Abstract
Sequence-based methods for typing Staphylococcus aureus, such as multilocus sequence typing (MLST) and spa typing, have increased interlaboratory reproducibility, portability, and speed in obtaining results, but pulsed-field gel electrophoresis (PFGE), remains the method of choice in many laboratories due to the extensive experience with this methodology and the large body of data accumulated using the technique. Comparisons between typing methods have been overwhelmingly based on a qualitative assessment of the overall agreement of results and the relative discriminatory indexes. In this study, we quantitatively assess the congruence of the major typing methods for S. aureus, using a diverse collection of 198 S. aureus strains previously characterized by PFGE, spa typing, MLST, and, in the case of methicillin-resistant S. aureus (MRSA), SCCmec typing in order to establish the quantitative congruence between the typing methods. The results of most typing methods agree in that MRSA and methicillin-susceptible S. aureus (MSSA) differ in terms of diversity of genetic backgrounds, with MSSA being more diverse. Our results show that spa typing has a very good predictive power over the clonal relationships defined by eBURST, while PFGE is less accurate for that purpose but nevertheless provides better typeability and discriminatory power. The combination of PFGE and spa typing provided even better results. Based on these observations, we suggest the use of the conjugation of spa typing and PFGE typing for epidemiological surveillance studies, since this combination provides the ability to infer long-term relationships while maintaining the discriminatory power and typeability needed in short-term studies.
Staphylococcus aureus is a leading human pathogen and remains a major cause of infections worldwide (16, 22, 38), with high rates of hospital-acquired infections in several countries (2, 12). Recently, the epidemiology of S. aureus, in particular for methicillin-resistant S. aureus (MRSA), has changed with the emergence of community-acquired MRSA, as reported by several studies (12, 13, 16, 22, 38). The epidemiology of infectious diseases relies on typing methods as tools for the characterization and discrimination of isolates based on either their genotypic or phenotypic characteristics, which may be used to establish clonal relationships between strains and to trace the geographic dissemination of bacterial clones. Nowadays, the classification of isolates is mostly based on molecular methods, which usually provide better discriminatory power than phenotypic methods. Pulsed-field gel electrophoresis (PFGE), after SmaI digestion of total bacterial DNA (33), is still regarded by many authors as the gold standard for benchmarking new typing methods, although it was originally proposed for outbreak investigation (37). Recently, due to the availability and affordability of DNA sequence technology, several sequenced-based typing methods have been developed and are now widely used, such as multilocus sequence typing (MLST) (23) and spa typing (34), which are the most frequently used for S. aureus. DNA sequence-based typing methods generate unambiguous and portable data, amenable to the creation of central databases, which enable the comparison of local data with data from previous studies in different geographical locations.
Apart from factors such as discriminatory indexes, reproducibility, and standardization, typing techniques differ dramatically in associated costs (32, 39), which may restrict the choice of typing methods due to budget limitations. For instance, MLST, which relies on the sequences of the internal fragments of seven housekeeping genes, is much more expensive than spa typing, which is based on the sequence of an internal fragment of a single gene. Although PFGE is labor-intensive and may be a more economical alternative, it has several drawbacks: it requires unique technical skills and has a high setup cost, and the interlaboratory comparison of results is not straightforward.
According to a proposal by Enright and colleagues (11) that was accepted by a subcommittee of the International Union of Microbiology Societies in Tokyo in 2002, MRSA clones are named according to their MLST and staphylococcal cassette chromosome (SCCmec) types (e.g., clone ST5-MRSA-II). However, the amount of sequencing required for MLST typing and the increasing number of primers need to define SCCmec types (25, 29) as new types and variants are found hamper the use of this combination of methods for clonal characterization of large collections, mainly due to cost-related reasons. Other combinations of methods that provide a similarly fine resolution of the accepted clonal group definition should be explored.
In line with this rationale, SeqNet (http://www.seqnet.org), the European Network of Laboratories for Sequence Based Typing of Microbial Pathogens, has proposed spa typing as the sequence-based method of choice to determine the genetic relatedness of S. aureus isolates. An online database is now available featuring automated curation of submitted sequences and assignment to spa types (3).
