Abstract
Quantification of Epstein-Barr virus (EBV) in peripheral blood is important for the diagnosis and management of serious EBV diseases, including posttransplant lymphoproliferative disorder. A variety of PCR-based methods are currently in use; however, there is little information on their comparability. This study assessed the relative performance of different quantitative assays. A multicenter comparative study was performed at eight sites using three panels consisting of serial dilutions of quantified EBV DNA and extracts from a total of 19 whole-blood specimens. Samples were distributed and tested blindly. Instrumentation, probe chemistries, amplification targets, and other test-related aspects varied considerably between laboratories. Each laboratory's calibration curve indicated strong evidence of a consistent log-linear relationship between viral load and cycle threshold, suggesting that intralaboratory tracking of a given patient would yield similar relative quantitative trends among the participating test sites. There was strong concordance among laboratories with respect to qualitative test results; however, marked quantitative discordance was seen. For most samples, the across-laboratory interquartile range of the reported viral load (in copies/μl) was roughly 0.6 log-units, and for one sample the overall range was approximately 4.2 log-units. While intralaboratory tracking of patients may yield similar results, these data indicate a need for caution when attempting to compare clinical results obtained at different institutions and suggest the potential value to be gained by more standardized testing methodology.
Epstein-Barr virus (EBV), like all members of the human herpesvirus family, persists in the host after primary infection and may reactivate at any time. EBV causes substantial morbidity especially among immunocompromised patients (2). Since EBV may replicate without causing apparent harm, it is important to be able to distinguish asymptomatic infection from impending or established EBV disease. Because of this, quantitative methods for measuring EBV are now widely utilized for the diagnosis, monitoring, and treatment of EBV-related diseases, particularly in the case of posttransplant lymphoproliferative disorder (1, 9). In addition, studies have indicated that preemptive treatment for EBV and reduction in immunosuppressive therapy can reduce the incidence of posttransplant lymphoproliferative disorder in immunocompromised patients (1, 7).
While the utility of molecular testing for EBV (most commonly PCR) is well accepted (16), and many have implemented it on a routine basis, there is a lack of uniformity and consistency among the currently available assays. The lack of standardized methods extends to all aspects of testing, including selection of specimen type, specimen collection methods, initial processing, nucleic acid extraction, molecular amplification, result interpretation, and reporting. Within the area of assay design, relevant issues include probe chemistry, target selection, cycling conditions, internal control (selection of target and mode of use), thermocycling platform, and quantitative calibration methods. Although many have moved to real-time methodologies, the implementation of these assays is by no means uniform or universal. Furthermore, and perhaps of greatest impact, there is no universally accepted quantitative standard for EBV, as has been adopted for other viruses (5, 12-14). All of these issues have grown in importance with the increased use of EBV quantitative analysis in patient management, which has become the standard-of-care in some clinical settings.
The relative effect of the different variables noted above on assay results and their impact on clinical utilization of test results when viewed over time within a given institution and when viewed across institutions have not been well defined. The optimal interpretation of studies performed at different centers using different test methodologies and the ability to monitor patients who transfer their care between different institutions becomes increasing dependent on a better understanding of these variables. Evaluating the sources of test variability should improve our ability to interpret such results and may also help establish a more uniform standard for performing these tests. This multicenter study is the first published evaluation of the variability of quantitative real-time PCR for EBV across a wide variety of institutions, testing platforms, and methodologies. Whole blood was chosen over other peripheral blood components for this analysis, based on data showing a higher degree of sensitivity in cellular compartments (whole blood or peripheral blood mononuclear cells) compared to plasma (3, 18). Also, whole blood was the most commonly used specimen type for this assay among participating laboratories.
(The results of this study were presented in part 30 April to 3 May 2006 at the 22nd annual meeting of the Pan American Society for Clinical Virology, Clearwater Beach, FL.)
MATERIALS AND METHODS
Study design.
