Abstract
We screened 457 Haemophilus influenzae strains isolated in Japan during 2002 to 2004 and identified 12 fluoroquinolone-resistant strains (2.6%). The resistant strains were divided into three genotypes (eight, three, and one of each type). These were isolated from patients over 58 years of age. Several fluoroquinolone-resistant clones appeared to have invaded the population of elderly patients in a particular area, Sapporo city.
Haemophilus influenzae, as well as Streptococcus pneumoniae, is a major causative agent of respiratory and otolaryngology infection, particularly community-acquired pneumonia in elderly persons and otitis media and sinusitis in children. Recently, there has been an increase worldwide in the prevalence of two types of β-lactam-resistant H. influenzae isolates: those that have acquired β-lactamases and those that carry mutations in the penicillin binding proteins. The latter are termed β-lactamase-negative ampicillin-resistant (BLNAR) strains. The recent increase in BLNAR strains has become a severe problem, leading to community-acquired infections (8, 19). Fluoroquinolone-resistant H. influenzae isolates have occasionally been reported worldwide (1, 4, 13, 15), but they have rarely been reported in Japan (9, 10). Fluoroquinolones are effective against the causative agents of atypical pneumonia, such as Chlamydophila pneumoniae and Mycoplasma pneumoniae, as well as those of typical bacterial pneumonia, S. pneumoniae and H. influenzae. The fluoroquinolones are considered to be candidates for the first choice of antimicrobial agents in cases of community-acquired pneumonia and otitis media in adults, so the emergence of fluoroquinolone-resistant H. influenzae and S. pneumoniae is a concern. Quinolone resistance is imparted by mutations in a particular domain referred to as the quinolone-resistance-determining region (QRDR) of one or both of the principal target enzymes, DNA gyrase (an A2B2 complex encoded by the gyrA and gyrB genes) and topoisomerase IV (a C2E2 complex encoded by the parC and parE genes).
In the present study, we screened and characterized fluoroquinolone-resistant H. influenzae strains isolated in clinical laboratories of general hospitals and a commercial clinical laboratory in Japan. Between 2002 and 2004, 457 clinical strains of H. influenzae were isolated at hospitals in the Hokkaido prefecture, Japan. The strains were collected and stored by Sapporo Clinical Laboratory, Inc. (Sapporo, Japan), Muroran General Hospital (Muroran, Japan), and Hokkaido University Hospital (Sapporo, Japan). Sapporo Clinical Laboratory, Inc., serves almost all areas of the Hokkaido prefecture. This study was approved by the institutional review boards of the organizations listed above. For each strain, we obtained information on the patient's age and sex, the clinical source, and the name of the city where the strain was isolated, in accordance with the Act on the Protection of Personal Information in Japanese law. The isolates were obtained from the following clinical specimens (with the numbers of samples in parentheses): sputum (190), rhinorrhea (178), nasal cavity (19), pharyngeal fluid (33), otopyorrhea (15), vaginal secreta (6), conjunctival discharge (5), bronchial lavage fluid (5), blood (2), urine (2), lochia (1), and cerebrospinal fluid (1). Cities where strains were isolated were Sapporo (228 strains), Muroran (112 strains), Asahikawa (61 strains), Tomakomai (23 strains), Obihiro (17 strains), Hakodate (14 strains), and Iwamizawa (2 strains). All isolates were grown at 37°C, in an atmosphere with 5% CO2, on chocolate agar II (Nippon Beckton-Dickinson, Tokyo, Japan).
Determination of MICs of fluoroquinolones was carried out by a microdilution method on Mueller-Hinton medium supplemented with 5% horse blood with hemolysis and 15 μg/ml NAD, according to the standard method approved by the Clinical and Laboratory Standards Institute (CLSI) (Wayne, PA) (2). Levofloxacin (LVX) (Daiichi Pharmaceuticals, Tokyo, Japan), ciprofloxacin (CIP) (Bayer, Osaka, Japan), sparfloxaxin (SPX) (Dainippon-Sumitomo Pharma, Osaka, Japan), tosufloxacin (TSX) (Abbott, Osaka, Japan), gatifloxacin (GAT) (Kyorin Pharmaceuticals, Tokyo, Japan), and NM394 (active form of prulifloxacin [PUFX]) (Meiji Seika Kaisha, Tokyo, Japan) were kindly provided by the manufacturers. Determination of the MICs of other antibiotics was performed using a MICroFAST 4J panel (Dade Behring, Tokyo, Japan).
