixed well for both extraction of antigens and virus inactivation. The solution was then filtered with the simplest disposable filter (1.5-μm pore size) to decrease its viscosity, and 50 μl of the solution was added to each well containing 50 μl blocking buffer. Samples were applied to each well, and the capture ELISA procedure was performed as described above.
Q-PCR.
The basic procedure of quantitative RT-PCR (Q-PCR) was reported previously (32). RNAs from experimental and clinical samples, cultured SARS CoV lysate, and NPA were extracted using a QIAamp virus RNA mini kit (Qiagen) as instructed by the manufacturer. Briefly, 140 μl of the sample was processed and the extracted RNA was eluted in 60 ml RNase-free water and stored at −80°C until use. The Q-PCR assay was performed as described previously (30). The cDNA was amplified in a 7000 sequence detection system (Applied Biosystems) by using a TaqMan PCR core reagent kit (Applied Biosystems) as reported previously (32).
RESULTS
Preparation of recombinant N protein of SARS CoV in E. coli.
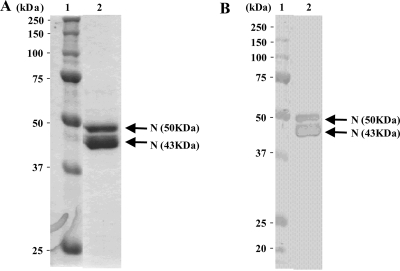
A His6-tagged recombinant N protein of SARS CoV was highly expressed in E. coli and purified from the soluble fraction of the bacterial extracts. As shown in Fig. 1A, the intact recombinant N protein of approximately 50 kDa was detected in the purified protein fraction. In addition, a 43-kDa protein was also detected. Since Western blot experiments shown in Fig. 1B illustrated that both bands were derived from SARS CoV protein, the lower-molecular-mass protein (43 kDa) is considered a partially degraded N protein. The recombinant N protein prepared had enough quality for preparing MAbs.
FIG. 1.
Purified recombinant N protein expressed in E. coli. (A) Results of SDS-PAGE (5 to 20% gradient gel) for recombinant N protein. (B) Results of Western blot analysis for recombinant N protein that reacted with anti-SARS CoV nucleocapsid MAb (catalog no. 654; IMGENEX Co. Ltd.). Lane 1, molecular mass marker; lane 2, purified recombinant His6-tagged N protein.
Evaluation of the reactivity and the specificity of MAbs against the SARS CoV N protein by ELISA and Western blotting.
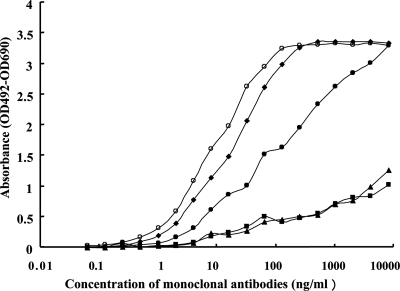
We classified some anti-N protein MAbs into three groups referencing the results of their affinities to immobilized N protein and selected five monoclonal antibodies from each group. Five different types of monoclonal antibodies (I-16, IgG1/κ; I-23, IgG2b/λ; II-3, IgG2a/κ; II-12, IgG2a/κ; and II-21, IgG2b/κ) were prepared from mice immunized with His6-tagged full-length recombinant N protein. Results of ELISA experiments using each MAb are shown in Fig. 2. MAbs, II-21 and II-12 gave a concentration of about 10 to 20 ng/ml at 50% maximum absorbance for immobilized full-length N protein, and they exhibited higher affinities to full-length N protein. MAb I-16 gave a concentration of about 200 ng/ml at 50% maximum absorbance for immobilized full-length N protein, and it exhibited a similar level of affinity to full-length N protein. On the other hand, two MAbs, I-23 and II-3, gave a concentration of over 8,000 ng/ml at 50% maximum absorbance for immobilized full-length N protein and had lower affinities to full-length N protein (Fig. 2). Five MAbs, I-16, I-23, II-3, II-12, and II-21, gave no significant absorbance (from 0.043 to 0.047 optical density at 492 nm), which was similar to the background absorbance with 0 μg/ml of each MAb (0.043 optical density at 492 nm) when they reacted to immobilized 0.5 μg/ml recombinant N protein of three human CoV strains including 229E, OC43, and NL63.
FIG. 2.
Immunoreactivities of MAbs. Immunoreactivity of anti-SARS CoV N protein MAbs. The immunoreactivity of each MAb to full-length recombinant His6-tagged N protein is shown: •, I-16; ▪, I-23; ▴, II-3; ⧫, II-12; and ○, II-21. The reactivities of serially diluted MAbs were measured in duplicates on a microtiter plate coated with full-length recombinant N protein. The mean value for 0 μg/ml of MAb was 0.042. Results show the means of net values (the mean value for each concentration of MAb/the mean value for 0 μg/ml of MAb).