Eight independent laboratories comprised the Working Group, each utilizing a different quantitative EBV PCR assay. Specimen preparation, including nucleic acid extraction, was performed prior to distribution, eliminating these factors as variables in the analysis. Prior to testing, each laboratory provided details of its respective testing procedure(s) for comparative evaluation; variation was seen in all aspects of assay methodology (Table 1). Three coded panels, consisting of serially diluted, commercially prepared EBV DNA calibrators and 19 whole-blood patient specimens, were prepared, nucleic acid was extracted, and extracts were distributed among the participating laboratories. Each site performed quantitative EBV testing on all panels using their own protocols, reagents, controls, and instrumentation. The composition of each panel, the quantitative calibrators used, and the number of replicate test runs performed are indicated in Table 2. The data were analyzed to determine both intra- and interlaboratory variability, concordance of quantitative values achieved, and evidence of a consistent quantitative relationship among laboratories using both common and differing quantitative calibrators. Variability in patient results was assessed independently and in relation to variability of linear regression calibration curves and calibrators. All human samples used in the study were assigned arbitrary identification designations prior to use. This research complied with relevant federal guidelines and institutional policies.
TABLE 1.
Comparison of selected assay and procedural characteristics among different test sites
| Lab | Platform | DNA input (μl) | Reaction vol (μl) | Target | Target length (bp) | Probe type/fluorescence | EBV calibration curve type | Dynamic range/vol (μl) | No. of patients/calibration curve |
|---|---|---|---|---|---|---|---|---|---|
| A | ABI 7900 | 10 | 50 | BNRF1 | 83 | FAM/NFQ | Plasmid | 1 × 10 to 1 × 106/10 | 6 |
| B | LightCycler 1.0 | 5 | 20 | EBNA1 | 251 | Fluor/Red 640 | Quantified viral DNA | 1 × 10 to 1 × 105/10 | 5 |
| C | ABI 7500 | 10 | 50 | BNRF1 | 74 | FAM/TAMRA | Plasmid | 8 × 102 to 4 × 108/10 | 5 |
| D1a | ABI 7700 | 10 | 50 | BNRF1 | 74 | FAM/TAMRA | Plasmid | 6.6 × 10 to 6.6 × 107/10 | 7 |
| D2b | ABI 7700 | 20 | 50 | EBNA1 | 97 | FAM/None | Cloned target region | 2 × 102 to 1 × 107/10 | 4 |
| E | ABI 7900 | 8 | 50 | BALF5 | 90 | FAM/TAMRA | Plasmid | 1.25 × 106 to 2.5 × 106/10 | 8 |
| F | ABI 7000 | 5 | 25 | EBNA1 | 71 | FAM/TAMRA | Plasmid | 2 × 10 to 2 × 106/10 | 6 |
| G | ABI 7700 | 5 | 25 | EBNA1 | 71 | FAM/TAMRA | Plasmid | 2 × 10 to 2 × 106/10 | 6 |
| H | LightCycler 1.0 | 5 | 20 | EBER | 319 | SYBR green | Plasmid | 1 × 10 to 5 × 106/5 | 4-5 |
Method D1 was used only for panel 1 testing.
Method D2 was used for testing of all panels.
TABLE 2.
Test panel characteristics
| Panela | No. of samples tested | Quantitative calibrator(s) used | No. of replicate runs performed by each laboratory |
|---|---|---|---|
| 1 | 4 | Independent analysis using both commercially prepared calibrators (same calibrators used by all labs) and “site-specific” calibrators (chosen and routinely used by each test site) | 1 |
| 2 | 6 | Commercially prepared: same calibrators used by all laboratories | 1 |
| 3 | 9 | Commercially prepared: same calibrators used by all laboratories | 3 |
Serially diluted commercial DNA preparation; panels 2 and 3 were prepared from the same DNA lot.
Control EBV DNA.
Two commercial lot numbers of fluorimetrically quantified EBV B95-8 DNA (Advanced Biotechnologies, Inc., Columbia, MA) were used to create a six-point tenfold serial dilution series, ranging from 2.62 copies/μl to 2.62 × 106 copies/μl to be used as quantitative calibrators. A separate dilution was prepared for each of the three test panels that were distributed. Lot 1 was used for panel 1; the serial dilution was centrally prepared in 0.1× Tris-EDTA buffer (TE) and shipped to participants at 4°C. Lot 2 of EBV DNA was used for both panels 2 and 3. For these panels, a single known concentration of EBV DNA was prepared in reagent-grade water, lyophilized at a concentration of 2.62 × 106 copies/μl, and distributed to each test site for reconstitution and serial dilution.