We carried out an initial screen for fluoroquinolone-resistant H. influenzae strains by using CIP and LVX. Twelve resistant strains were identified among 457 isolates (frequency of occurrence, 2.6%). All fluoroquinolone-resistant isolates were derived from patients that were over 58 years of age, and none were isolated from children (Table 1). The frequency of occurrence of fluoroquinolone-resistant strains among patients above 50 years of age was 10.2%. Similar age distributions were observed for patients with S. pneumoniae isolates in our previous reports (21, 22). We reported that fluoroquinolone-resistant S. pneumoniae strains were present in over 20% of elderly patients, but their overall frequency is low (2.4% of 670 strains tested) because they have not been identified in children. The most probable reason for this is that the use of fluoroquinolones other than norfloxacin has not been approved for children in Japan. In Japan, rare, sporadic occurrence (0 to 0.5%) of fluoroquinolone-resistant H. influenzae has been reported (6, 9, 23). In this study, fluoroquinolone-resistant strains were found with higher frequency (2.6%). The clinical samples obtained from adults were predominantly sputa, while those from children were mainly rhinorrhea. Ten of the resistant strains (83.3%) were isolated from sputa. All resistant strains were isolated in Sapporo city, which is the largest city in the Hokkaido prefecture, and 11 of 12 strains were isolated in the private clinical laboratory. However, these strains were isolated in more than one hospital in this city. More-detailed information on these hospitals and patients have not been available due to the Act on the Protection of Personal Information in Japanese law.
TABLE 1.
Relationship between patient age and prevalence of fluoroquinolone-resistant H. influenzae clinical isolates
| Patient age group (yr) | No. of strains | No. of resistant strains | Frequency of resistant strains (%) |
|---|---|---|---|
| 0-9 | 285 | 0 | 0.0 |
| 10-19 | 10 | 0 | 0.0 |
| 20-29 | 7 | 0 | 0.0 |
| 30-39 | 20 | 0 | 0.0 |
| 40-49 | 11 | 0 | 0.0 |
| 50-59 | 26 | 1 | 3.8 |
| 60-69 | 41 | 2 | 4.9 |
| 70-79 | 42 | 5 | 11.9 |
| ≥80 | 15 | 4 | 26.7 |
| Total | 457 | 12 | 2.6 |
We determined the nucleotide sequences of the QRDRs of the quinolone target genes parC, gyrA, parE, and gyrB (Table 2). Genomic DNA was isolated from cells by using a DNeasy kit (QIAGEN, Hilden, Germany). PCR amplification was performed using HotStarTaq DNA polymerase (QIAGEN). The primer sets used to obtain DNA fragment of the parC, gyrA, parE, and gyrB QRDRs were previously described by Pérez-Vázquez et al. (14). Direct sequencing of the PCR products by the primers used for the PCR was carried out with an ABI PRISM3100 (Applied Biosystems, Foster City, CA). We examined the status of β-lactam resistance as follows. The presence of the genes for TEM-1 and ROB-1 β-lactamases and resistance-associated mutations in penicillin binding protein 3 (PBP3) was determined by PCR according to the method of Hasegawa et al. (5). In comparison to a fluoroquinolone-sensitive standard strain, Rd, all 12 resistant strains carried three to five mutations in these genes. We classified the 12 strains into three types, termed types I, II, and III, based on the patterns of mutations.
TABLE 2.
