Abstract
Opisthorchis viverrini is an important food-borne trematode in Southeast Asia. The infection causes significant morbidity in terms of hepatobiliary diseases and cholangiocarcinoma. The aim of this study was to improve the sensitivity of the PCR-based diagnosis of O. viverrini infection. A new fecal DNA extraction protocol for the detection of O. viverrini DNA using cetyltrimethyl-ammoniumbromide to remove PCR inhibitor was used and compared with the commercial stool kit method. The sensitivity of the new test was 79.3%, compared with the 44.8% of the previous method (P < 0.01). PCR-positive tests identified several cases judged parasite negative by the parasitological method (28.6%), indicating the new test's advantage in the diagnosis of individuals with light infections.
Opisthorchis viverrini is a carcinogenic food-borne trematode known to cause hepatobiliary disease and cholangiocarcinoma in Southeast Asia, particularly Thailand, the Lao People's Democratic Republic (PDR), Cambodia, and Vietnam (3, 4, 14). Humans become infected with O. viverrini through the consumption of raw or undercooked cyprinoid fish containing metacercariae (4). Hence, parasite control should, in part, reduce the risk of morbidity and cancer development (12). To date, standard diagnoses of O. viverrini by parasitological methods are increasingly unreliable because of low sensitivity in people with light infections (12) and difficulties in discriminating a mixed infection with intestinal trematodes (7, 8, 16). Alternatively, a PCR-based assay that offers high specificity and sensitivity was recently described for the detection of O. viverrini DNA in fecal samples. High sensitivity is achieved in cases of moderate-to-severe infections (more than 1,000 eggs per g [EPG] of feces), but the sensitivity is low in light infections (<1,000 EPG) (17, 18). We recently applied an identical PCR system for the diagnosis of opisthorchiasis and observed a sensitivity of 45% in samples of human stool from the Lao PDR (15). In order to solve the problem of low sensitivity of the PCR-based diagnosis in opisthorchiasis, the present study was carried out to improve the DNA extraction and clean-up protocol and evaluate the efficacy of the new PCR method for the reliable detection of O. viverrini.
Fecal samples collected from an area of the Lao PDR where opisthorchiasis is endemic that was originally described in a previous study (15) were analyzed. A sample of 79 fecal specimens was available for this study, and parasite infections determined by the quantitative formalin-ethyl acetate concentration technique (FECT) consisted of O. viverrini (65.8%), Strongyloides stercoralis (17.7%), minute intestinal flukes (5.1%), hookworms (21.5%), Ascaris lumbricoides (5.1%), and Trichuris trichiura (5.1%).
