Abstract
Candida nivariensis was isolated from an Indonesian human immunodeficiency virus-infected patient who suffered from oropharyngeal candidiasis and was identified with molecular tools. Our isolate demonstrated low MICs to amphotericin B, flucytosine, posaconazole, caspofungin, and isavuconazole and was susceptible to fluconazole, itraconazole, and voriconazole.
CASE REPORT
In this article, we report on the isolation of Candida nivariensis from a human immunodeficiency virus (HIV)-infected patient in Indonesia who suffered from oropharyngeal candidiasis (OPC). In the course of a molecular epidemiology study of yeasts isolated from Indonesian HIV-infected patients, we noted a yeast isolate that did not fit any conventional clinically relevant yeast species using amplified fragment length polymorphism (AFLP) typing (4), randomly amplified polymorphic DNA (RAPD) fingerprinting, and sequence analysis of the D1/D2 domains of the 26S ribosomal DNA (rDNA) and internal transcribed spacer (ITS) 1 and 2 regions. Only when C. nivariensis was described (1) could the Indonesian isolate be identified with this species, which consequently appears to have a wider geographical and clinical distribution than previously known. In addition, we compared partial rDNA sequences of the Indonesian isolate with those of the type strains of C. nivariensis and Candida bracarensis (8) and we performed DNA-DNA reassociation experiments (17) between the genomic DNAs of these species in order to validate their taxonomic status. In addition, in vitro susceptibility to eight antifungal agents was determined.
In December 2004, a 28-year-old male who used drugs intravenously by sharing needles and who had needle tracks on both arms was admitted to the hospital due to fever and cough from which he had suffered for a month. He was complaining about difficulties with the intake of food due to pain, and he had had a burning sensation in his mouth for the last 2 weeks. He suffered from diarrhea and lost weight during these 2 months. At the time of admission, he was diagnosed as infected with HIV.
Physical examination revealed a severely ill, but still fully alert and very thin, cachectic patient with liver enlargement and pneumonia suspected to be tuberculosis. Temperature on admission was 39°C, respiration rate was 20 breaths per min, and heart rate was 120 beats per min, with pale conjunctivae and unicteric sclerae. Examination of the oral cavity revealed diffuse erythematous lesions with white plaque occurring especially around the edge of the tongue, with pus-like excreta. Further physical examination revealed rales in both lungs, with no wheezing and no lymph node enlargement. Laboratory investigations revealed values for hemoglobin of 6.7 g/dl, a leukocyte count of 6,500/mm3, a platelet count of 220,000/mm3, and a CD4 count of 104 cells/mm3. The patient died before undergoing complete examination and having proper treatment. This patient suffered from sepsis that might be due to unidentified pneumonia with differential diagnosis of lung tuberculosis. Moreover, OPC, wasting syndrome with chronic diarrhea, HIV infection, and liver enlargement of unidentified cause were observed.
On CHROMagar Candida (Paris, France), yeasts from the oral rinses grew in a concentration of 720 colonies/ml and were consistent with Candida glabrata. Identification was further done with the Auxacolor2 colorimetric sugar assimilation test (Bio-Rad, Marnes-la-Coquette, France). The Indonesian isolate and the type strain of C. nivariensis (CBS 9983), however, showed a profile (10000-04) consistent with Candida inconspicua. Both C. bracarensis (CBS 10154) and C. glabrata (CBS 138) gave the profile 10400-04, consistent with C. glabrata. Candida glabrata may be identified quickly with the Glabrata RTT test (Fumouze Diagnostics, Levallois-Perret, France). Strains with a positive signal for trehalase combined with negative maltose utilization are identified as C. glabrata. By using the RTT test, the type strains of C. nivariensis and C. bracarensis and the clinical isolate from Indonesia were negative, whereas the type strain of C. glabrata was positive. Because it was claimed that C. glabrata is a trehalose-nonfermenting species (1), this feature was tested using standard 2% trehalose Durham tubes at 25°C and using glucose fermentation as a positive control. In short, the type strain of C. glabrata CBS 138 fermented trehalose after approximately 2 days at 25°C, the type strain of C. bracarensis CBS 10154 showed trehalose fermentation after 6 to 7 days, and the two investigated strains of C. nivariensis, namely the type strain CBS 9983 and the Indonesian isolate CBS 10161, took 7 to 8 days for a clear fermentation reaction. Thus, all of the species in the C. glabrata complex are able to ferment trehalose, albeit at different rates.