Molecular epidemiology studies of clinical microorganisms often rely on the application of typing methods that produce different type assignments. From the comparison and analysis of these assignments, a classification of the isolate in terms of clonal type or lineage is generated. What is more, since different laboratories may use different combinations of methods and, over time, implement new typing schemes, the definition of clones is neither universal nor static. Since different typing schemes analyze different phenotypic and genotypic properties of bacteria, if a congruent result is obtained between different methods, it suggests that a phylogenetic signal is being recovered by both methods, allowing high confidence in the assignment of clonal types. Therefore, the quantification of congruence between different methods, with an assessment of the confidence for predicting an unknown character from another typing method, can be a useful tool in epidemiology (6), enabling an informed choice between typing methods for a given study, taking into account the degree of discrimination needed and the available budget.
At the Molecular Genetics Laboratory, Instituto de Tecnologia Química e Biológica, Oeiras, Portugal, different S. aureus strains from different worldwide locations have been collected since the late 1980s. These strains have been analyzed by different typing methods over time. PFGE has been the standard typing technique during this period due to its high discriminatory power and relatively low cost per isolate. However, with the introduction of spa typing and MLST schemes, the characterization of S. aureus isolates now involves a combination of different techniques (including the SCCmec type for the characterization of MRSA strains [20, 27, 28]). We previously extended the work of Robinson et al. (31) by proposing the use of measures of clustering concordance—adjusted Rand (AR) and Wallace (W) coefficients—to compare type assignments, allowing a quantitative approach for exploring the concordance between typing methods (6). In this study, we have implemented the use of that methodological framework for a set of 198 S. aureus strains, which were previously characterized by PFGE, spa typing, MLST, and SCCmec typing (for MRSA), in order to quantify the congruence between methods and the discriminatory power of each method and combination of methods.
MATERIALS AND METHODS
Strain collection.
A collection of 198 S. aureus strains (116 MRSA and 82 methicillin-susceptible S. aureus [MSSA] strains) was included in this study (see Tables S1 and S2 in the supplemental material). These strains were chosen from a large (>5,000) international collection of S. aureus isolates isolated in several parts of the world, mainly in hospitals in southern and eastern Europe, Latin America, and the United States, since the late 1980s and included representatives of early isolates from the United Kingdom and Denmark isolated between 1957 and 1972. Overall, among the 198 selected S. aureus strains, 19 countries are represented: Argentina (n = 9; 4.5%), Brazil (n = 1; 0.5%), Cabo Verde (n = 12; 6%), Chile (n = 1; 0.5%), Colombia (n = 2; 1%), Czech Republic (n = 3; 1.5%), Denmark (n = 46; 23%), Egypt (n = 10; 5%), Greece (n = 5; 2.5%), Hungary (n = 18; 9%), Italy (n = 2; 1%), Japan (n = 7; 3.5%), Mexico (n = 4; 2%), Poland (n = 4; 2%), Portugal (n = 63; 32%), Spain (n = 1; 0.5%), Taiwan (n = 5; 2.5%), United Kingdom (n = 8; 4.0%), and United States (n = 6; 3%). The criteria used in the strain selection process excluded duplicate outbreak strains in order to minimize sampling bias and tried to maximize the diversity represented in the analyzed collection relative to that present in the >5,000 isolates screened. All strains included in this study were characterized by PFGE (7), MLST (9, 10), and spa typing (21, 34). MRSA strains were also characterized by SCCmec typing (27, 28).
PFGE data analysis.
PFGE patterns were analyzed in Bionumerics version 4.61 from Applied Maths (Sint-Martens-Latem, Belgium). Gel photographs were acquired using Polaroid black-and-white instant pack film 667, and the negatives were digitized as 8-bit grayscale TIFF images for use with the above-mentioned software. Each image was then analyzed using the resources of the Bionumerics software. A spectral analysis was performed for each image in order to obtain the background subtraction (background scale) and the cutoff threshold for least-squares filtering (Wiener cutoff scale). After this process, intra- and intergel PFGE runs were normalized using S. aureus strain NCTC8325 loaded in each gel as a reference. Band assignments were manually curated after automatic band detection for all gel images; bands ranging from 10 kb to ∼674 kb were considered for analysis in the study. For band pattern comparisons within and between different gels, the following settings were used: optimization of 0.5% and position tolerance of 1.25%. PFGE types and subtypes were defined by groups formed at 80% and 95% Dice similarity cutoffs on a dendrogram constructed by the unweighted-pair group method using average linkages (UPGMA). The groups defined at these thresholds were previously shown to approximate those defined using Tenover's criteria for visual PFGE type definition (5, 24, 37).