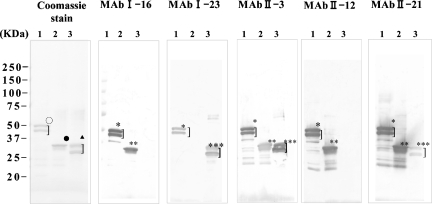
Results of Western blot analysis of these anti-N protein MAbs are shown in Fig. 3. The finding that I-16 and II-12 exhibited no reactivity to truncated NΔ1-141 protein indicates that the epitopes for these MAbs appear to reside within the amino-terminal region of N protein spanning from the 1st to 141st amino acid residues. In addition, I-23 had no reactivity to truncated NΔ283-422 protein, suggesting that its epitope might exist in the carboxyl-terminal region of N protein from the 283rd to 422nd position. Although II-21 strongly reacted to NΔ283-422, similar to I-16 and II-12, it reacted to NΔ1-141 protein only weakly. II-3 reacted to NΔ283-422 protein more weakly than with NΔ1-141 protein. These findings clearly demonstrate the distinct immunoreactivities against SARS CoV N protein.
FIG. 3.
Western blot detection of MAb reactivity to SARS CoV N protein. (Coomassie stain panel) Coomassie blue staining of purified recombinant SARS CoV N protein. The recombinant viral protein was loaded at 1.0 μg/lane and analyzed on a 5 to 20% SDS-PAGE and stained by Coomassie blue dye. (MAb I-16, MAb I-23, MAb II-3, MAb II-12, and MAb II-21 panels) Western blot detection of recombinant SARS CoV N protein by MAbs. Recombinant N proteins were transferred to polyvinylidene difluoride membrane after SDS-PAGE, and each membrane was reacted to different MAbs (I-16, I-23, II-3, II-12, and II-21). Lane 1, full-length N protein was applied; lane 2, truncated N protein (NΔ283-422) was applied; and lane 3, truncated N protein of SARS CoV (NΔ1-141) was applied. ○, completely stained and partially degraded N protein (full length); •, completely stained N protein ΔN283-422; ▴, completely stained and partially degraded N protein (NΔ1-141); *, anti-N MAb reacted to complete and partially degraded N protein (full length); **, anti-N protein MAb reacted to complete N protein (NΔ283-422); ***, anti-N protein MAb reacted to complete and partially degraded N protein (NΔ1-141).
Development of a CLEIA for detecting N protein.
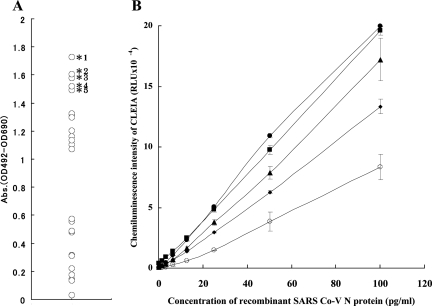
In order to assess the best combination of these MAbs for N protein detection by CLEIA, twenty-five (52) different combinations of capture MAb and detecting MAb were examined with conventional EIA using a color detection system by labeling one of the MAbs (I-16, I-23, II-3, II-12, and II-21) with biotin and screening for their reactivity with 20 ng/ml of recombinant viral N protein as antigen. Among these combinations, five sets of MAbs showing high reactivities to the N protein were selected and further examined (Fig. 4A). Twofold serial dilutions of recombinant full-length N protein of SARS CoV (1.56 to 100 pg/ml) were used to assess the sensitivities and specificities in CLEIA (Fig. 4B). We found that the minimum detection limits ranged from 1.56 to 6.25 pg/ml. The CLEIA using the combination of I-23 (capture MAb) and II-12 (detecting MAb), from hereon termed as the “best” CLEIA, gave the highest reactivity to recombinant N protein, detecting as low as 1.56 pg/ml (Table 3). The best CLEIA had no cross-reactivity to the full-length recombinant N protein of three common human coronaviruses (229E, OC43, and NL63) even at the highest antigen concentration of 100 ng/ml (relative light unit [RLU] sensitivities of 565 ± 149, 843 ± 80, and 593 ± 94, respectively [blank control, 782 ± 283; means ± standard deviations, n = 3]), whereas it exceeded the upper limit of detection (greater than 200,000 RLU) with the N protein of SARS CoV used as a target antigen (data not shown).
FIG. 4.
Evaluation of sandwich ELISA system with various MAbs. (A) Evaluation of sandwich ELISA by color detection system using 25 different combinations of anti-SARS CoV N protein MAbs. Twenty nanograms of recombinant full-length N protein was measured by different combination of MAbs with sandwich ELISA systems. Asterisks indicate the best five results among the 25 ELISAs with different combinations of MAbs. *1, ELISA with I-23/II-12 (capture MAb/detection MAb); *2, ELISA with II-21/II-3; *3, ELISA with II-12/I-23; *4, ELISA with II-21/I-23; and *5, ELISA with I-16/I-23. (B) Standard antigen ELISA curves determined with purified recombinant N protein by sandwich ELISA (CLEIA) using five kinds of MAb pairs (capture MAb/detecting MAb): •, I-23/II-12; ▪, II-21/I-23; ▴, II-12/I-23; ⧫, II-21/II-3; and ○, I-16/I-23. Serial dilutions of recombinant N protein were measured in triplicate by CLEIA. Results show the means of triplicate results. Error bars indicate the range of standard deviations.
TABLE 3.