Fluoroquinolone MICs and QRDR mutations in topoisomerase IV and DNA gyrase of fluoroquinolone-resistant H. influenzae
| Type | Strain | Patient age (yr) | Sexa | Material | MIC (μg/ml)
|
Mutation(s)b
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LVX | GAT | CIP | TSX | SPX | NM394 | ParC | GyrA | ParE | GyrB | |||||
| IA | SRI68 | 67 | M | Sputum | 32 | 8 | 16 | ≥32 | 16 | 4 | Ser84Ile** (Asn138Ser) | Ser84Leu** Asp88Asn** | Asp420Asn* | |
| SRI182 | 70 | M | Sputum | 16 | 8 | 8 | ≥32 | 8 | 2 | Ser84Ile** (Asn138Ser) | Ser84Leu** Asp88Asn** | Asp420Asn* | ||
| IB | SRI131 | 72 | M | Rhinorrhea | 16 | 8 | 16 | ≥32 | 8 | 2 | Ser84Ile** (Asn138Ser) | Ser84Leu** Asp88Asn** | Asp420Asn* | |
| SRI126 | 76 | M | Sputum | 16 | 8 | 16 | ≥32 | 8 | 2 | Ser84Ile** (Asn138Ser) | Ser84Leu** Asp88Asn** | Asp420Asn* | ||
| IC | SRI97 | 83 | M | Sputum | 16 | 4 | 8 | ≥32 | 8 | 2 | Ser84Ile** (Asn138Ser) | Ser84Leu** Asp88Asn** | Asp420Asn* | |
| SRI26 | 76 | M | Sputum | 16 | 8 | 16 | ≥32 | 8 | 2 | Ser84Ile** (Asn138Ser) | Ser84Leu** Asp88Asn** | Asp420Asn* | ||
| SRI183 | 88 | F | Sputum | 16 | 8 | 16 | ≥32 | 8 | 2 | Ser84Ile** (Asn138Ser) | Ser84Leu** Asp88Asn** | Asp420Asn* | ||
| SRI186 | 88 | M | Sputum | 16 | 8 | 8 | ≥32 | 8 | 2 | Ser84Ile** (Asn138Ser) | Ser84Leu** Asp88Asn** | Asp420Asn* | ||
| IIA | SRI38 | 84 | M | Sputum | 2 | 1 | 4 | 0.5 | 1 | 2 | Gly82Cys* Ser84Arg** | Asp88Asn** Pro118Tyr? | Ser467Tyr* | |
| SRI181 | 58 | F | Sputum | 2 | 1 | 4 | 0.5 | 1 | 2 | Gly82Cys* Ser84Arg** | Asp88Asn** Pro118Tyr? | Ser467Tyr* | ||
| IIB | SRI177 | 65 | F | Conjunctival discharge | 1 | 1 | 2 | 0.5 | 2 | 1 | Ser84Arg** | Asp88Asn** Pro118Tyr? | Ser467Tyr* | |
| III | HUI313 | 70 | F | Sputum | 0.5 | 0.5 | 1 | 0.25 | 1 | 0.5 | (Asn138Ser) | Ser451Tyr? Pro452Leu? | ||
M, male; F, female.
**, resistance mutation previously seen in H. influenzae; *, resistance mutation previously seen in other bacterial species; ?, mutation with unknown contribution to resistance. Mutations in parentheses do not contribute resistance.
Type I (eight strains) showed extremely high MICs, which were four or eight times higher than cutoff values of nonsusceptible MICs proposed by CLSI (2). Type I commonly carried two mutations in ParC and GyrA and a single mutation in ParE. Mutations of Ser84Ile in ParC and Ser84Leu and Asp88Asn in GyrA were previously reported to contribute to quinolone resistance in H. influenzae (4, 12, 14, 23). Asp420Asn in ParE corresponds to resistance-associated mutation sites in Staphylococcus aureus ParE (Asp432) and in S. pneumoniae ParE (Asp435) (16, 17). The Asn138Ser mutation in ParC was also found in susceptible H. influenzae strains (data not shown), which indicates that it does not contribute to fluoroquinolone resistance. Within the same genotype, strains had some different phenotypes, so we subdivided type I into types IA, IB, and IC (Tables 2 and 3). Strains SRI68 and SRI182 (type IA) carried the gene for TEM-1-type β-lactamase, did not have β-lactam resistance-associated mutations in PBP3, and had a high level of resistance to tetracycline and chloramphenicol (Table 3). Strains SRI131 and SRI126 (type IB) were BLNAR but showed ampicillin susceptibility, and they carried low-grade β-lactam resistance mutations in PBP3. The remaining four type I strains carried no β-lactam resistance mutations in PBP3 (thus, β-lactamase negative and ampicillin sensitive).
TABLE 3.