DNA sequence data analysis. (i) spa typing.
Ridom StaphType software, version 1.4 (Ridom GmbH, Würzburg, Germany) was used for spa type analyses. The new spa type assignments were provided automatically through the Ridom SpaServer (http://spa.ridom.de/index.shtml). The BURP algorithm (Ridom StaphType software) was used to calculate spa clonal complexes (CC) with the following default parameters for the group/cluster definition: “exclude spa types that are shorter than five repeats” and “spa types are clustered if cost is less than or equal to 6” (30).
(ii) MLST.
MLST sequence types (ST) were assigned through the MLST database (http://www.mlst.net). The e-BURST algorithm was used to assign MLST CC (http://eburst.mlst.net).
(iii) SCCmec typing.
SCCmec types were determined by a multiplex PCR strategy, which generated a specific amplification pattern for each SCCmec structural type (28). SCCmec type assignments were confirmed by ccrAB typing, as previously described by Okuma et al. (27).
(iv) Diversity indexes.
Hunter and Gaston (18) proposed the use of Simpson's index of diversity (SID) (35) to measure the discriminatory abilities of typing systems. This index indicates the probability of two strains sampled randomly from a population belonging to two different types. Grundmann et al. (14a) introduced a method for determining confidence intervals of SID, thereby improving the objective assessment of the discriminatory powers of typing techniques.
Comparison of typing methods.
A framework for assessing the quantitative correspondence between typing methods was proposed by Carriço et al. (6). It is based on two coefficients developed to compare two ways to partition a given data set: AR (17) and W (40).
The AR coefficient corrects Rand's coefficient, commonly used for quantifying the congruence of typing methods (31), for the presence of chance agreement, i.e., that the two sets of results match by chance alone. The use of the Rand coefficient, which is formally equivalent to the concordance measure proposed by Robinson et al. (31), leads to overestimation of the agreement between two typing methods and should be avoided. (For a more detailed discussion of the use of these indexes in the context of microbial typing, see Carriço et al. [6].) The W coefficient can provide an even finer comparison between two methods, since the value indicates the probability that two strains classified as the same type by one method are also classified as the same type by the other method. A high value of the W coefficient (it can assume any value from 0 to 1) indicates that partitions defined by a given method could have been predicted from the results of another method, suggesting that the use of both methodologies could be redundant. The combined use of the two coefficients can provide further information: two methods can have a low global agreement (assessed by AR) and yet one of those methods can predict very well the results of another, which can be assessed by the W coefficient. To facilitate the use of these coefficients, a web page with the Bionumerics scripts used in this study has been made available at http://www.comparingpartitions.info.
RESULTS
Our strain collection can be divided into two distinct groups, MRSA and MSSA, that differ in the “broad-spectrum” resistance to β-lactams as a consequence of the acquisition of the SCCmec element. The following analysis was always applied to the two groups and to the overall collection, since the MRSA and MSSA populations were expected to differ. For each typing method, two levels of discrimination were considered, one corresponding to the direct result of the method itself (MLST ST, spa type, PFGE subtype, and, for MRSA, SCCmec type) and another resulting from the application of an algorithm that generates groups of related isolates from these primary data (eBURST for MLST data, BURP for spa, and the 80% cutoff for defining the PFGE type). In order to explore whether there was an improvement in discriminatory power compared with classifications obtained with individual typing method results, classifications based on the combination of typing methods were also considered. One such conjugation of typing methods evaluated was the currently accepted ST-SCCmec combination for the definition of MRSA lineages. We also considered the conjugation of PFGE type and subtype with spa type, the two methods recently shown to be suitable for long range epidemiological studies (15), and in the MRSA subset, PFGE type and subtype together with SCCmec type. The results of the number of groups found for each method or conjugation of methods are presented in Table 1.