Sensitivity of CLEIA for detecting recombinant SARS CoV N protein
| Capture MAb/detecting MAb | Sensitivity (RLU ± SD) with the following concns of SARS CoV recombinant N protein (pg/ml)a
|
|||
|---|---|---|---|---|
| 0 | 1.56 | 3.13 | 6.25 | |
| I-16/I-23 | 375 ± 154 | 532 ± 118 | 1,219 ± 344b | 3,297 ± 991c |
| II-21/I-23 | 1,858 ± 401 | 2,412 ± 96b | 4,327 ± 322c | 7,531 ± 141c |
| II-12/I-23 | 1,306 ± 548 | 2,592 ± 874 | 3,563 ± 205c | 7,321 ± 116c |
| II-21/II-3 | 3,963 ± 473 | 6,164 ± 465c | 9,266 ± 521c | 13,906 ± 618c |
| I-23/II-12 | 782 ± 283 | 2,040 ± 66c | 4,561 ± 252c | 10,859 ± 183c |
All data are expressed as means ± SD (n = 3).
The value of the mean minus 1.0 SD is higher than that of the mean plus 1.0 SD of the control (0 pg/ml).
The value of the mean minus 2.0 SD is higher than that of the mean plus 2.0 SD of the control (0 pg/ml).
To assess the reproducibility of the best CLEIA, intra- and interassay variabilities were obtained with five fixed dilutions of recombinant N protein, namely 6.25, 12.5, 25, 50, and 100 pg/ml. The intra-assay variability was determined with replicate tests 20 times with five samples in the same CLEIA. The RLU values, means, and standard deviations for 6.25, 12.5, 25, 50, and 100 pg/ml were 10,864 ± 399, 23,208 ± 1,332, 48,410 ± 1,690, 106,482 ± 6,003, and 199,484 ± 718, respectively, with corresponding coefficients of variation of 3.6, 5.7, 3.4, 5.6, and 0.4. The interassay variability was determined by subjecting four samples to 10 different assays performed repeatedly 10 times on different days. The RLU values, means, and standard deviations for 6.25, 12.5, 25, 50, and 100 pg/ml were 10,839 ± 1,017, 24,292 ± 2,432, 51,139 ± 4,308, 110,387 ± 9,320, and 199,685 ± 791, respectively, and their coefficients of variation were 9.4, 10.0, 8.4, 8.4, and 0.4.
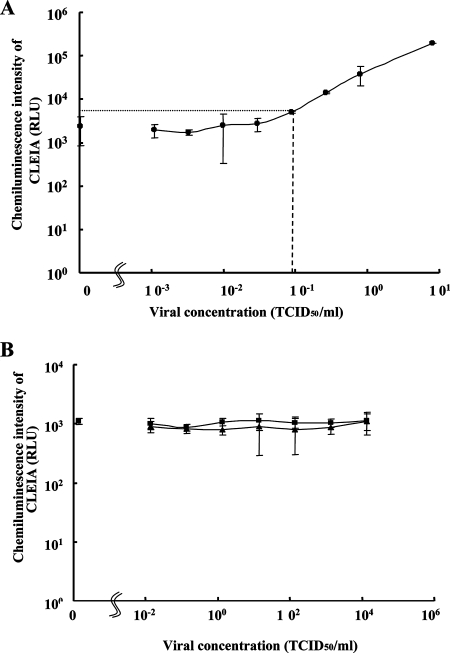
To examine the actual reactivity of the best CLEIA to cell culture SARS CoV and common human CoV strains (229E and OC43), serial dilutions of cell culture lysates from SARS CoV, 229E, and OC43 were used. As shown in Fig. 5A, the lowest positive result for detecting the SARS CoV using the cell lysate from cells infected with SARS CoV (HKU-39849) was with a viral concentration of 0.087 TCID50/ml, which corresponded to 111 copies/ml by Q-PCR (44). Neither 229E nor OC43 cell culture lysates even at the highest (10000 TCID50/ml) virus content gave detectable signals by the CLEIA with SARS CoV N antigen (Fig. 5B).
FIG. 5.
Sensitivity of CLEIA in detecting SARS CoV N antigen. (A) Limiting dilution experiment with cultured SARS CoV (HKU-39849) for detecting SARS CoV N antigen by CLEIA using I-23 as capture MAb and II-12 as detecting MAb. Twofold dilutions of cell culture lysates of SARS CoV were measured in triplicate by the CLEIA. Results show mean values of the triplicates. Error bars express the range of 2× standard deviation values. The limit of detection of CLEIA is 0.087 TCID50/ml (a coarse dotted line), which is the minimum concentration (TCID50/ml) of cultured SARS CoV (HKU-39849) giving the signal value of the mean minus 2.0 standard deviations, which is higher than that of the mean plus 2.0 standard deviations of 0.0 TCID50/ml (a fine dotted line). (B) Limiting dilution experiment with cell culture lysates of common human CoV (229E [▪] or OC43 [▴]). Serial dilutions of recombinant N protein were measured in triplicate by the CLEIA. Results show mean values of the triplicates. Error bars indicate 2× standard deviation values.
Detection of the viral N protein in NPA from SARS patients.