Molecular identification of the isolate was performed by sequence analysis of the D1/D2 domains and the ITS 1 and 2 spacer regions of the rDNA according to Okoli et al. (14). To further assess the identification of our isolate with the species belonging to the C. glabrata complex, molar percentages of G+C of the nuclear DNA of these species and DNA-DNA reassociation values between the DNAs of the species were compared. Therefore, the DNA of C. glabrata CBS 138, C. nivariensis CBS 9983, C. bracarensis CBS10154, and the Indonesian isolate, C-127 (CBS 10161), was isolated and analyzed as described previously (17). Molecular identification by sequence analysis of the D1/D2 domains of the 26S rDNA and the ITS 1 and 2 regions of the rDNA revealed that the Indonesian yeast was identical to C. nivariensis (accession numbers for strain CBS 10154 are as follows: D1/D2 25S rDNA, EF056323; and ITS 1 and 2, EF056322). The ITS sequence completely matched that of the type strain of this species, and D1/D2 showed 99.5% similarity. The type strains of C. glabrata and C. bracarensis agreed at 99.5 and 97.3% for the D1/D2 domains and 70.0 and 80.2% for the ITS 1 and 2, respectively. Therefore, the identification of representatives belonging to these closely related species requires analysis of the ITS 1 and 2 regions as the D1/D2 domains showed too little nucleotide divergence. The G+C values of the strains investigated were as follows: C. glabrata CBS 138, 34.4 mol%; C. nivariensis CBS 9983, 33.5 mol%; C. bracarensis CBS 10154, 35.5 mol%; and Indonesian isolate C-127 (CBS 10161), 34.7 mol%. Hence, it is clear that this compositional genome parameter cannot be used to discriminate among these species. DNA-DNA reassociation experiments revealed the following data. DNA of the Indonesian isolate C-127 showed 8.1, 22.7, and 97.6% DNA-DNA similarity to those of the type strains of C. bracarensis, C. glabrata, and C. nivariensis, respectively. Candida nivariensis showed 11.5 and 26.9% similarity to C. glabrata and C. bracarensis, and C. bracarensis showed 15.7% similarity to C. glabrata. These data clearly support the recognition of C. nivariensis and C. bracarensis as distinct species next to C. glabrata and, moreover, indicate that the Indonesian isolate belonged to C. nivariensis.
Intraspecific genetic variation was assessed by AFLP as described by Borst et al. (4) and RAPD typing and PCR fingerprinting (3, 11). The PCR primers used were GTG(5) and M13 (5′-GAGGGTGGCGGTTCT) for the PCR fingerprinting and OPA-18 (5′-AGCTGACCGT), OPE-04 (5′-GTGACATGCC), and OPE-18 (5′-GGACTGCAGA) for the RAPD analysis. PCRs were performed as described previously for GTG(5) and M13 (3) and for the other three primers according to reference 3. PCR products were run on a 1.5% agarose gel with Massruler Express DNA ladder mix (Fermentas International, Inc., St. Leon-Rot, Germany) as a size marker. The AFLP and RAPD data, however, clearly showed that the Indonesian isolate of C. nivariensis is genotypically distinct from the European strains (Fig. 1 and 2).
FIG. 1.
AFLP banding patterns of Candida nivariensis, C. bracarensis, and C. glabrata. The Indonesian isolate (CBS 10161 = C-127) shows the highest resemblance to the type strain of C. nivariensis and not to that of C. glabrata or C. bracarensis. However, the Indonesian isolate of C. nivariensis differs considerably genotypically from that of the type strain, CBS 9983, of the species.
FIG. 2.