TABLE 1.
Resolution of typing methods for the three data sets analyzed
| Data set | Typing technique | No. of types | Typeability (%) | SID | 95% CIa |
|---|---|---|---|---|---|
| Entire collection (n = 198) | PFGE type | 56 | 100 | 95.92 | (94.89-96.95) |
| PFGE subtype | 153 | 100 | 99.69 | (99.55-99.82) | |
| spa type | 98 | 99 | 97.31 | (96.30-98.33) | |
| BURP (spa) | 29 | 97 | 87.35 | (84.88-89.81) | |
| ST (MLST) | 61 | 100 | 94.82 | (93.42-96.21) | |
| e-BURST (MLST) | 21 | 100 | 82.24 | (78.19-86.29) | |
| PFGE type + spa type | 129 | 100 | 98.88 | (98.29-99.47) | |
| PFGE subtype + spa type | 175 | 100 | 99.85 | (99.74-99.95) | |
| MRSA strains (n = 116) | PFGE type | 32 | 100 | 94.27 | (92.70-95.84) |
| PFGE subtype | 92 | 100 | 99.51 | (99.20-99.81) | |
| spa type | 51 | 98.3 | 95.85 | (94.34-97.35) | |
| BURP (spa) | 14 | 97.4 | 78.62 | (74.98-82.26) | |
| ST (MLST) | 34 | 100 | 91.36 | (88.63-94.10) | |
| e-BURST (MLST) | 12 | 100 | 70.84 | (63.99-77.69) | |
| SCCmec | 11 | 97.4 | 83.13 | (79.89-86.38) | |
| PFGE type +spa type | 72 | 100 | 98.32 | (97.47-99.17) | |
| PFGE subtype + spa type | 101 | 100 | 99.67 | (99.38-99.96) | |
| PFGE type + SCCmec | 56 | 100 | 97.32 | (96.21-98.43) | |
| PFGE subtype + SCCmec | 96 | 100 | 99.57 | (99.26-99.87) | |
| SCCmec + ST (MLST) | 50 | 100 | 96.42 | (95.18-97.65) | |
| MSSA strains (n = 82) | PFGE type | 30 | 100 | 94.85 | (92.85-96.86) |
| PFGE subtype | 63 | 100 | 99.28 | (98.83-99.72) | |
| spa type | 55 | 100 | 97.83 | (96.27-99.39) | |
| BURP (spa) | 26 | 96.3 | 93.83 | (91.73-95.92) | |
| ST (MLST) | 35 | 100 | 96.18 | (94.78-97.58) | |
| e-BURST (MLST) | 17 | 100 | 90.88 | (88.19-93.56) | |
| PFGE type + spa type | 59 | 100 | 98.01 | (96.43-99.59) | |
| PFGE subtype + spa type | 74 | 100 | 99.76 | (99.55-99.97) |
CI, confidence interval.
PFGE.
The type and subtype definitions in PFGE are commonly obtained by determining a cutoff value on a dendrogram of distances constructed with the Dice coefficient and UPGMA. Several cutoff values and parameters for dendrogram construction have been proposed (5, 24, 26, 36). Ideally, the determination of the cutoff level should be supported by other epidemiologically relevant data referring to the strains. To evaluate the groups defined by PFGE, two cutoff levels were considered—80% to define PFGE types and 95% to define PFGE subtypes—which were shown to be adequate for the analysis of large and diverse collections of strains (24). Considering the entire collection of 198 strains, 56 PFGE types (80% cutoff) and 153 PFGE subtypes (95% cutoff) were detected, which can be split into 32 types and 92 subtypes if only MRSA strains are considered and 30 types and 63 subtypes for MSSA.
spa typing.
spa types were assigned using Ridom StaphType software, and the BURP algorithm was run on all the datasets using the settings described in Materials and Methods. For the 198 strains, 98 distinct spa types were found. For the MRSA and MSSA subsets, 51 and 55 types were determined, respectively. Some spa types were shared by both subsets: t002, t008, t012, t018, t021, t024, t127, and t148.