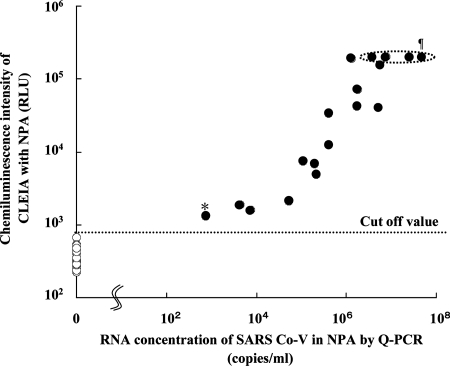
To evaluate the performance of the best CLEIA, 18 NPA samples from patients previously diagnosed as SARS positive by one-step Q-PCR and 20 SARS-negative NPA samples from SARS negative donors were measured by the best CLEIA. As shown in Fig. 6, the mean RLU and standard deviation for SARS-negative NPA was 363 ± 118. Thus, the cutoff value of the best CLEIA, defined as the mean plus three standard deviations, was 717. All eighteen SARS-positive NPA samples confirmed by Q-PCR were positive by the best CLEIA. Fourteen of them were above the maximum limit (200,000 RLU) of detection of the luminometer. In addition, a statistically significant (P < 0.01) correlation between the results of Q-PCR and those of the best CLEIA was observed with the 18 SARS-positive NPA (Fig. 6). Thus, our CLEIA had a sensitivity high enough to detect the minimum concentration (757 copies/ml by Q-PCR) of SARS-positive NPA. On the other hand, none of the twenty negative NPA from SARS-negative donors was positive by the best CLEIA. Thus, both the specificity and sensitivity of this CLEIA were 100%.
FIG. 6.
Correlation between CLEIA and Q-PCR in detecting SARS CoV N antigen using clinical samples obtained from SARS patients. In CLEIA, I-23 and II-12 MAbs are used as capture and detecting Abs, respectively. For the Q-PCR detection of SARS CoV RNA, Q-PCR was performed as previously reported (32). Eighteen clinical NPA samples from 18 SARS-positive patients (•) and 20 SARS-negative NPA samples (○) were analyzed. RNA was prepared from each NPA sample, and the SARS CoV RNA concentration was quantitatively evaluated by Q-PCR. The amount of SARS CoV N protein was measured by CLEIA as described above. Results show the means of triplicates for each specimen. Cutoff value of CLEIA, 718 RLU (as measured by the mean of negative specimens plus 3 standard deviations). *, the results with SARS-positive NPA with the minimum concentration of viral RNA by Q-PCR (757 copies/ml); ¶, the four dots in the ellipse drawn with a dotted line indicate the chemiluminescence intensity for four NPA samples which exceeded the maximum limit of detection by luminometer.
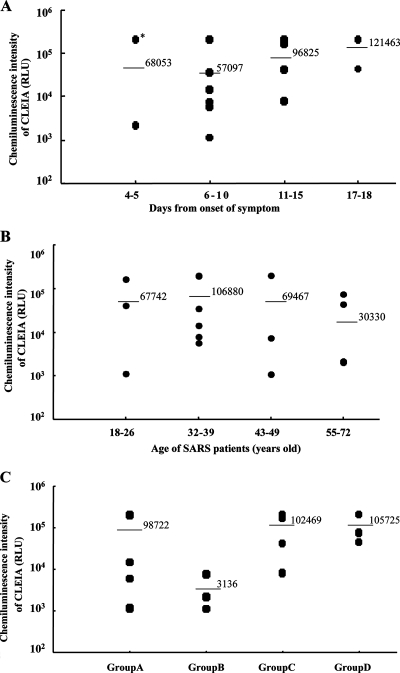
Statistical analysis with results of CLEIA on difference between data of onset and age.
In order to examine the relationship between the history of the NPA samples and the signals of CLEIA, some statistical analyses were performed with the results of the 18 SARS-positive patients. No significant difference was found between CLEIA data for the difference of age groups or the days from onset of symptoms. In spite of the days from the onset or the age group, the range of plotted CLEIA signals on diagrams was very wide in every group of SARS patients (Fig. 7A and B). But, in the results of samples from 4 to 10 days from the onset of symptoms, the mean of N protein levels in NPA from patients under 40 years old (group B) was 10 times higher than that for patients over 40 years old (group A). In the results of samples from 11 to 18 days from the onset of symptoms, there was no significant difference in N protein levels between samples from patients under 40 years old (group C) and over 40 years old (group D) (Fig. 7C). One NPA specimen (P16) in the patient group of the early stage (4 to 5 days from onset of symptoms) converted both a high level of N protein and a high level of viral load (2.43E + 07 copies/ml). The specimens from patients from 11 to 15 and 17 to 18 days from onset of symptoms in infection still gave the same high CLEIA signals as those from patients in the early stage (Fig. 7A). Those specimens also converted high levels of viral load (from 1.09E + 5 to 4.56E + 07 copies/ml) which were measured by Q-PCR.
FIG. 7.