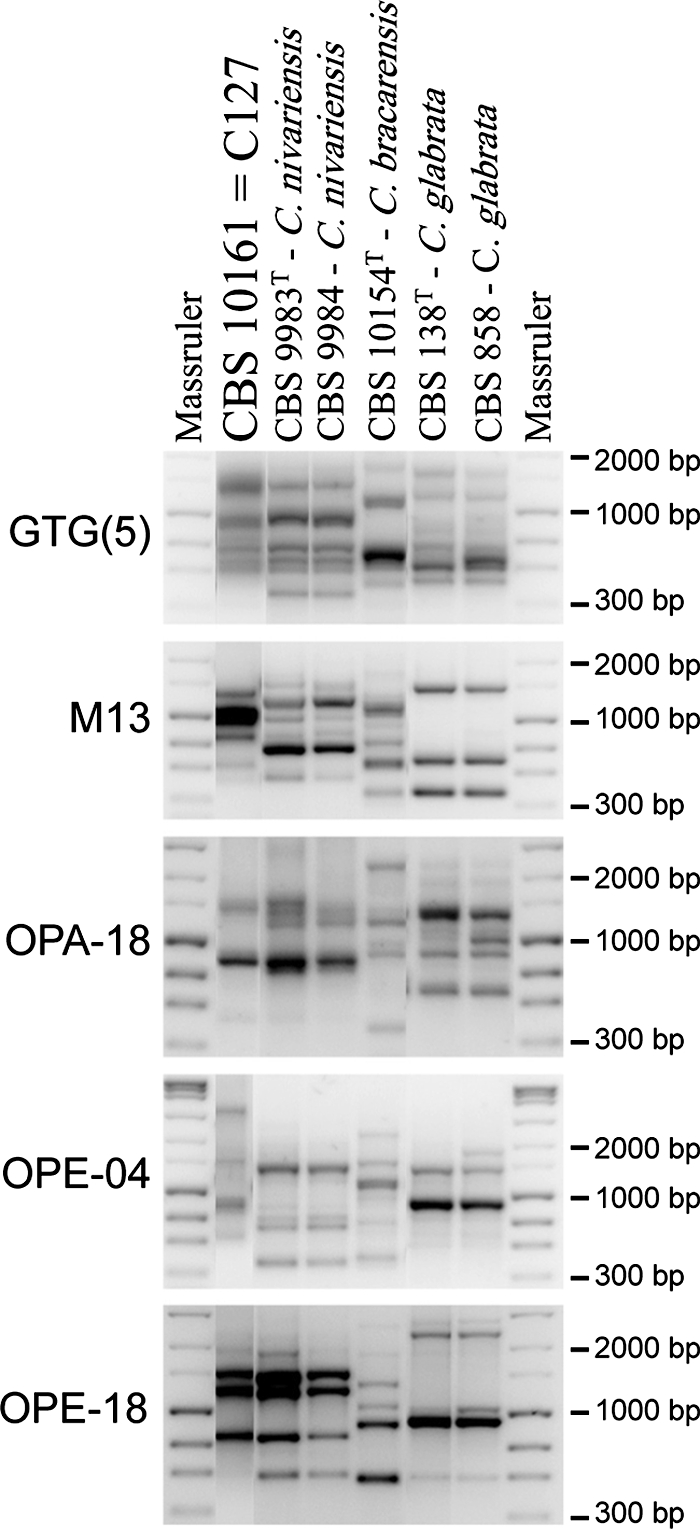
PCR fingerprint [GTG(5) and M13] and RAPD (OPA-18, OPE-04, and OPE-18) banding patterns of the Indonesian isolate of C. nivariensis (CBS 10161 = C-127), two European strains of C. nivariensis (CBS 9983T and CBS 9984), C. bracarensis (CBS 10154T), and two strains of C. glabrata (CBS 138T and CBS 858). It can be seen that the banding pattern of the Indonesian isolate is closer to that of the two strains of C. nivariensis than to that of C. bracarensis and C. glabrata. However, the banding patterns from the three C. nivariensis isolates indicate the presence of considerable genotypic diversity in this species.
Tests to determine the in vitro susceptibility of the strains to amphotericin B (AMB; Bristol-Myers Squibb, Woerden, The Netherlands), 5-flucytosine (5FC; Valeant Pharmaceuticals, Zoetermeer, The Netherlands), itraconazole (Itra; Janssen Research Foundation, Beerse, Belgium), fluconazole (Fluco) and voriconazole (Vori) (both Pfizer Central Research, Sandwich, United Kingdom), posaconazole (Posa; Schering-Plough, Kenilworth, NJ), caspofungin (Caspo; Merck Sharp & Dohme BV, Haarlem, The Netherlands), and isavuconazole (Isavu; BAL 4815) (Basilea Pharmaceuticals, Basel, Switzerland) were conducted in accordance with the guidelines in CLSI (formerly NCCLS) document M27-A2 (12). In brief, RPMI 1640 medium with l-glutamine without bicarbonate (GIBCO BRL, Life Technologies, Woerden, The Netherlands), buffered to pH 7 with 0.165 M 3-N-morpholinepropanesulfonic acid (Sigma-Aldrich Chemie, Steinheim, Germany), was used for microdilution studies. The inocula were prepared from 24-h fresh Sabouraud dextrose agar cultures; the cells were harvested in medium and diluted to about 1 × 103 to 5 × 103 cells/ml determined by spectrophotometer. The plates were incubated at 35°C for 24 h. The final concentrations of the antifungal agents were 0.016 to 8 μg/ml for AMB, Itra, Vori, Posa, and Caspo; 0.063 to 32 μg/ml for 5FC and Fluco; and 0.004 to 4.00 μg/ml for Isavu. Drug-free and yeast-free controls were included. Following incubation, the MIC at 24 h was determined visually and spectrophotometrically as the lowest concentration of drug showing absence of growth or a ≥50% reduction of growth compared with that of a growth control (drug-free well) for AMB and 5FC and azoles, respectively. Caspo was read visually as minimal effective concentration. Candida krusei ATCC 6258 and C. parapsilosis ATCC 22019 strains were used for quality control (12). Using broth microdilution testing, MICs (μg/ml) for C. bracarensis (CBS 10154) were as follows: AMB, 0.5 to 1; 5FC, 0.063; Fluco, 4; Itra, 0.25; Vori, 0.063 to 0.125; Posa, 0.25; Caspo, 1; and Isavu, 0.016. For the type strain of C. nivariensis (CBS 9983), the MICs (μg/ml) were as follows (in duplicate tests): AMB, 1; 5FC, 0.25; Fluco, 8; Itra, 0.5; Vori, 0.125; Posa, 0.5; Caspo, 0.5; and Isavu, 0.031. The MICs (μg/ml) for the Indonesian isolate of C. nivarienses (CBS 10161) were as follows: AMB, 1; 5FC, 0.125; Fluco, 2; Itra, 0.125; Vori, 0.063; Posa, 0.25; Caspo, 1; and Isavu, 0.008. According to the interpretive guidelines of the CLSI (formerly NCCLS) (12), our isolate was susceptible to Fluco, Vori, and Itra and had low MICs for the other drugs, for which no breakpoints have been established yet. Compared to C. albicans, C. glabrata is known to be less sensitive to triazoles, with about 10 to 15% resistance to Fluco. Importantly, geographic differences in susceptibility have been observed to occur among C. glabrata strains from the United States and the Asia Pacific area, with the strains originating from the United States being more resistant to Fluco (15). More strains of C. nivariensis need to be analyzed to get a complete picture of its resistance profile.