Presence of the gene for TEM-1 type β-lactamase, PBP3 mutation phenotypes, and MICs for fluoroquinolone-resistant H. influenzae
| Type | Strain | TEM-1 gene presenta | Genotypeb | MIC (μg/ml)
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicil-lin | Cefaclor | Cefixime | Cefditoren pivoxil | Cefotiam | Cefotaxime | Ceftriax-one | Cefozo-pran | Cefepime | Mero-penem | Ampicillin-sulbactam | Amoxicillin-clavulanic acid | Clarithro-mycin | Tetracy-cline | Chloram-phenicol | Rifam-pin | ||||
| IA | SRI 68 | + | BLPACS | ≥8 | 4 | <0.25 | <0.03 | 2 | <0.12 | <0.12 | <0.5 | <0.25 | <0.12 | 1 | <1 | 16 | ≥8 | ≥8 | <0.5 |
| SRI182 | + | BLPACS | ≥8 | 2 | <0.25 | <0.03 | 1 | <0.12 | <0.12 | <0.5 | <0.25 | <0.12 | 1 | <1 | 16 | ≥8 | ≥8 | <0.5 | |
| IB | SRI131 | − | Low BLNAR | 0.25 | 2 | <0.25 | <0.03 | 1 | <0.12 | <0.12 | <0.5 | <0.25 | <0.12 | <0.5 | <1 | 32 | <0.5 | 1 | <0.5 |
| SRI126 | − | Low BLNAR | 0.25 | 2 | <0.25 | <0.03 | 1 | <0.12 | <0.12 | <0.5 | <0.25 | <0.12 | <0.5 | <1 | 32 | <0.5 | <0.5 | <0.5 | |
| IC | SRI97 | − | BLNAS | 0.25 | 1 | <0.25 | <0.03 | 2 | <0.12 | <0.12 | <0.5 | <0.25 | <0.12 | <0.5 | <1 | 32 | <0.5 | <0.5 | <0.5 |
| SRI26 | − | BLNAS | 0.25 | 1 | <0.25 | <0.03 | 1 | <0.12 | <0.12 | <0.5 | <0.25 | <0.12 | <0.5 | <1 | 32 | <0.5 | <0.5 | 1 | |
| SRI183 | − | BLNAS | 0.25 | 1 | <0.25 | <0.03 | 1 | <0.12 | <0.12 | <0.5 | <0.25 | <0.12 | <0.5 | <1 | 32 | <0.5 | <0.5 | <0.5 | |
| SRI186 | − | BLNAS | 0.25 | 1 | <0.25 | <0.03 | 1 | <0.12 | <0.12 | <0.5 | <0.25 | <0.12 | <0.5 | <1 | 16 | <0.5 | <0.5 | <0.5 | |
| IIA | SRI38 | + | Low BLPACR | ≥8 | 32 | 1 | 0.12 | ≥16 | ≥4 | <0.12 | 1 | 0.5 | <0.12 | 2 | <1 | 16 | <0.5 | <0.5 | <0.5 |
| SRI181 | + | Low BLPACR | ≥8 | 8 | 0.5 | 0.12 | <0.12 | <0.12 | <0.12 | 1 | 0.5 | <0.12 | 2 | <1 | 16 | <0.5 | <0.5 | <0.5 | |
| IIB | SRI177 | + | BLPACS | ≥8 | 2 | <0.25 | <0.03 | 1 | <0.12 | <0.12 | <0.5 | <0.25 | <0.12 | 1 | <1 | 16 | <0.5 | <0.5 | <0.5 |
| III | HUI313 | − | High BLNAR | 0.5 | 2 | <0.25 | 0.06 | 2 | <0.12 | <0.12 | <0.5 | <0.25 | <0.12 | <0.5 | <1 | 32 | <0.5 | 1 | <0.5 |
+, TEM-1 β-lactamase gene is present; −, TEM-1 β-lactamase gene is absent.
Status of β-lactam resistance was determined by the PCR method as described in the text. BLNAS, β-lactamase negative, ampicillin sensitive; high BLNAR, β-lactamase negative, highly ampicillin resistant; low BLNAR, β-lactamase negative, weakly ampicillin resistant; BLPACS, β-lactamase positive, amoxicillin-clavulanic acid sensitive; low BLPACR, β-lactamase positive, weakly amoxicillin-clavulanic acid resistant.
Type II (three strains) showed nonsusceptible MICs (≥2 μg/ml) to CIP and LVX MICs of 1 or 2 μg/ml, lower than the cutoff value of nonsusceptible MICs. Type II carried two mutations in GyrA, one or two mutations in ParC, and one mutation in GyrB (Table 2). Ser84Arg in ParC and Asp88Asn in GyrA are previously known mutations associated with quinolone resistance in H. influenzae (4, 12, 14, 23). Gly82Cys in ParC corresponds to Gly78 in Escherichia coli ParC and Gly85 in Neisseria gonorrhoeae ParC. Mutations of these residues have been shown to cause fluoroquinolone resistance (7, 11, 18). Ser467Tyr in GyrB corresponds to a resistance-associated mutation site in Salmonella enterica serovar Typhimurium ParE (Ser464) (3). It is unclear that Pro118Tyr in GyrA is involved in resistance. All type II strains carried the gene for TEM-1-type β-lactamase (Table 3). Type II strains were subdivided into type IIA (two strains) and IIB (one strain). Type IIA had low-grade resistance mutations in PBP3 and carried an additional mutation (Gly82Cys) in ParC. On the other hand, the two type IIA strains (SRI38 and SRI181) exhibited markedly different susceptibilities to cefaclor, cefotiam, and cefotaxime.