Patient samples.
Nineteen de-identified whole blood specimens (totals of 4, 6, and 9 for the first, second, and third test panels, respectively) were studied. Nucleic acid was extracted from multiple 200-μl aliquots of each sample using a QIAamp DNA blood minikit (Qiagen, Inc., Valencia, CA), with elution into 100 μl of 10 mM Tris-Cl-0.5 mM EDTA buffer (pH 9.0). All eluates from each sample were subsequently pooled and divided again into aliquots prior to distribution to ensure the uniformity of the samples among participant laboratories. All samples were centrally prepared, de-identified, coded, and then stored at −80°C until distribution and testing.
Test panel instructions and result reporting.
Each laboratory was instructed to test samples using their usual standard operating procedures, reagents, and instrumentation. All samples were tested blindly, with each laboratory testing their usual number of replicates (one to three sample replicates and two to three calibrator replicates). The results were reported in “copies EBV per μl of input DNA” and submitted to an independent facility for tabulation prior to unblinding. Additional assay run data, such as cycle threshold (CT) values and calibration curve equations, were solicited for all panels, as were key facets of each site's EBV quantification protocols. To assess inter-run variability, sites performed three independent runs with panel 3. All testing was performed within a 3-day time window in order to limit the effects of specimen stability on interlaboratory result variability.
Statistical methods and data analysis.
Classical least-squares regression with CT as the y variable and log10 copies/μl as the x variable was used to obtain each standard curve. The root mean square error (RMSE) was used to summarize the deviation of the calibrator CT values from the fitted line. The reported viral load result for each unknown sample was determined by averaging the CT values across replicates and mapping the result against the calibration curve. For each unknown sample, the range and interquartile range were used to summarize the variability of reported results across laboratories.
To statistically compare the interlaboratory deviation of regression parameter estimates (slope, intercept, or RMSE) of one panel to those of another panel, the absolute deviation of each lab's regression estimate from the across-lab mean for that parameter was determined. Next, for each lab, the difference between the two panels' estimates was computed, and the Wilcoxon signed-rank test was applied to the set of differences. Pagano and Gauvreau (10) describe all of these statistical methods. No adjustments for multiple testing were performed. S-Plus software (Insightful Corp, Seattle, WA) was used to perform all analyses.
RESULTS
Calibration curves.
All calibration curves showed strong consistent linear relationships between viral load and cycle threshold (CT). However, differences were noted both between and within testing sites with respect to slope, intercept, and RMSE. Panel 1 results reflected nine assays conducted at eight sites (one site tested twice, using two distinct methods). Panels 2 and 3 were each tested using eight quantification methods (one method per test site).
Calibration curves for panel 1 were generated using both site-specific calibrators (site-specific calibrator curves) and using calibrators common to all testing sites (common calibrator curves). Summary statistics showing calibration curve variability for both calibrator types are shown in Table 3. The intercept estimates, slope estimates, and RMSE values showed less variability across laboratories when common calibrators were used than when site-chosen calibrators were used (P = 0.0742, P = 0.0273, and P = 0.0039). For instance, the intercept estimates across laboratories ranged from 28.84 to 49.36 with a standard deviation of 5.91 when site-chosen calibrators were used; but when common calibrators were used, the intercept estimates ranged from 33.2 to 40.6 with a standard deviation of only 2.19. Regression curves generated using common calibrators also demonstrated improved interlaboratory consistency compared to those generated with site-specific reagents (Fig. 1, panel 1 plots).
TABLE 3.