When the BURP algorithm was applied for the inference of clusters of related isolates, 192 strains (96.7%) were distributed in 29 groups, where 12 of those groups were singletons (groups represented by a single type) and six strains (three MRSA and three MSSA strains) were excluded due to the rules described in Materials and Methods.
MLST.
ST were assigned through the MLST database (http://www.mlst.net), and MLST CC were calculated using the e-BURST algorithm. Overall, the 198 strains were distributed among 61 ST belonging to 21 CC. Thirty-two ST, belonging to 11 CC, were found among the 116 MRSA strains. MSSA strains were more diverse, since among the 82 MSSA strains, 35 ST belonging to 17 CC were detected.
Comparing discriminatory powers of different methods.
SID provides a measure of the discriminatory powers of the different typing methods or conjugations of typing methods within a confidence interval. If the confidence intervals of any two methods overlap, one cannot exclude the hypothesis that they have similar discriminatory powers at a 95% confidence level. The SID values obtained for our data sets are presented in Table 1. When either the entire collection or only the MRSA subset was analyzed, PFGE had the highest SID whenever PFGE subtype assignments were compared with spa types or ST or PFGE types or subtypes were compared to eBURST or BURP. However when the PFGE type assignment was compared with the spa types, as is usually done in the literature, similar levels of discriminatory power were found (15). This observation was valid for the entire data set or any of the MRSA or MSSA subsets. As expected for MRSA strains, SCCmec typing was the least discriminatory technique due to the restricted number of variants generated by the method. Nevertheless, if SCCmec types were conjugated with ST, the level of discrimination was similar to that of spa typing or conjugation of PFGE type and SCCmec type. Among all conjugations of two typing methods, the highest SIDs were obtained for the PFGE subtype with either spa or SCCmec type. It is interesting that for MSSA, the three typing methods (PFGE, MLST, and spa typing) showed similar SIDs, in contrast to MRSA, where ST do not perform so well. Also, when the MLST discriminatory powers at the ST or eBURST level in MRSA and MSSA were compared, the MSSA always had higher values. This was also true when the discriminatory powers of groups made by the BURP algorithm were compared, further confirming the more diverse genetic structure of the MSSA subset.
Global concordance of typing method groupings: the AR coefficient.
By analyzing the values of the AR coefficients in all typing methods, an overall concordance value could be reached (Tables 2, 3, and 4). For comparison with other published values for concordance between typing methods (31), tables with Rand coefficient values are provided in the supplemental material (see Tables S3 to S5 in the supplemental material), although these values clearly overestimate the concordance between typing methods, as discussed in Materials and Methods and previous publications (6). For the entire collection (Table 2), the highest AR value found between two single typing methodologies was between eBURST and BURP groups: 0.551. This value suggests that clustering ST in CC using eBURST or spa types in groups using BURP produces roughly similar phylogenetic signals. All other methods and combinations of methods presented AR values lower than 0.5, except, as expected, one comparing a method with a conjugation of itself with another method (e.g., PFGE type plus spa type versus spa type: 0.581 concordance).
TABLE 2.
Adjusted Rand values for the entire collection (n = 198)
| Typing technique | Adjusted Rand value
|
||||||
|---|---|---|---|---|---|---|---|
| PFGE type | PFGE subtype | spa type | BURP (spa) | ST (MLST) | e-BURST (MLST) | PFGE type + spa type | |
| PFGE subtype | 0.1373 | ||||||
| spa type | 0.3079 | 0.0975 | |||||
| BURP (spa) | 0.3215 | 0.0376 | 0.3201 | ||||
| ST (MLST) | 0.3372 | 0.0823 | 0.3915 | 0.4915 | |||
| e-BURST (MLST) | 0.2719 | 0.0286 | 0.2189 | 0.5507 | 0.4041 | ||
| PFGE type + spa type | 0.4198 | 0.2112 | 0.5810 | 0.1448 | 0.2266 | 0.0990 | |
| PFGE subtype + spa type | 0.0699 | 0.6586 | 0.1057 | 0.0210 | 0.0471 | 0.0142 | 0.2399 |
TABLE 3.