Methods used for sample preparation for DNA extraction, DNA clean-up, and PCR were modified from previous reports (5, 17). Briefly, eggs were isolated from fecal samples (approximately 500 mg) by mixing the samples with 4 ml of sterilized normal saline and 400 μl of ethyl acetate, shaking the mixture vigorously, and centrifuging it at 4,000 rpm for 10 min. The pellet was incubated in 1 ml of 0.5 N NaOH for 60 min at room temperature, autoclaved for 60 min (121°C), mixed, and centrifuged for 10 min at 4,000 rpm. The supernatant was transferred to a 1.5-ml microcentrifuge tube, 56 μl of 10× Tris-EDTA (TE; pH 8.0) was added, and the pH was adjusted to 7 to 8. A 30-μl volume of 10% sodium dodecyl sulfate and 3 μl of 20 mg/ml proteinase K (60 μg) were added to the supernatant, and the mixture was incubated at 65°C for 1 h. One hundred microliters of 5 M NaCl and 80 μl of cetyltrimethyl-ammoniumbromide-NaCl were added, and the mixture was incubated at 65°C for 30 min (0.7 M NaCl and 0.1 volume of cetyltrimethyl-ammoniumbromide). The mixture was extracted twice with an equal volume of chloroform-isoamyl alcohol (24:1), followed by phenol-chloroform-isoamyl alcohol (25:24:1). DNA was precipitated by adding 0.6 volume of isopropanol and pelleted by centrifugation, washed once with 70% ethanol, dried, and dissolved in 50 μl of 1× TE buffer. Two microliters of the final DNA solution in TE buffer (4%) was used in the PCR. O. viverrini-specific primers OV-6 (5′-CTGAATCTCTCGTTTGTTCA-3′ and 5′-GTTCCAGGTGAGTCTCTCTA-3′) were used for PCR (17). The primers were designed to amplify a repetitive DNA fragment specific for O. viverrini which showed no significant homology to other parasites, and the amplification yielded a specific product with a size of 330 bp (10, 17). PCR assays were carried out in a final volume of 25 μl consisting of PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl, 1.5 mM MgCl2), 200 μΜ each deoxynucleoside triphosphate, and 1 μM Taq DNA polymerase. PCR assays were performed in a DNA thermal cycler (MJ PTC-200) and adjusted to the following cycling conditions: an initial denaturation step at 94°C for 5 min, denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 45 s (30 cycles). Amplified products were analyzed by electrophoresis in a 2% agarose gel with 1× Tris-borate-EDTA buffer, pH 8.0. The amplicons obtained from the sample analyses were sequenced to compare with that of the specific product with genomic DNA prepared from adult O. viverrini as the template. For PCR-negative samples, further tests were performed with ITS-2 general primers to identify parasites other than O. viverrini as previously described (2). In order to ascertain the presence of an inhibitor as the cause of a failure to find parasite DNA, O. viverrini DNA was added to the negative samples and the PCR test was repeated. A positive PCR test result confirmed that the sample was free of inhibitory substances and the specimen was regarded as negative for O. viverrini DNA. Typical examples of the PCR analyses of O. viverrini egg-positive and parasite-negative samples are shown in Fig. 1.
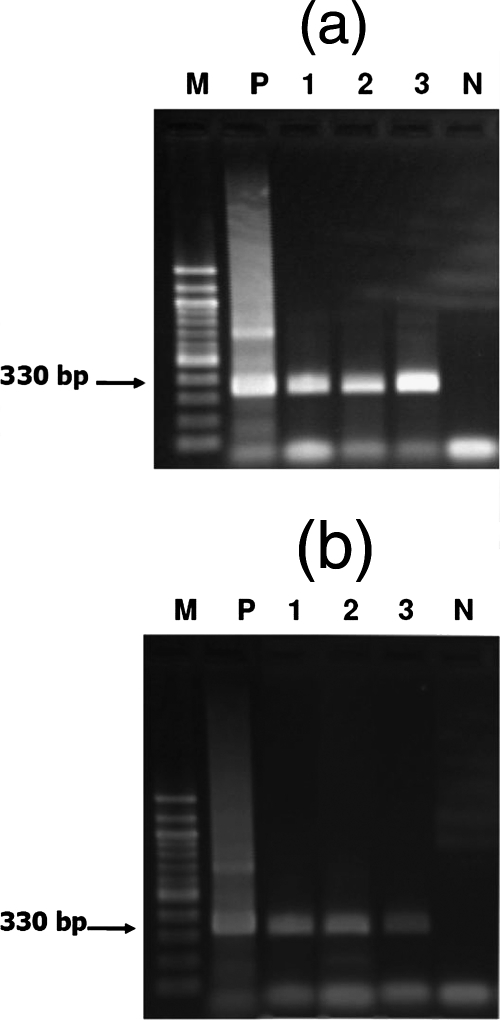
FIG. 1.
Detection of O. viverrini DNA in human fecal samples by PCR. Ethidium bromide staining of PCR products separated in a 2% agarose gel is shown. (a) O. viverrini egg-positive specimens. Lane M is a 100-bp marker, lane P is a positive control. Lane 1, 442 EPG; lane 2, 1,120 EPG; lane 3, 180 EPG; lane N, negative control. (b) Parasite-negative specimens. Lane M, 100-bp marker; lane P, positive control. Lanes 1 to 3 contain DNAs from parasite-negative specimens. Lane N, negative control.