Interlaboratory summaries of calibration regression estimates
| Panela | Regression estimate
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercept (CT)
|
Slope
|
RMSE
|
||||||||||||||||
| Mean | Median | SD | Min | Max | Range | Mean | Median | SD | Min | Max | Range | Mean | Median | SD | Min | Max | Range | |
| 1* | 39.91 | 39.10 | 5.91 | 28.84 | 49.36 | 20.53 | −3.56 | −3.50 | 0.31 | −4.14 | −3.08 | 1.06 | 0.42 | 0.32 | 0.24 | 0.13 | 0.82 | 0.69 |
| 1† | 36.93 | 37.00 | 2.19 | 33.20 | 40.58 | 7.38 | −3.51 | −3.52 | 0.08 | −3.60 | −3.35 | 0.24 | 0.28 | 0.22 | 0.13 | 0.09 | 0.45 | 0.36 |
| 2† | 39.32 | 39.49 | 2.07 | 35.60 | 42.68 | 7.08 | −3.57 | −3.59 | 0.20 | −3.95 | −3.30 | 0.64 | 0.42 | 0.45 | 0.27 | 0.10 | 0.86 | 0.76 |
| 3.1† | 41.61 | 41.37 | 1.33 | 39.29 | 43.43 | 4.14 | −3.66 | −3.58 | 0.25 | −4.23 | −3.45 | 0.78 | 0.34 | 0.31 | 0.08 | 0.25 | 0.47 | 0.22 |
| 3.2† | 42.16 | 41.41 | 2.99 | 39.24 | 48.52 | 9.28 | −3.67 | −3.53 | 0.43 | −4.44 | −3.12 | 1.32 | 0.49 | 0.35 | 0.42 | 0.14 | 1.34 | 1.20 |
| 3.3† | 42.48 | 42.28 | 3.00 | 37.39 | 47.75 | 10.36 | −3.75 | −3.71 | 0.20 | −4.00 | −3.51 | 0.49 | 0.41 | 0.38 | 0.15 | 0.16 | 0.60 | 0.44 |
*, site-specific calibrators were used; †, for all remaining panels common calibrators were used. Min, minimum; Max, maximum.
FIG. 1.
Comparison of calibration curves for all test panel runs.
The finding of improved quantitative consistency with the use of common calibration standards was reproduced with a second panel of EBV DNA. Calibration curve characteristics for panel 2 were analyzed using a single set of common calibrators. Calibration curve characteristics for panel 2 were compared to those of panel 1 generated from common and site specific calibrators (Table 3 and Fig. 1). The interlaboratory variability of intercept estimates, slope estimates, and the RMSE of panel 2 did not differ significantly from those of panel 1 using common calibrators (P = 1.00, P = 0.25, and P = 0.1093, respectively). The interlaboratory variability of the intercept and slope of panel 2 did not differ significantly from those of panel 1 with site-chosen calibrators (P = 0.1953 and P = 0.1484, respectively). However, panel 2 had significantly less interlaboratory variation of the RMSE than did panel 1 using site-chosen calibrators (P = 0.0078).
Panel 3 was analyzed using a single set of common calibrators but differed from panels 1 and 2 in that each laboratory ran the panel (calibrators and patient samples) on three separate occasions. Intralaboratory variability of calibration curve slope, y intercept, and RMSE differed markedly among the eight test sites (data not shown). There was further evidence of this variability when cross-panel comparisons were performed. For example, the intercept estimates for one laboratory ranged from 37.00 to 41.33 across panels, while the intercept estimates for another laboratory ranged from 28.84 to 41.70 across panels (Table 4). While expected discrepancies were seen between calibration curves generated from the site-specific and common calibrators curves in panel 1, calibration curve plots generated by some laboratories were more widely splayed than others, when all runs from all panels (Fig. 2, laboratories C and H) or when replicates from panel 3 test runs were compared (data not shown).
TABLE 4.