Adjusted Rand values for the MRSA strains (n = 116)
| Typing technique | Adjusted Rand value
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PFGE type | PFGE subtype | spa type | BURP (spa) | ST (MLST) | e-BURST (MLST) | SCCmec | PFGE type + spa type | PFGE subtype + spa type | PFGE type + SCCmec | PFGE subtype + SCCmec | |
| PFGE subtype | 0.1513 | ||||||||||
| spa type | 0.3065 | 0.1343 | |||||||||
| BURP (spa) | 0.2862 | 0.0345 | 0.2749 | ||||||||
| ST (MLST) | 0.1996 | 0.0801 | 0.3529 | 0.4897 | |||||||
| e-BURST (MLST) | 0.1935 | 0.0239 | 0.1798 | 0.5310 | 0.3735 | ||||||
| SCCmec | 0.1663 | 0.0409 | 0.1579 | 0.1599 | 0.1883 | 0.0571 | |||||
| PFGE type + spa type | 0.4389 | 0.2981 | 0.5654 | 0.1182 | 0.1804 | 0.0726 | 0.1002 | ||||
| PFGE subtype + spa type | 0.1033 | 0.7992 | 0.1419 | 0.0240 | 0.0576 | 0.0160 | 0.0286 | 0.3246 | |||
| PFGE type + SCCmec | 0.6244 | 0.2675 | 0.3245 | 0.1698 | 0.2087 | 0.0998 | 0.2393 | 0.5333 | 0.1943 | ||
| PFGE subtype + SCCmec | 0.1341 | 0.9352 | 0.1238 | 0.0316 | 0.0750 | 0.0210 | 0.0421 | 0.2787 | 0.7835 | 0.2734 | |
| SCCmec + ST (MLST) | 0.2605 | 0.1767 | 0.3873 | 0.2264 | 0.5644 | 0.1656 | 0.3096 | 0.3206 | 0.1250 | 0.4175 | 0.1802 |
TABLE 4.
Adjusted Rand values for the MSSA strains (n = 82)
| Typing technique | Adjusted Rand value
|
||||||
|---|---|---|---|---|---|---|---|
| PFGE type | PFGE subtype | spa type | BURP (spa) | ST (MLST) | e-BURST (MLST) | PFGE type + spa type | |
| PFGE subtype | 0.2365 | ||||||
| spa type | 0.5288 | 0.1575 | |||||
| BURP (spa) | 0.6055 | 0.1639 | 0.5039 | ||||
| ST (MLST) | 0.6560 | 0.2424 | 0.4160 | 0.5764 | |||
| e-BURST (MLST) | 0.5890 | 0.1352 | 0.3395 | 0.5623 | 0.5674 | ||
| PFGE type + spa type | 0.5439 | 0.1690 | 0.9556 | 0.4712 | 0.4094 | 0.3361 | |
| PFGE subtype + spa type | 0.0852 | 0.4982 | 0.1965 | 0.0708 | 0.0996 | 0.0470 | 0.2128 |
A similar situation was found when only the MRSA subset was considered (Table 3).
For the MSSA subset (Table 4), AR values were higher, although always below 0.66. The PFGE type had the highest agreement values among the methods: 0.53 with spa type, 0.61 with BURP groups, 0.66 with ST, and 0.59 with e-BURST CC. Similarly to the entire collection and the MRSA subset, for the MSSA subset, an agreement of 0.56 was found between BURP and eBURST and 0.58 between ST and BURP groups.
Directional agreement between typing method groupings: the W coefficient.