With the new PCR system, 52 (65.8%) out of 79 specimens showed positive results (Table 1). The prevalence of O. viverrini by PCR was not significantly different from that by parasitological diagnosis (FECT) (P > 0.05). Of the samples negative for O. viverrini eggs (n = 21), 6 (28.6%) were positive by PCR. When only those samples positive for O. viverrini eggs were considered (n = 58), 46 (79.3%) were positive by PCR. With reference to FECT, the overall sensitivity of the modified PCR was 79.3% and the specificity was 71.4%. The positive and negative predictive values were 88.5% and 55.6%, respectively.
TABLE 1.
Relationship between diagnoses of O. viverrini infection by PCR and the FECT method in 79 fecal samples
| PCR test result | No. (%) of samples with FECT result of:
|
Total no. (%) | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 46 | 6 | 52 (65.8) |
| Negative | 12 | 15 | 27 (34.1) |
| Total | 58 (73.4) | 21 (26.6) | 79 (100) |
As shown in Table 2, within different O. viverrini intensity groups, the rates of PCR positivity by the modified protocol used in this study were consistently higher than that observed in the previous report (15). In total, the O. viverrini prevalence of 65.8% obtained by our PCR was significantly greater than that obtained by the previous protocol (35.4%, 28 out 79 fecal samples) (P < 0.05). More positive tests (up to 50%) were discovered in the egg-negative group, as well as those with egg counts in the range of 1 to 6,700 EPG. A case with 4,050 EPG gave negative tests by both PCR protocols. Overall, the sensitivity of PCR diagnosis in this study (79.3%) was significantly greater than that observed in the previous study (44.8%).
TABLE 2.
Diagnosis of O. viverrini infection by PCR in different intensity groups based on the previous protocola and the new protocol described in this study
| No. of EPG | No. of samples | No. (%) PCR positive by:
|
|
|---|---|---|---|
| Previous protocol | New protocol | ||
| 0 | 21 | 3 (14.3) | 6 (28.6) |
| 1-99 | 5 | 3 (60) | 4 (80) |
| 100-199 | 10 | 4 (40) | 9 (90) |
| 200-999 | 28 | 13 (46.4) | 21 (75) |
| 1,000-1,999 | 10 | 2 (20) | 8 (80) |
| >2,000 | 5 | 3 (60) | 4 (80) |
| Total | 79 | 28 (35.4) | 52 (65.8) |
Described in reference 15.
Since the primers and PCR conditions of the new and previous PCR systems are identical, the reason for the improvement in sensitivity of the modified method in this study is likely due to more efficient removal of a PCR inhibitor(s) from the specimen in the DNA extraction procedure. Substances typically present in human feces such as proteases, bilirubin, bile salt, fat, and complex polysaccharides may interfere with the PCR (1). In particular, polysaccharides may originate from the consumption of a high-carbohydrate diet, mostly grains and vegetables, by the sample population in the Lao PDR.
A correlation of PCR diagnosis for O. viverrini and infection status is shown in Table 3. In the parasite-negative group, 5.1% (four) of the samples were positive by PCR, and it is not unexpected that PCR is more sensitive than FECT. Since multiple parasite infections were common in the samples obtained from the Lao PDR, the effects of mixed infections with O. viverrini and other parasites on diagnosis by PCR was assessed. Samples with O. viverrini infection alone showed a PCR positivity rate of 30.4%, which is comparable to that of mixed infections with O. viverrini and other parasites such as hookworms, S. stercoralis, Taenia, T. trichiura, minute intestinal flukes, and A. lumbricoides (27.8%, P > 0.05). Two PCR-positive results were found in a case with a mixed infection of hookworms and S. stercoralis and another case with T. trichiura infection. No positive PCR was seen in three cases of hookworm infection. Because fecal examination is not an ideal “gold standard” and the number of samples with single-parasite infections in this study was small, it is not permissible to perform a comprehensive analysis of cross-reactivity of the PCR diagnosis.
TABLE 3.