Calibration curve parameter estimates summarized across panels for each lab
| Laboratory | Regression estimatea
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercept
|
Slope
|
RMSE
|
||||||||||||||||
| Mean | Median | SD | Min | Max | Range | Mean | Median | SD | Min | Max | Range | Mean | Median | SD | Min | Max | Range | |
| A | 42.80 | 42.41 | 2.51 | 40.58 | 47.37 | 6.79 | −3.57 | −3.49 | 0.29 | −4.14 | −3.30 | 0.84 | 0.36 | 0.33 | 0.20 | 0.16 | 0.70 | 0.54 |
| B | 39.73 | 40.11 | 2.14 | 36.70 | 41.91 | 5.21 | −3.57 | −3.59 | 0.11 | −3.72 | −3.41 | 0.31 | 0.28 | 0.33 | 0.12 | 0.09 | 0.39 | 0.30 |
| C | 44.62 | 45.27 | 4.49 | 39.09 | 49.36 | 10.27 | −3.69 | −3.60 | 0.29 | −4.11 | −3.35 | 0.76 | 0.42 | 0.41 | 0.14 | 0.27 | 0.60 | 0.33 |
| D | 38.83 | 38.90 | 1.53 | 37.00 | 41.33 | 4.33 | −3.46 | −3.49 | 0.21 | −3.71 | −3.08 | 0.62 | 0.39 | 0.30 | 0.27 | 0.13 | 0.86 | 0.73 |
| E | 41.53 | 41.94 | 2.94 | 37.19 | 44.53 | 7.34 | −3.79 | −3.78 | 0.14 | −3.97 | −3.60 | 0.38 | 0.32 | 0.23 | 0.24 | 0.10 | 0.67 | 0.57 |
| F | 40.16 | 40.30 | 1.82 | 37.61 | 42.64 | 5.03 | −3.54 | −3.58 | 0.10 | −3.64 | −3.39 | 0.25 | 0.39 | 0.40 | 0.13 | 0.22 | 0.55 | 0.33 |
| G | 39.72 | 40.17 | 2.30 | 36.39 | 42.68 | 6.29 | −3.50 | −3.52 | 0.28 | −3.95 | −3.12 | 0.83 | 0.51 | 0.44 | 0.21 | 0.32 | 0.88 | 0.55 |
| H | 36.27 | 36.49 | 4.84 | 28.84 | 41.70 | 12.86 | −3.88 | −3.82 | 0.41 | −4.44 | −3.40 | 1.04 | 0.42 | 0.27 | 0.45 | 0.14 | 0.34 | 1.20 |
Min, minimum; Max, maximum.
FIG. 2.
Comparison of calibration curves by laboratory, comparing all test panel runs.
Patient sample results.
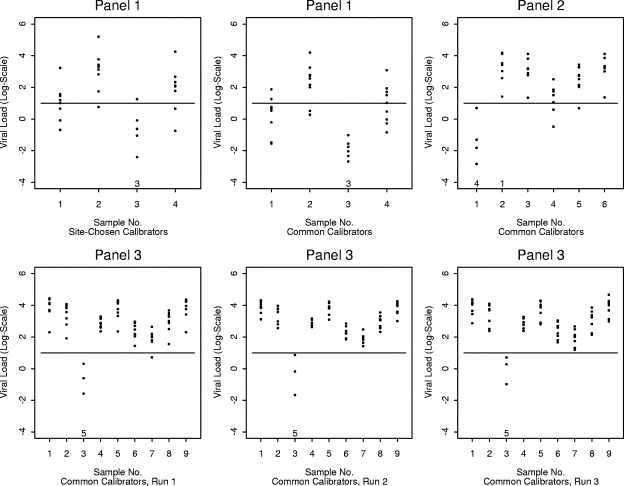
As shown in Fig. 3, the interlaboratory variability of quantitative results for individual samples was tremendous. The range of viral load results exceeded 1 log-unit in most cases; in one case the range was almost 5 log-units. The lowest interlaboratory range was 0.54 log-units (Fig. 3, sample 4, bottom-center plot). The plots include quantitative results for samples that had a reported CT value; samples yielding results of less than 1 on the log-scale were generally interpreted as negative results by the laboratories. Based on the latter criterion, there was complete agreement among all participants for all negative samples in all three panels when common calibrators were used (Fig. 3, samples 3, 1, and 3 in panels 1, 2, and 3, respectively). Using its own calibrators, one laboratory would report a positive result with a low viral load for sample 3 in panel 1 (Fig. 3). In a few cases, some labs would report negative results, while most labs would report positive results (Fig. 3). Interpreting viral loads less than 2 on the log-scale as negative findings would introduce substantially greater disagreement in interpretation across laboratories for some samples with lower viral loads (Fig. 3). The lowest and highest viral loads were reported by the same labs for many negative or low-viral-load samples (Fig. 4).
FIG. 3.