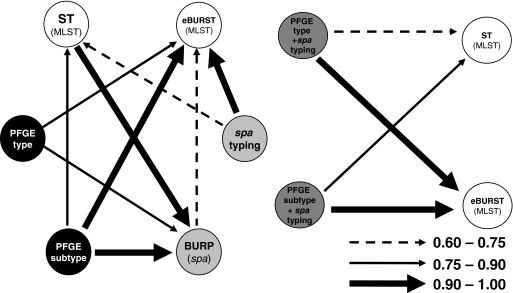
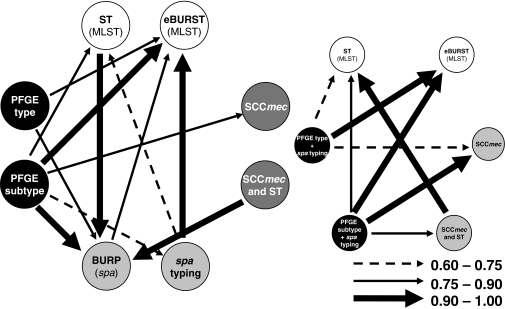
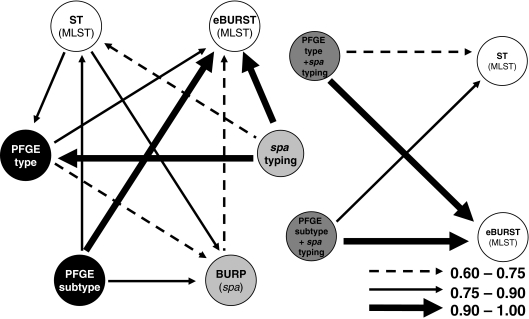
In order to determine how the results of one method map onto the results of the other methods, we calculated the W coefficients. The results are presented in Tables S6 to S8 in the supplemental material and are summarized in Fig. 1, 2, and 3. Overall, W coefficients showed that MRSA and MSSA data sets differed in the confidence of the predictions of the results of other typing methods when those of a single method were known.
FIG. 1.
Representation of correspondences between typing methods and combinations of typing methods used in the 198-strain collection, calculated by W coefficients. The arrows represent W coefficients of >0.60, excluding the obvious relationships.
FIG. 2.
Representation of correspondences between typing methods and combinations of typing methods used in the 116-strain MRSA subset, calculated by W coefficients. The arrows represent W coefficients of >0.60, excluding the obvious relationships.
FIG. 3.
Representation of correspondences between typing methods and combinations of typing methods used in the 82-strain MSSA subset, calculated by W coefficients. The arrows represent W coefficients of >0.60, excluding the obvious relationships.
Concerning spa type performance, it was found that in all datasets, if any two strains shared the same spa type, they had a high probability (over 94%) of belonging to the same eBURST group. spa typing was also able to predict the PFGE type in MSSA strains with a 92% probability, while for MRSA strains, the agreement was only about 40% (Fig. 2 and 3). In all data sets, spa typing was also not able to clearly predict the ST (W = 0.61 for the whole data set; W = 0.60 for MRSA, and W = 0.52 for MSSA).
PFGE was able to predict the BURP group much better for MRSA than for MSSA: W was 0.83 for PFGE type and 0.97 for PFGE subtype in MRSA strains versus 0.69 and 0.83, respectively, in MSSA strains. Concerning the ability of PFGE (type or subtype) to predict eBURST complexes, the W values were similar for MRSA and MSSA, as well as for the overall data set: around 0.84 for PFGE type and 1.0 for PFGE subtype. Similar results, at a lower level of agreement, were found for the PFGE subtype's capability to predict ST: W = 0.82 for the MRSA data set and W = 0.79 for MSSA and the entire data set. Conversely, if two strains had the same ST, when the whole data set was considered, they had only 33% probability of sharing the same PFGE type. This value was lower in the MRSA data set (W = 0.21) but, interestingly, higher for the MSSA data set (W = 0.79). In summary, knowing the ST information, one could only predict the PFGE type with some certainty if an isolate was MSSA. In the case of MRSA strains, PFGE subtypes also predicted quite well the SCCmec type (W = 0.88).
When the results of PFGE and spa typing were conjugated, it was found that if two strains were classified together in the same PFGE-spa type, there was 100% probability of sharing the same eBURST for MSSA or 99.5% for the entire collection (it was not 100% because there were two strains classified together in PFGE type 6/spa type t030, but one belonged to CC 8 while the other was a singleton). For MRSA, the agreement between PFGE-spa types and eBURST complexes was slightly lower (W = 0.94), which is explained by the fact that five strains grouped together in PFGE type 18/spa type t001 were divided into CC 228 (two strains) and CC 5 (three strains) and two strains grouped together in PFGE type 4-spa type t030 were classified as CC8 and as a singleton.