N protein detection and the difference between the days from onset and age groups of SARS patients. (A) Chemiluminescence intensity of CLEIA with NPA from SARS patients who were classified into four groups by their days from the onset of symptoms (4 to 5, 6 to 10, 11 to 15, and 16 to 18 days). *, the CLEIA signal of patient P16 in the early stage (4 days from onset of symptoms) whose viral load is 2.43E + 07 copies/ml. (B) Chemiluminescence intensity of CLEIA with NPA from SARS patients who were classified into four groups by their ages (18 to 26, 32 to 39, 43 to 49, and 55 to 72 years). Chemiluminescence intensity (RLU) of the NPA from each SARS patient is expressed in its proper lane of panel A or B. Bars and numbers in each diagram indicate mean values of signals with specimens in each group. (C) Chemiluminescence intensity of CLEIA with NPA from SARS patients who were classified into four groups by a combination of their age and their days from the onset of symptoms. Group A, NPA from patients 18 to 39 years old and at 4 to 10 days from the onset of symptoms; group B, NPA from patients 44 to 57 years old and at 4 to 10 days from the onset of symptoms; group C, NPA from patients 24 to 35 years old and at 11 to 17 days from the onset of symptoms; group D, NPA from patients 43 to 71 years old and at 12 to 18 days from the onset of symptoms.
DISCUSSION
In this study, we have established a highly sensitive CLEIA for detecting SARS CoV N protein. Although a similar EIA system was already reported previously (2, 10, 20, 34), our CLEIA system using newly developed MAbs can detect as low as 1.56 pg/ml and 0.087 TCID50/ml of the recombinant N protein of SARS CoV and the cell culture lysate of virus-infected cells (111 copies/ml of viral RNA), respectively, whereas the limits of detection in previous studies using the recombinant N protein were 50 (2), 37.5 (10), 200 (20), and 1,000 (34) pg/ml, and those using the cell culture lysate of virus-infected cells were 15 (20) and 50 (10) TCID50/ml. Our novel SARS CLEIA gave higher sensitivity than those previous EIA tests. For example, the detection limit for SARS CoV in the infected cells using our method exhibited approximately 172-fold higher sensitivity than that of a previous EIA (20). In addition, the sensitivity of our CLEIA was comparable to the results with other methods using RT-PCR, such as the results reported from the World Health Organization (0.1 TCID50/ml) (13), results from a Realart HPA CoV assay (100 copies/ml of viral RNA), and appeared to be even better than results from a Roche LightCycler SARS CoV quantification assay (10,000 copies/ml of viral RNA) (13). Moreover, it showed no cross-reactivity even at higher concentrations of recombinant protein of conventional human CoV (229E, OC43, and NL63) and high-titer cell culture lysates from human CoV, 229E, and OC43 (Fig. 5B). The sensitivity and specificity are 100% when NPA were tested with our CLEIA. The sensitivity is 100%, but the number of negative samples tested was relatively small. However, considering the presence of a large discrepancy in the value of chemiluminescence (the y axis in Fig. 6 is in logarithmic scale), it is considered that the number of negative samples was sufficient.
In sharp contrast to the previously reported EIA systems using cocktail MAbs (2) and polyclonal antisera from immunized animals (20), our CLEIA characteristically uses a combination of two newly developed MAbs and has exhibited much higher sensitivity yet still with specificity. In order to achieve this goal, we have carefully evaluated the combination of various MAbs by testing all 25 different combinations and we identified the best combination of two such MAbs, I-23 and II-12: capture Ab I-23 reacted to the linear epitope located at the 283-to-422 amino acid residues of SARS CoV N protein, and detecting Ab II-12 reacted to another independent linear epitope located within the 1-to-141 amino acid residues of N protein. We consider the epitope is presumably linear because this recombinant nucleocapsid protein was denatured during SDS-polyacrylamide gel electrophoresis (PAGE) in Western blot analysis (Fig. 3). These epitopes are distinct from those reported previously (34), wherein an anti-N protein MAb that reacted to a peptide from the 249th to 317th amino acid residues of N protein and another MAb that reacted to a peptide from the 317th to 395th amino acid residues were used in a sandwich ELISA for detecting N protein at ng/ml orders (34). Identification of the MAb recognizing the C-terminal regions of N protein was reported by others (1); however, the sensitivity in a practical EIA system was not demonstrated. Thus, in our system, either better epitopes are reacted by MAbs I-23 and II-12 or this combination is superior to those of other previous reports (1, 34). The fact that a combination of MAbs I-23 and II-12 in conventional EIA by color detection could detect around 12 pg/ml of recombinant N protein at the detection limit using a color detection system (data not shown) suggests that the immunoreactivities of our newly developed MAbs are potent enough to develop a more-sensitive CLEIA system.
A number of epitope mapping studies on the N protein of human Abs from SARS patients have been conducted (3, 11, 22, 24, 39). The major epitopes of human Abs induced after SARS infection were found in the carboxyl-terminal region of N protein (3). Li et al. (22) reported that the linear epitope located within the carboxyl-terminal region of N protein spans from the 371st to 407th amino acid residue, as identified by protein structure analysis (22). In our study, approximately 27% of all MAbs recognized the amino-terminal region, from the 1st to 143rd amino acid residues, of N protein. On the other hand, approximately 60% of all MAbs recognized the carboxyl-terminal region, from the 283rd to 422nd amino acid residue, of N protein. Our MAb II-12 appears to have reactivity to that linear epitope within the amino-terminal region. It is noted that there has not been any report of a MAb reacting to the amino-terminal region of SARS CoV N protein. Therefore, our success in establishing a highly sensitive CLEIA system might be ascribed to the development of such a MAb reacting to the amino-terminal region of SARS CoV N protein. More detailed information about the epitopes of our five MAbs reported here should provide us with further information on how our MAbs recognize the viral N protein (7, 14, 16, 45).