Classification of PCR test results for the diagnosis of O. viverrini infection according to infection status determined by the FECT method in 79 samples
| Parasite(s) | No. (%) of samples PCR positive |
|---|---|
| None | 4 (5.1) |
| O. viverrini | 24 (30.4) |
| O. viverrini and othersa | 22 (27.8) |
| Hookworms and Strongyloides stercoralis | 1 (1.2) |
| Trichuris trichiura | 1 (1.2) |
| Hookworms | 0 (0) |
Hookworms, S. stercoralis, Taenia, Trichuris trichiura, minute intestinal flukes, and Ascaris lumbricoides.
Among the samples that were positive for O. viverrini by both egg and PCR analyses, 34 (79%) fell within the light infection group (<1,000 EPG) and 12 (80%) fell within the moderate-to-heavy infection group (>1,000 to 9,999 EPG). However, discrepancies in test results were obtained with 12 specimens that were positive for eggs but negative by PCR. Eight samples (75%) had egg counts of <1,000 EPG, while four samples (25%) had egg counts of >1,000 EPG. The trend of a positive correlation between PCR test results and egg counts was similarly observed in a previous report (15). However, the possibility that the eggs observed in some specimens were not O. viverrini but other small intestinal flukes cannot be ruled out (6, 7).
In terms of cost, the new PCR protocol established in this study is more economical than the previous protocol since no commercial kit is needed (15). Regarding the sample-processing time required to complete the PCR test, the protocol took approximately 8 h while the previous protocol using the commercial stool kit took approximately 7 h. The new protocol needed an hour longer for proteinase K digestion and phenol-chloroform extraction.
An alternative clean-up method using KOH and a combination of sucrose and KCl, followed by either autoclaving or homogenization to break the eggs, has been reported, and the detection limit was 10 EPG in the PCR diagnosis of Clonorchis sinensis (9). In our study, the lowest egg count detected was 31 EPG in the case of egg-positive samples and probably lower than this in the case of egg-negative samples. The finding of PCR positive results in egg-negative samples is in concordance with the monoclonal-antibody-based coproantigen detection method, which also demonstrated positive reactions in egg-negative cases (11). Moreover, in an autopsy study, only 30% of the people who harbored fewer than 10 adult O. viverrini worms excreted eggs that were detectable in their feces while the majority of them (70%) were egg negative (13). That we obtained PCR-positive results with apparently egg-negative samples suggests that diagnosis by our PCR method is more sensitive than the fecal examination method, particularly in the light infection group.
Acknowledgments
This research was supported by the Liver Fluke and Cholangiocarcinoma Research Center (LFCRC); the Faculty of Medicine, Graduate School, Khon Kaen University, Khon Kaen, Thailand; and the European Commission through the TREMKIT project (ICA4-2000-10163).
Footnotes
Published ahead of print on 14 November 2007.
REFERENCES
- 1.Abu Al-Soud, W., and P. Radstrom. 2000. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol. 384463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando, K., P. Sithithaworn, C. Nuchjungreed, S. Tesana, T. Srisawangwong, W. Limviroj, and Y. Chinzei. 2001. Nucleotide sequence of mitochondrial CO I and ribosomal ITS II genes of Opisthorchis viverrini in northeast Thailand. Southeast Asian J. Trop. Med. Public Health 32(Suppl. 2)17-22. [PubMed] [Google Scholar]
- 3.Anonymous. 1994. Infection with liver flukes (Opisthorchis viverrini, Opisthorchis felineus and Clonorchis sinensis). IARC Monogr. Eval. Carcinog. Risks Hum. 61121-175. [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 1995. Control of foodborne trematode infection. WHO Tech. Rep. Ser. 8491-157. [PubMed] [Google Scholar]
- 5.Ausubel, M. F., R. Brent, E. R. Kingston, D. D. Moore, G. J. Seidman, A. J. Smith, and K. Struhl. 2002. Short protocols in molecular biology, 5th ed., vol. 1. John Wiley & Sons, Inc., New York, NY.