Type III (one strain) showed significantly higher MICs (1 μg/ml to CIP and 0.5 μg/ml to LVX) than other susceptible strains (<0.12 μg/ml), but these were lower than the nonsusceptible cutoff value. Type III carried a single mutation in ParC and two mutations in ParE. Asn138Ser is not related to resistance as described above. The type III strain contained only two mutations (Ser451Tyr and Pro452Leu) in QRDRs of ParE, and there is no evidence to date that these mutations are associated with fluoroquinolone resistance (Table 2). Additional studies are needed to determine whether mutations Pro118Tyr in GyrA (found in genotype II) and Ser451Tyr and Pro452Leu in ParC (found in genotype III) contribute to resistance.
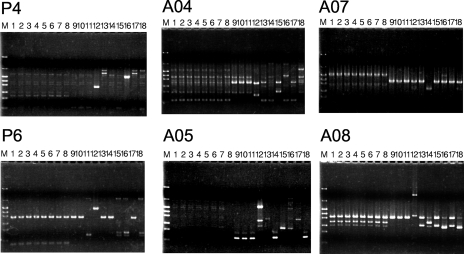
These genotypes were confirmed by randomly amplified polymorphic DNA PCR (RAPD-PCR). RAPD-PCR was carried out as described previously (20). The sequences of the RAPD-PCR primers for H. influenzae were as follows: P4, 5′-AAGAGCCCGT-3′; P6, 5′-CCCGTCAGCA-3′; A04, 5′-ATCAGCGCACCA-3′; A05, 5′-AGCAGCGCCTCA-3′; A07, 5′-TGCCTCGCACCA-3′; and A08, 5′-GCCCCGTTAGCA-3′. PCR amplification was performed using HotStarTaq DNA polymerase (QIAGEN) and GeneAmp PCR system 9700 (Applied Biosystems). DNA (20 ng) was used for each PCR. The PCR consisted of 95°C for 15 min, 35 cycles consisting of 94°C for 2 min, 38°C for 2 min, and 72°C for 2 min, and 72°C for 10 min. The products were analyzed by 1.5% (wt/vol) agarose gel electrophoresis. The results of RAPD-PCR analysis also correlated with this classification scheme (Fig. 1). Within each type, the RAPD pattern was indistinguishable, but there were clearly different RAPD patterns across types, except that one strain of type II gave a RAPD pattern that was different from those of the other two strains by primer P6 (Fig. 1, panel P6, lanes 9 to 11). This line of evidence suggested that the strains of each genotype were derived from a common ancestor.
FIG. 1.
RAPD-PCR analysis of fluoroquinolone resistance of H. influenzae clinical isolates. Lane M, size marker (pHY); lanes 1 to 8, genotype I strains (1, SRI68; 2, SRI182; 3, SRI131; 4, SRI126; 5, SRI97; 6, SRI26; 7, SRI183; 8, SRI186); lanes 9 to 11, genotype II strains (9, SRI38; 10, SRI181; 11, SRI177); lane 12, genotype III strain HUI313; lanes 13 to 18, strains susceptible to fluoroquinolones (13, SRI2; 14, SRI3; 15, SRI4; 16, SRI12; 17, SRI13; 18, SRI18). PCR primers used are indicated the top of each panel.
We examined the resistance to other fluoroquinolones, such as TSX, SPX, GAT, and NM394 (active form of PUFX) (Table 1). The MICs of TSX, SPX, and GAT were similar to those of LVX and CIP. The order of resistance, from highest to lowest, was type I > type II > type III. The MICs of NM394 were similar for types I and II. With the exception of that of NM394, the MICs of all the fluoroquinolones tested against type I strains were extremely high. The MIC of NM394 was moderately high (1 to 2 μg/ml) for type I, similar to that for type II. For types II and III, the MICs indicated moderate resistance to all the fluoroquinolones, including NM394. One possible explanation for this is that the dominant target molecules of PUFX differ from those of the other fluoroquinolones. Another possibility is that there are other mechanisms of resistance that do not involve mutations in the QRDRs and that PUFX is refractory to these mechanisms of resistance. However, additional studies are needed to clarify this issue.