Previous serological tests of NPA samples collected at 1 to 5 days and 6 to 10 days after onset of symptoms analyzed with EIA exhibited very low sensitivities of about 0% and 27%, respectively (20). In contrast, our CLEIA could detect lower concentrations of SARS CoV in NPA samples obtained at the early phase of infection. This feature is especially important when detection of SARS patients is urgent in order to isolate those who are infected with the virus for containment in epidemic areas.
The current conventional EIA tests could detect 80 to 90% of positive SARS cases with serum samples collected 1 to 5 days after onset of symptoms (4, 23), possibly because of the higher level of N proteins in serum during the early stage of infection. But a similar EIA test could detect the virus in NPA but not in serum from SARS patients in early infection, possibly because of higher viral load of SARS CoV in NPA (20). This is because the concentration of N protein in sera in the early phase could be higher than in NPA, urine, and stool (29). However, our newly established CLEIA could detect 3/3 (100%) and 8/8 (100%) NPA samples obtained at 1 to 5 days and 6 to 10 days after onset of symptoms, respectively. Although the sample sizes were small and all donors were Asian people (and thus a larger scale and extensive screening study is needed in the near future), it is clear that our CLEIA should be useful in detecting SARS infection using NPA samples obtained from patients.
In the results of statistical analyses of the differences between data from the onset and age and the detection, no significant difference was found between the levels of N protein in NPA by the differences of the age groups or the days from the onset of symptoms. But, a SARS patient in the early stage in infection (4 to 5 days from the onset of symptoms) has already converted high levels of both N protein and viral load in NPA (Fig. 7). According to a previous report (20), the levels of N protein in NPA from patient groups in the early stage in infection (1 to 5 days from the onset of symptoms) are thought to be very low on the whole. But, our results could suggest that there are some patients who convert high levels of N protein in NPA during the early stage in infection.
On the other hand, two SARS patients in the late stage in infection (17 to 18 days from the onset of symptoms) still converted high levels of both N protein and viral load in NPA in our results (Fig. 7). According to previous reports (8, 15), the viral loads, which have good correlation with levels of N protein (Fig. 6.), in all serum/plasma from patients in the late stage in infection (after 10 or 13 days from the onset of symptoms) are very low (under 1E + 03 copies/ml). Considering those facts, the ability of our simple CLEIA system to detect the N protein with NPA samples might be useful in monitoring the clinical efficacies of therapy and the risk of viral transmission from patients. It could also provide important information to decide when the patients should be isolated and discharged from the isolation ward.
In the results of statistical analyses with NPA from patients in the early stage (4 to 10 days from the onset of symptom), the mean of N protein levels in NPA from patients under 40 years old was 10 times higher than that from patients over 40 years old (Fig. 7C). As the World Health Organization reported, death rates are increased in accordance with the increase of the ages of SARS patients (43). The lower levels of viral loads in NPA from older patients in the early stage of infection might relate to the higher death rates of older patients.
Acknowledgments
We thank Marni Cueno and Ann-Florence Victoriano for their proofreading.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Bussmann, B. M., S. Reiche, L. H. Jacob, J. M. Braum, and C. Jassoy. 2006. Antigenic and cellular location analysis of the severe acute respiratory syndrome coronavirus nucleocapsid protein using monoclonal antibodies. Virus Res. 122119-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Che, X. Y., L. W. Qie, Y. X. Pan, K. Wen, W. Hao, L. Y. Zhang, Y. D. Wang, Z. Y. Liao, C. C. Cheng, and K. Y. Yuen. 2004. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J. Clin. Microbiol. 422629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow, S. C. S., C. Y. S. Ho, T. T. T. Tam, C. Wu, T. Cheung, P. K. S. Chan, M. H. Ng, P. K. Heu, H. K. Ng, D. M. Y. Au, and A. W. I. Lo. 2006. Specific epitopes of the structural and hypothetical proteins elicit variable humoral response in SARS patients. J. Clin. Pathol. 59468-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di, B., W. Hao, Y. Gao, M. Wang, Y. D. Wang, L. W. Qiu, K. Wen, D. H. Zhou, X. W. Wu, E. J. Lu, Z. Y. Liao, Y. B. Mei, B. J. Zheng, and X. Y. Che. 2005. Monoclonal antibody-based antigen capture enzyme-linked immunosorbent assay reveals high sensitivity of the nucleocapsid protein in acute-phase sera of severe acute respiratory syndrome patients. Clin. Diagn. Lab. Immunol. 12135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant, P. R., J. A. Garson, R. S. Tedder, P. K. Chan, J. S. Tam, and J. J. Sung. 2003. Detection of SARS coronavirus in plasma by real-time RT-PCR. N. Engl. J. Med. 3492468-2469. [DOI] [PubMed] [Google Scholar]