OPC is a common opportunistic fungal infection in HIV-infected patients. The introduction of highly active antiretroviral therapy has reduced its incidence significantly, but OPC remains important as it may be indicative of infection with HIV (5, 7, 9). Candida albicans is the predominant cause of OPC, even though the non-albicans Candida species, such as C. glabrata, C. tropicalis, C. parapsilosis, C. krusei, and C. dubliniensis can cause the disease (13, 16). Furthermore, Candida dubliniensis, which has similar phenotypic characteristics to C. albicans, is also reported as a cause of OPC in HIV-infected patients (6, 16).
Recently, two new species have been described that are closely related to C. glabrata. These are C. nivariensis (1) and C. bracarensis (8). Candida nivariensis was isolated from three patients with deep-seated infections in Spain and phenotypically resembled C. glabrata. The species can be differentiated from C. glabrata by sequence differences in the D1/D2 domains of the 26S rDNA and the ITS 1 and 2 regions. In addition, colony color on CHROMagar was found to be different and the species is able to ferment trehalose (1). A set of ITS-based primers was developed to differentiate the species from other clinically important Candida yeasts, especially C. glabrata (2). Recently, C. nivariensis was also reported from a catheter-related infection in a Japanese woman (10). The second species, C. bracarensis, was isolated from patients suffering from vulvovaginal candidiasis and from blood samples from Portugal and the United Kingdom (8). Candida bracarensis could be differentiated from C. glabrata by assimilation of l-lysine and from Kluyveromyces delphensis by assimilation of l-lysine and α,α-trehalose, the ability to ferment α,α-trehalose, growth at temperatures above 40°C, and its inability to assimilate ethanol (8).
C. glabrata seems to represent a species complex. A correct identification of the species is important for appropriate treatment and may be important for the management of the patients infected by the various C. glabrata look-alike species. Due to limited phenotypic differences, it is preferable to use molecular diagnostics to identify these species (2). As the D1/D2 domains of the 26S rDNA showed limited nucleotide divergence, analysis of the ITS 1 and 2 regions is required for a reliable identification. Moreover, as indicated above, all species are able to ferment trehalose.
In conclusion, we isolated C. nivariensis from an Indonesian HIV-infected patient who suffered from OPC. The isolate proved to be C. nivariensis by sequence analysis of the D1/D2 domains of the 26S rDNA and ITS 1 and 2 regions of the rDNA, as well as DNA-DNA reassociation experiments. In vitro susceptibility testing showed that this C. nivariensis isolate had low MICs to eight antifungal drugs.