Lines of evidence indicated that the fluoroquinolone-resistant H. influenzae strains expanded clonally in a particular area, namely, Sapporo city. Strains that were assigned to the same genotype based on their mutation profiles and RAPD-PCR analysis had varied phenotypes. This suggested that a fluoroquinolone-resistant clone invaded in a city for a time required for the acquisition of genetic changes, such as the presence of the TEM-1 β-lactamase gene, mutations in PBP3, and resistance to tetracycline and chloramphenicol. Recently, Pérez-Vázquez et al. showed that fluoroquinolone resistance in H. influenzae occurs mainly in hypermutable strains (15). The fluoroquinolone-resistant strains found in the present study could be hypermutable, leading to phenotypic changes within genotypes. This clonal expansion of fluoroquinolone-resistant H. influenzae does not appear to be a transient outbreak, based on the fact that genetic variations were observed in the strains of the same genotype and the strains were not isolated from a single hospital.
The results of our current analysis are quite different from results of studies of S. pneumoniae (21, 22). Fluoroquinolone-resistant S. pneumoniae strains had marked differences in the patterns of mutation in their QRDRs and RAPD-PCR results and were isolated from various areas in the Hokkaido prefecture. Thus, it appears that fluoroquinolone-resistant H. influenzae strains expanded clonally, while fluoroquinolone-resistant S. pneumoniae strains were generated sporadically. Both strains of fluoroquinolone-resistant bacteria were found only in elderly patients because fluoroquinolones other than norfloxacin are not applicable to children in Japan.
Acknowledgments
We thank Michitoshi Kimura (Laboratory of Cell and Tissue, Department of BioMedical Engineering, Sapporo Medical University School of Medicine) for analysis of DNA sequences. We thank Tasuku Hayashi and Keiko Matsuda (Clinical Laboratory, Muroran General Hospital), Hirotsugu Akizawa (Clinical Laboratory, Hokkaido University Hospital), and Osamu Kuwahara (Sapporo Clinical Laboratory, Inc.) for provision of H. influenzae clinical isolates.
Footnotes
Published ahead of print on 31 October 2007.
REFERENCES
- 1.Biedenbach, D. J., and R. N. Jones. 2003. Five-year analysis of Haemophilus influenzae isolates with reduced susceptibility to fluoroquinolones: prevalence results from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 4655-61. [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M07-A7, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Gensberg, K., Y. F. Jin, and L. J. Piddock. 1995. A novel gyrB mutation in a fluoroquinolone-resistant clinical isolate of Salmonella typhimurium. FEMS Microbiol. Lett. 13257-60. [DOI] [PubMed] [Google Scholar]
- 4.Georgiou, M., R. Munoz, F. Roman, R. Canton, R. Gomez-Lus, J. Campos, and A. G. De La Campa. 1996. Ciprofloxacin-resistant Haemophilus influenzae strains possess mutations in analogous positions of GyrA and ParC. Antimicrob. Agents Chemother. 401741-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasegawa, K., K. Yamamoto, N. Chiba, R. Kobayashi, K. Nagai, M. R. Jacobs, P. C. Appelbaum, K. Sunakawa, and K. Ubukata. 2003. Diversity of ampicillin-resistance genes in Haemophilus influenzae in Japan and the United States. Microb. Drug Resist. 939-46. [DOI] [PubMed] [Google Scholar]
- 6.Hasegawa, M., Y. Sato, A. Kanayama, K. Matsuzaki, H. Muraoka, A. Amano, T. Saika, and I. Kobayashi. 2006. Antibacterial activity of tosufloxacin against major organisms detected from patients with respiratory or otorhinological infections: comparison with the results obtained from organisms isolated about 10 years ago. J. Infect. Chemother. 12152-156. [DOI] [PubMed] [Google Scholar]
- 7.Heisig, P. 1996. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 40879-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoban, D. J., G. V. Doern, A. C. Fluit, M. Roussel-Delvallez, and R. N. Jones. 2001. Worldwide prevalence of antimicrobial resistance in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2)S81-S93. [DOI] [PubMed] [Google Scholar]