- 6.Guan, Y., J. S. Peiris, B. Zheng, L. L. Poon, K. H. Chan, F. Y. Zeng, C. W. Chan, M. N. Chan, J. D. Chen, K. Y. Chow, C. C. Hon, K. H. Hui, J. Li, V. Y. Li, Y. Wang, S. W. Leung, K. Y. Yuen, and F. C. Leung. 2004. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet 36399-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo, J. P., M. Petric, W. Campbell, and P. L. McGreer. 2004. SARS corona virus peptides recognized by antibodies in the sera of convalescent cases. Virology 324251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haibin, W., Y. Mao., L. Ju, J. Zhang, Z. Liu, X. Zhou, Q. Li, Y. Wang, S. Kim, and L. Zhang. 2004. Detection and monitoring of SARS coronavirus in the plasma and peripheral blood lymphocyte of patients with severe acute respiratory syndrome. Clin. Chem. 501237-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamre, D., and J. J. Procknow. 1966. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121190-193. [DOI] [PubMed] [Google Scholar]
- 10.He, Q., Q. Du, S. Lau, I. Manopo, L. Lu, S. W. Chan, B. J. Fenner, and J. Kwang. 2005. Characterization of monoclonal antibody against SARS coronavirus nucleocapsid antigen and development of an antigen capture ELISA. J. Virol. Methods 12746-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, Y., Y. Zhou, H. Wu, Z. Kou, S. Liu, and S. Jiang. 2004. Mapping of antigeneic sites on the nucleocapsid protein of the severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 425309-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoek, L., K. Pyrc., M. Jebbink, W. Vermeulen-Oost, R. J. M. Berkhoust, K. C. Wolthers, P. M. E. W. Dillen, J. Kaapdrop, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hourfar, M. K., W. K. Roth, E. Seifried, and M. Schmidt. 2004. Comparison of two real-time quantitative assays for detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 422094-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, Q., L. Yu, A. M. Petros, A. Gunasekera, Z. Liu, N. Xu, P. Hajduk, J. Mark, S. W. Fesik, and E. T. Olejniczak. 2004. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry 436059-6063. [DOI] [PubMed] [Google Scholar]
- 15.Hung, I. F. N., V. C. C. Cheng, A. K. L. Wu, B. S. F. Tang, K. H. Chan, C. M. Chu, M. M. L. Woung, W. T. Hui, L. L. M. Poon, D. M. W. Tse, K. S. Chan, P. C. Y. Woo, S. K. P. Lau, J. S. M. Peiris, and K. Y. Yuen. 2004. Viral loads in clinical specimens and SARS manifestations. Emerg. Infect. Dis. 101550-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayaram, H., H. Fan, B. R. Bowman, A. Ooi, J. Jayaram., E. W. Collisson, J. Lescar, and B. V. V. Prasad. 2006. X-ray structure of the N- and C-terminal domains of coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 806612-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kogai, H., Y. Uchida, N. Fujii, Y. Kurano, K. Miyake, Y. Kido, H. Kariwa, I. Takashima, H. Tamashiro, A. E. Ling, and M. Okada. 2005. Novel rapid immunochromatographic test based on enzyme immunoassay for detecting nucleocapsid antigen in SARS-associated coronavirus. J. Clin. Lab. Anal. 19150-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256495-497. [DOI] [PubMed] [Google Scholar]
- 19.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 3481953-1966. [DOI] [PubMed] [Google Scholar]
- 20.Lau, S. K. P., P. C. Y. Woo, B. H. L. Wong, H. W. Tsoi, G. K. S. Woo, R. W. S. Poon, K. H. Chan, W. I. Wei, J. S. M. Peiris, and K. Y. Yuen. 2004. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in SARS patients by enzyme-linked immunosorbent assay. J. Clin. Microbiol. 422884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung, D. T. M., C. H. Tam, C. H. Ma, P. K. S. Chen, J. L. K. Cheung, H. Niu, J. S. L. Tam, and P. L. Lim. 2004. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 190379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, S., L. Lin, H. Wang, J. Yin, Y. Ren, Z. Zhao, J. Wen, C. Zhou, X. Zhang, X. Li, J. Wang, Z. Zhou, J. Liu, J. Shao, T. Lei, J. Fang, N. Xu, and S. Liu. 2003. The epitope study on the SARS-CoV nucleocapsid protein. Genomics Proteomics Bioinformatics 1198-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Y. H., J. Li, W. E. Liu, L. Wang, T. Li, Y. H. Zhou, and H. Zhuaung. 2005. Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: comparison with results of other viral makers. J. Virol. Methods 13045-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, Y., Y. Wan, L. W. Qiu, J. Zhou, B. Ni, B. Geo, Q. Zou, L. Zou, W. Zhou, Z. Jia, X. Y. Che, and Y. Wu. 2005. Comprehensive antibody epitope mapping of the nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus: insight into the humoral immunity of SARS. Clin. Chem. 511382-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, X., Y. Shi, P. Li, L. Li, Y. Yi, Q. Ma, and C. Cao. 2004. Profile of antibodies to the nucleocapsid protein of the severe acute respiratory syndrome (SARS)-associated coronavirus in probable SARS patients. Clin. Diagn. Lab. Immunol. 11227-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 3001399-1404. [DOI] [PubMed] [Google Scholar]