Variation in viral load values for patient samples. Each point gives the result from a specific laboratory for the indicated panel and sample number. The horizontal line at y = 1 corresponds to the threshold used to qualitatively interpret the findings as positive or negative. The numbers at the bottom of each plot indicate the number of laboratories that did not report a CT value and interpreted the result as negative (zeroes are not shown).
FIG. 4.
Consistency of results for negative and low viral load samples. Each solid line shows the results of a negative or low viral sample for one panel across labs. The plot includes all instances with a mean viral load less than or equal to 2 on the log-scale. The results deemed negative by the lab are shown as −4 on this plot. A dashed horizontal line at y = 1 is included for purposes of qualitative interpretation.
DISCUSSION
Quantitative data showed substantial differences both between and within laboratory test sites; it is likely that even greater variability would have been seen if different nucleic acid extraction methods been included in the study. This variability was apparent in linear regression calibration curves, as shown by differences in slope, y intercept, and RMSE. Differences were similarly reflected in results from patient samples. Although the use of common calibration materials had substantial benefit in reducing variability across test sites, marked inter- and intralaboratory discrepancies remained.
Standardized calibration materials have been developed for a number of commonly tested analytes (5, 6, 11-15). These international standards, combined with assay improvements, including broadly available commercial reagents and automated systems for specimen preparation/processing, have played important roles in the development of quantitative tests and have greatly facilitated their broad implementation (8, 17). In addition, for some analytes, the availability of international standards and test systems with excellent reproducibility has allowed the development of international consensus guidelines on quantitative thresholds to guide patient management (4). Finally, standardized diagnostics with excellent reproducibility can be critical for the management of individual patients across geographical boundaries and in the accurate interpretation of data generated at various sites.
The data presented here suggest that similar improvements in quantitative precision can be achieved for EBV PCR using real-time methods. The generation of internationally agreed upon calibration standards would be a first step toward such a goal. However, these data also show substantial remaining variability. That variability can be ascribed to differences in assay design and laboratory technique. The latter aspect of test performance likely explains much of the intralaboratory variation seen here. Although such run-to-run changes may be mitigated somewhat by increased training or experience, it is likely that automated technologies will be more effective in reducing interlaboratory variability.
The present study was not designed to assess the relative contributions of various assay characteristics to result variability. Nor could certain variables such as the effects of transportation on test panel samples be totally obviated. However, the distribution of lyophilized material for common calibrators, the use of a single extraction method, and in panel 3 the specification of testing dates and procedures for storing and thawing specimens were all measures intended to minimize the effects of specimen handling on interlaboratory variability.
It is hoped that our findings will stimulate efforts to further standardize quantitative assays for EBV and other viruses. The problems demonstrated here exemplify the challenges in this still-developing diagnostic field. The implementation of a replicable paradigm for developing quantitative viral controls or calibration standards is clearly an unmet need. Furthermore, the availability of commercially produced and marketed assays and the introduction of automation and reagents produced using good manufacturing practices should contribute to assay precision and should allow more widespread implementation of quantitative testing.
Acknowledgments
The EBV Working Group thank C.-H. Webb (University of Minnesota, Minneapolis, MN), B. Lembke (Labcorp, ViroMed Laboratories, Minnetonka, MN), and S. Verma (St. Jude Children's Research Hospital, Memphis, TN) for assistance with this work.
This study was supported in part by the American Lebanese Syrian Associated Charities, the Minnesota Medical Foundation, and the University of Minnesota International Center for Antiviral Research and Epidemiology.
Footnotes
Published ahead of print on 7 November 2007.
REFERENCES
- 1.Bakker, N. A., E. A. Verschuuren, M. E. Erasmus, B. G. Hepkema, N. J. Veeger, C. G. Kallenberg, and W. van der Bij. 2007. Epstein-Barr virus-DNA load monitoring late after lung transplantation: a surrogate marker of the degree of immunosuppression and a safe guide to reduce immunosuppression. Transplantation 83433-438. [DOI] [PubMed] [Google Scholar]
- 2.Cohen, J. I. 2005. Clinical aspects of Epstein-Barr virus infection, p. 35-54. In E. Robertson (ed.), Epstein-Barr virus. Caister Academic Press, Norfolk, England.