- 6.Chai, J. Y., J. H. Park, E. T. Han, S. M. Guk, E. H. Shin, A. Lin, J. L. Kim, W. M. Sohn, T. S. Yong, K. S. Eom, D. Y. Min, E. H. Hwang, B. Phommmasack, B. Insisiengmay, and H. J. Rim. 2005. Mixed infections with Opisthorchis viverrini and intestinal flukes in residents of Vientiane Municipality and Saravane Province in Laos. J. Helminthol. 79283-289. [DOI] [PubMed] [Google Scholar]
- 7.Ditrich, O., T. Scholz, and M. Giboda. 1990. Occurrence of some medically important flukes (Trematoda: Opisthorchiidae and Heterophyidae) in Nam Ngum water reservoir, Laos. Southeast Asian J. Trop. Med. Public Health 21482-488. [PubMed] [Google Scholar]
- 8.Kaewkes, S., D. B. Elkins, P. Sithithaworn, and M. R. Haswell-Elkins. 1991. Comparative studies on the morphology of the eggs of Opisthorchis viverrini and lecithodendriid trematodes. Southeast Asian J. Trop. Med. Public Health 22623-630. [PubMed] [Google Scholar]
- 9.Müller, B., J. Schmidt, and H. Mehlhorn. 2007. PCR diagnosis of infections with different species of Opisthorchiidae using a rapid clean-up procedure for stool samples and specific primers. Parasitol. Res. 100905-909. [DOI] [PubMed] [Google Scholar]
- 10.Sermswan, R., S. Mongkolsuk, S. Panyim, and S. Sirisinha. 1991. Isolation and characterization of Opisthorchis viverrini specific DNA probe. Mol. Cell. Probes 5399-407. [DOI] [PubMed] [Google Scholar]
- 11.Sirisinha, S., R. Chawengkirttikul, M. R. Haswell-Elkins, D. B. Elkins, S. Kaewkes, and P. Sithithaworn. 1995. Evaluation of a monoclonal antibody-based enzyme linked immunosorbent assay for the diagnosis of Opisthorchis viverrini infection in an endemic area. Am. J. Trop. Med. Hyg. 52521-524. [DOI] [PubMed] [Google Scholar]
- 12.Sithithaworn, P., and M. Haswell-Elkins. 2003. Epidemiology of Opisthorchis viverrini. Acta Trop. 88187-194. [DOI] [PubMed] [Google Scholar]
- 13.Sithithaworn, P., S. Tesana, V. Pipitgool, S. Kaewkes, C. Pairojkul, B. Sripa, A. Paupairoj, and K. Thaiklar. 1991. Relationship between faecal egg count and worm burden of Opisthorchis viverrini in human autopsy cases. Parasitology 102(Pt. 2)277-281. [DOI] [PubMed] [Google Scholar]
- 14.Sripa, B., S. Kaewkes, P. Sithithaworn, E. Mairiang, T. Laha, M. Smout, C. Pairojkul, V. Bhudhisawasdi, S. Tesana, B. Thinkamrop, J. M. Bethony, A. Loukas, and P. J. Brindley. 2007. Liver fluke induces cholangiocarcinoma. PLoS Med. 4e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stensvold, C. R., W. Saijuntha, P. Sithithaworn, S. Wongratanacheewin, H. Strandgaard, N. Ornbjerg, and M. V. Johansen. 2006. Evaluation of PCR based coprodiagnosis of human opisthorchiasis. Acta Trop. 9726-30. [DOI] [PubMed] [Google Scholar]
- 16.Tesana, S., T. Srisawangwonk, S. Kaewkes, P. Sithithaworn, P. Kanla, and C. Arunyanart. 1991. Eggshell morphology of the small eggs of human trematodes in Thailand. Southeast Asian J. Trop. Med. Public Health 22631-636. [PubMed] [Google Scholar]
- 17.Wongratanacheewin, S., W. Pumidonming, R. W. Sermswan, and W. Maleewong. 2001. Development of a PCR-based method for the detection of Opisthorchis viverrini in experimentally infected hamsters. Parasitology 122175-180. [DOI] [PubMed] [Google Scholar]
- 18.Wongratanacheewin, S., W. Pumidonming, R. W. Sermswan, V. Pipitgool, and W. Maleewong. 2002. Detection of Opisthorchis viverrini in human stool specimens by PCR. J. Clin. Microbiol. 403879-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]