- 27.McIntosh, K., J. H. Dees, W. B. Becker, A. L. Kapikian, and R. M. Chanock. 1967. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc. Natl. Acad. Sci. USA 57933-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peiris, J. S., S. T. Lai, L. L. M. Poon, Y. Guan, Y. L. Y. C. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, K. Y. Yuen, and the SARS Study Group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 3611319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peiris, J. S. M., C. M. Chu, V. C. C. Cheng, K. S. Chan, I. F. N. Hung, L. L. M. Poon, K. I. Law, B. S. F. Tang, T. Y. W. Hon, C. S. Chan, K. H. Chan, J. S. C. Ng, B. J. Zheng, R. W. M. Lai, Y. Guan, K. Y. Yuen, and the HKU/UCH SARS Study Group. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 3611767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon, L. L. L., K. H. Chan, O. K. Wong, W. C. Yam, K. Y. Yuen, Y. Guan, Y. M. D. Lo, and J. S. M. Peiris. 2003. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J. Clin. Virol. 28233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poon, L. L. M., B. W. Y. Wong, K. H. Chan, S. S. F. Ng, K. Y. Yuen, Y. Guan, and J. S. M. Peiris. 2005. Evaluation of real-time reverse transcriptase PCR and real-time loop-mediated amplification assays for severe acute respiratory syndrome coronavirus detection. J. Clin. Microbiol. 433457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poon, L. L. M., K. H. Chan, O. K. Wong, T. K. W. Cheung, I. Ng, B. Zheng, W. H. Set, K. Y. Yuen, Y. Guan, and J. S. M. Peiris. 2004. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin. Chem. 5067-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogel, S. Penaranda, B. Bankamp, K. Maher, M.-H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, T. G. Erdman, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of novel coronavirus associated with severe acute respiratory syndrome. Science 3001394-1398. [DOI] [PubMed] [Google Scholar]
- 34.Shang, B., X. Y. Wang, J. W. Yuan, A. Vabret, X. D. Wu, R. F. Yang, R. L. Tian, Y. Y. Ji, V. Deuble, and B. Sun. 2005. Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus. Biochem. Biophys. Res. Commun. 336110-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, Y., Y. Yi, P. Li, T. Kuang, L. Li, M. Dong, M. Ma, and C. Cao. 2004. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 415781-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thai, H. T. C., M. Q. L. Le, C. D. Vuong, M. Parida, H. Minekawa, T. Notori, F. Hasebe, and K. Morita. 2004. Development and evaluation of novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 421956-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timani, K. A., L. Ye., L. Ye, Y. Zhu, Z. Wu, and Z. Gong. 2004. Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J. Clin. Virol. 30309-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchiya, Y., K. Morioka, J. Shirai, K. Yoshida, and S. Inumaru. 2006. Comparison of artificial synthesis methods of gene. Nucleic Acids Symp. Ser. 50275-276. [DOI] [PubMed] [Google Scholar]
- 39.Wang, J., J. Ji, J. Ye, X. Zhao, J. Wen, W. Li, J. Hu, D. Li, M. Sun, H. Zeng, Y. Hu, X. Tian, X. Tan, N. Xu, C. Zeng, J. Wang, S. Bi, and H. Yang. 2003. The structure analysis and antigenicity study of N protein of SARS-CoV. Genomics Proteomics Bioinformatics 1145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woo, P. C. Y., S. K. P. Lau, B. H. L. Wong, K. H. Chan, C. M., H. W. Tsoi, Y. Huang, J. S. M. Peiris, and K. Y. Yuen. 2004. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin. Diagn. Lab Immunol. 11665-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo, P. C. Y., S. K. P. Lau, B. H. L. Wong, H. W. Tsoi, A. M. Y. Fung, K. H. Chan, K. P. Tam, J. S. M. Peiris, and K. Y. Yuen. 2004. Detection of specific antibodies to severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 422306-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woo, P. C. Y., S. K. P. Lau, B. H. L. Wong, H.-W. Tsoi, A. M. Y. Fung, R. Y. T. Kao, K.-H. Chan, J. S. M. Peiris, and K.-Y. Yuen. 2005. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 433054-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.World Health Organization. 2003. Consensus document on the epidemiology of severe acute respiratory syndrome (SARS). World Health Organization, Geneva, Switzerland.
- 44.Yam, W. C., K. H. Chan, L. L. M. Poon, Y. Guan, K. Y. Yuen, W. H. Seto, and J. S. M. Peiris. 2003. Evaluation of reverse transcription-PCR assay for rapid diagnosis of severe acute respiratory syndrome associated with novel coronavirus. J. Clin. Microbiol. 414521-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu, I. M., M. L. Oldham, J. Zhang, and J. Chen. 2006. Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona- and arteriviridae. J. Biol. Chem. 28117134-17139. [DOI] [PMC free article] [PubMed] [Google Scholar]