- 3.Hakim, H., C. Gibson, J. Pan, K. Srivastava, Z. Gu, M. J. Bankowski, and R. T. Hayden. 2007. Comparison of various blood compartments and reporting units for the detection and quantification of Epstein-Barr virus in peripheral blood. J. Clin. Microbiol. 452151-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammer, S. M., M. S. Saag, M. Schechter, J. S. Montaner, R. T. Schooley, D. M. Jacobsen, M. A. Thompson, C. C. Carpenter, M. A. Fischl, B. G. Gazzard, J. M. Gatell, M. S. Hirsch, D. A. Katzenstein, D. D. Richman, S. Vella, P. G. Yeni, and P. A. Volberding. 2006. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society-USA panel. Top. HIV Med. 14827-843. [PubMed] [Google Scholar]
- 5.Holmes, H., C. Davis, A. Heath, I. Hewlett, and N. Lelie. 2001. An international collaborative study to establish the first international standard for HIV-1 RNA for use in nucleic acid-based techniques. J. Virol. Methods 92141-150. [DOI] [PubMed] [Google Scholar]
- 6.Jorgensen, P. A., and P. D. Neuwald. 2001. Standardized hepatitis C virus RNA panels for nucleic acid testing assays. J. Clin. Virol. 2035-40. [DOI] [PubMed] [Google Scholar]
- 7.Lee, T. C., B. Savoldo, C. M. Rooney, H. E. Heslop, A. P. Gee, Y. Caldwell, N. R. Barshes, J. D. Scott, L. J. Bristow, C. A. O'Mahony, and J. A. Goss. 2005. Quantitative EBV viral loads and immunosuppression alterations can decrease PTLD incidence in pediatric liver transplant recipients. Am. J. Transplant. 52222-2228. [DOI] [PubMed] [Google Scholar]
- 8.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 301292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda, A., H. Wakiguchi, W. Yokoyama, H. Hisakawa, T. Tomoda, and T. Kurashige. 1999. Persistently high Epstein-Barr virus (EBV) loads in peripheral blood lymphocytes from patients with chronic active EBV infection. J. Infect. Dis. 1791012-1015. [DOI] [PubMed] [Google Scholar]
- 10.Pagano, M., and K. Gauvreau. 2000. Principles of biostatistics. Duxbury, Pacific Grove, CA.
- 11.Robertson, J. S. 1998. International standardization of gene amplification technology. Biologicals 26111-113. [DOI] [PubMed] [Google Scholar]
- 12.Saldanha, J., W. Gerlich, N. Lelie, P. Dawson, K. Heermann, and A. Heath. 2001. An international collaborative study to establish a World Health Organization international standard for hepatitis B virus DNA nucleic acid amplification techniques. Vox Sang 8063-71. [DOI] [PubMed] [Google Scholar]
- 13.Saldanha, J., A. Heath, N. Lelie, G. Pisani, and M. Y. Yu. 2005. A World Health Organization International standard for hepatitis A virus RNA nucleic acid amplification technology assays. Vox Sang 8952-58. [DOI] [PubMed] [Google Scholar]
- 14.Saldanha, J., N. Lelie, A. Heath, et al. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang 76149-158. [DOI] [PubMed] [Google Scholar]
- 15.Saldanha, J., N. Lelie, M. W. Yu, and A. Heath. 2002. Establishment of the first World Health Organization International standard for human parvovirus B19 DNA nucleic acid amplification techniques. Vox Sang 8224-31. [DOI] [PubMed] [Google Scholar]
- 16.Stevens, S. J., E. A. Verschuuren, S. A. Verkuujlen, A. J. Van Den Brule, C. J. Meijer, and J. M. Middeldorp. 2002. Role of Epstein-Barr virus DNA load monitoring in prevention and early detection of post-transplant lymphoproliferative disease. Leuk. Lymphoma 43831-840. [DOI] [PubMed] [Google Scholar]
- 17.Vernet, G. 2004. Molecular diagnostics in virology. J. Clin. Virol. 31239-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wadowsky, R. M., S. Laus, M. Green, S. A. Webber, and D. Rowe. 2003. Measurement of Epstein-Barr virus DNA loads in whole blood and plasma by TaqMan PCR and in peripheral blood lymphocytes by competitive PCR. J. Clin. Microbiol. 415245-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]