- 9.Inoue, M., S. Kohno, M. Kaku, K. Yamaguchi, J. Igari, and K. Yamanaka. 2005. PROTEKT 1999-2000: a multicentre study of the antimicrobial susceptibility of respiratory tract pathogens in Japan. Int. J. Infect. Dis. 927-36. [DOI] [PubMed] [Google Scholar]
- 10.Inoue, M., N. Y. Lee, S. W. Hong, K. Lee, and D. Felmingham. 2004. PROTEKT 1999-2000: a multicentre study of the antibiotic susceptibility of respiratory tract pathogens in Hong Kong, Japan and South Korea. Int. J. Antimicrob. Agents 2344-51. [DOI] [PubMed] [Google Scholar]
- 11.Kumagai, Y., J. I. Kato, K. Hoshino, T. Akasaka, K. Sato, and H. Ikeda. 1996. Quinolone-resistant mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob. Agents Chemother. 40710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, X., N. Mariano, J. J. Rahal, C. M. Urban, and K. Drlica. 2004. Quinolone-resistant Haemophilus influenzae in a long-term-care facility: nucleotide sequence characterization of alterations in the genes encoding DNA gyrase and DNA topoisomerase IV. Antimicrob. Agents Chemother. 483570-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nazir, J., C. Urban, N. Mariano, J. Burns, B. Tommasulo, C. Rosenberg, S. Segal-Maurer, and J. J. Rahal. 2004. Quinolone-resistant Haemophilus influenzae in a long-term care facility: clinical and molecular epidemiology. Clin. Infect. Dis. 381564-1569. [DOI] [PubMed] [Google Scholar]
- 14.Pérez-Vázquez, M., F. Román, B. Aracil, R. Cantón, and J. Campos. 2004. Laboratory detection of Haemophilus influenzae with decreased susceptibility to nalidixic acid, ciprofloxacin, levofloxacin, and moxifloxacin due to gyrA and parC mutations. J. Clin. Microbiol. 421185-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez-Vázquez, M., F. Román, S. García-Cobos, and J. Campos. 2007. Fluoroquinolone resistance in Haemophilus influenzae is associated with hypermutability. Antimicrob. Agents Chemother. 511566-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perichon, B., J. Tankovic, and P. Courvalin. 1997. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 411166-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi, H., T. Kikuchi, S. Shoji, S. Fujimura, A. B. Lutfor, Y. Tokue, T. Nukiwa, and A. Watanabe. 1998. Characterization of gyrA, gyrB, grlA and grlB mutations in fluoroquinolone-resistant clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 4149-57. [DOI] [PubMed] [Google Scholar]
- 18.Trees, D. L., A. L. Sandul, W. L. Whittington, and J. S. Knapp. 1998. Identification of novel mutation patterns in the parC gene of ciprofloxacin-resistant isolates of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 422103-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ubukata, K. 2003. Problems associated with high prevalence of multidrug-resistant bacteria in patients with community-acquired infections. J. Infect. Chemother. 9285-291. [DOI] [PubMed] [Google Scholar]
- 20.Yokota, S., A. Harimaya, K. Sato, Y. Somekawa, T. Himi, and N. Fujii. 2007. Colonization and turnover of Haemophilus influenzae, and Moraxella catarrhalis in otitis-prone children. Microbiol. Immunol. 51223-230. [DOI] [PubMed] [Google Scholar]
- 21.Yokota, S., K. Sato, O. Kuwahara, S. Habadera, N. Tsukamoto, H. Ohuchi, H. Akizawa, T. Himi, and N. Fujii. 2002. Fluoroquinolone-resistant Streptococcus pneumoniae strains occur frequently in elderly patients in Japan. Antimicrob. Agents Chemother. 463311-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokota, S., K. Sato, S. Yoshida, and N. Fujii. 2004. Molecular epidemiology of fluoroquinolone-resistant Streptococcus pneumoniae in Japan. Kansenshogaku Zasshi 78428-434. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 23.Yoshizumi, S., Y. Takahashi, Y. Watanabe, E. Okezaki, Y. Ishii, and K. Tateda. 2004. In vitro antibacterial activities of new fluoroquinolones against clinical isolates of Haemophilus influenzae with ciprofloxacin-resistance-associated alterations in GyrA and ParC. Chemotherapy 50265-275. [DOI] [PubMed] [Google Scholar]