Acknowledgments
R.W. was supported by a SPIN mobility grant (03-MP-09) received from the Royal Netherlands Academy of Arts and Sciences.
Footnotes
Published ahead of print on 14 November 2007.
REFERENCES
- 1.Alcoba-Flórez, J., S. Méndez-Álvarez, J. Cano, J. Guarro, E. Pérez-Roth, and M. del Pilar Arévalo. 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 434107-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcoba-Flórez, J., M. del Pilar Arévalo, F. J. González-Paredes, J. Cano, J. Guarro, E. Pérez-Roth, and S. Méndez-Álvarez. 2005. PCR protocol for specific identification of Candida nivariensis, a recently described pathogenic yeast. J. Clin. Microbiol. 436194-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bautista-Muñoz, C., X. M. Boldo, L. Villa-Tanaca, and C. Hernández-Rodríguez. 2003. Identification of Candida spp. by randomly amplified polymorphic DNA analysis and differentiation between Candida albicans and Candida dubliniensis by direct PCR methods. J. Clin. Microbiol. 41414-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, A., B. Theelen, E. Reinders, T. Boekhout, A. C. Fluit, and P. H. M. Savelkoul. 2003. Use of amplified fragment length polymorphism analysis to identify medically important Candida spp., including C. dubliniensis. J. Clin. Microbiol. 411357-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bravo, I. M., M. Correnti, L. Escalona, M. Perrone, A. Brito, V. Tovar, and H. Rivera. 2006. Prevalence of oral lesions in HIV patients related to CD4 cell count and viral load in a Venezuelan population. Med. Oral. Patol. Oral. Cir. Bucal. 11E1-E5. [PubMed] [Google Scholar]
- 6.Coleman, D. C., D. J. Sullivan, D. E. Bennet, G. P. Moran, H. J. Barry, and D. B. Shanley. 1997. Candidisasis: the emergence of novel species, Candida dubliniensis. AIDS 11557-566. [DOI] [PubMed] [Google Scholar]
- 7.Coogan, M. M., J. Greenspan, and S. J. Challacombe. 2005. Oral lesion in infection with human immunodeficiency virus. Bull. W. H. O. 83700-706. [PMC free article] [PubMed] [Google Scholar]
- 8.Correia, A., P. Sampaio, S. James, and C. Pais. 2006. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int. J. Syst. Evol. Microbiol. 56313-317. [DOI] [PubMed] [Google Scholar]
- 9.de Repentigny, L., D. Lewandowski, and P. Jolicoeur. 2004. Immunopathogenesis of oropharyngeal candidiasis in human immunodeficiency virus infection. Clin. Microbiol. Rev. 17729-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita, S., Y. Senda, T. Iwagami, H. Miyazaki, Y. Oota, H. Takada, K. Yamada, and M. Kawano. 2007. Catheter-related fungemia due to fluconazole-resistant Candida nivariensis. J. Clin. Microbiol. 453459-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer, W., and T. G. Mitchell. 1995. Polymerase chain reaction fingerprinting in fungi using single primers specific to minisatellites and simple repetitive DNA sequences: strain variation in Cryptococcus neoformans. Electrophoresis 161648-1656. [DOI] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A2. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 13.Odds, F. C. 1988. Candida and candidiasis, 2nd ed. Bailliere Tindall, London, United Kingdom.
- 14.Okoli, I., C. A. Oyeka, K. J. Kwon-Chung, B. Theelen, V. Robert, J. Z. Groenewald, D. C. McFadden, A. Casadevall, and T. Boekhout. 2006. Cryptotrichosporon anacardii gen. nov., sp. nov., a new trichosporonoid capsulate basidiomycetous yeast from Nigeria that is able to form melanin on Niger seed agar. FEMS Yeast Res. 7339-350. [DOI] [PubMed] [Google Scholar]
- 15.Pfaller, M. A., S. A. Messer, L. Boyken, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2004. Geographic variation in the susceptibilities of invasive isolates of Candida glabrata to seven systematically active antifungal agents: a global assessment from the ARTEMIS antifungal surveillance program conducted in 2001 and 2002. J. Clin. Microbiol. 423142-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruhnke, M. 2002. Skin and mucous membrane infections, p. 307-325. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, DC.
- 17.Smith, M. T., A. W. A. M. de Cock, G. A. Poot, and H. Y. Steensma. 1995. Genome comparisons in the yeastlike fungal genus Galactomyces Redhead et Malloch. Int. J. Syst. Bacteriol. 45826-831. [DOI] [PubMed] [Google Scholar]