The combination of PFGE subtype with spa type does not add to the PFGE subtype alone for the prediction of eBURST groups, although it slightly improves the prediction of ST (Fig. 1). However, in MRSA strains, the PFGE subtype-spa type combination was found to perform better in the prediction of the SCCmec type (W = 0.91) than the PFGE subtype alone (W = 0.88).
We also analyzed the performances of PFGE type-SCCmec type and PFGE subtype-SCCmec type combinations for MRSA strains, and in both cases, there was no significant difference compared with the results of the PFGE type-spa type conjugation, although the latter performed better for the prediction of eBURST CC (W = 0.85 versus 0.94). It is also interesting that if two MRSA strains shared the same PFGE type-SCCmec type, they had a 94% probability of sharing the same BURP group, which was a higher probability than that of sharing the same eBURST group (W = 0.85).
DISCUSSION
Currently, a wide variety of genotype-based typing methods are available for classifying S. aureus isolates for epidemiological studies. Molecular methods based on the analysis of band patterns, such as PFGE, are now being replaced by more portable sequence-based methods, such as spa typing and MLST. However, the advantages of these newer methods in terms of discriminatory power and the relationships between the groups defined by the once-dominant typing methods and the new ones, now more frequently used, have not been fully explored.
In this study, we analyzed a collection of 198 epidemiologically unrelated S. aureus strains, composed of 82 MSSA and 116 MRSA strains, in order to quantify the congruence between the most frequently used genotyping methods for the characterization of S. aureus strains: PFGE subtypes (defined at a 95% cutoff on the Dice/UPGMA dendrogram), MLST, spa typing, and, for MRSA strains, SCCmec typing. We also evaluated the congruence between techniques in assigning strains to larger groups, using eBURST for MLST, BURP for spa type, and PFGE type defined as the groups formed at an 80% cutoff on the Dice/UPGMA dendrogram. For PFGE, the type definition at the 80% cutoff approximates Tenover's criteria for possibly related isolates (up to six bands difference) (5), while the 95% cutoff for subtype definition is a more discriminatory cutoff usually allowing up to a one- or two-band difference (depending on the total number of bands for each isolate), which would correspond to indistinguishable or closely related strains in Tenover's classification.
We evaluated not only the overall congruence of results by AR but also the capability of one method to predict the results of any other in terms of W coefficients. Our goal was to determine, among the methods used or any combination of them, which was the best and most cost-effective method to infer genetic relatedness.
Discriminatory power is an important parameter for the evaluation of any typing method's performance. Our results show that, in contrast with some previous studies (8), PFGE at the subtype level is the most discriminatory technique, with SID values over 99.69% (Table 1). When the MRSA and MSSA sub-data sets were compared, it was found that all methods had higher discriminatory power for the latter (always above over 90%), supporting the notion that MSSA strains have a more diverse genetic structure than MRSA strains and in agreement with the hypothesis that MRSA derived recently from a limited number of MSSA lineages by the acquisition of the SCCmec element (4, 19).
When the overall concordance between typing methods was assessed using the AR coefficient, a distinction between MRSA and MSSA was also apparent. Although overall the levels of concordance were not high (below 80%), they were higher for the MSSA subset than for MRSA. Assuming that MRSA strains are largely confined to the clinical setting, intense antibiotic selective pressure may favor the exchange of genetic material. Since different methods probe different areas of the genome, the higher levels of concordance for MSSA strains may indicate that the MSSA subpopulation of strains is comprised of more stable clones, whereas among MRSA strains, recombination is a more frequent event. This is further supported by the analysis of W coefficients, where, for instance, it was found that if two MSSA strains shared the same spa type they had a 92% probability of sharing the same PFGE type, whereas for MRSA strains, this probability dropped to 40%. This does not exclude the fact that recombination is an infrequent event in S. aureus (14).
spa typing and BURP analysis have been proposed as the sequence-based methods of choice to determine the genetic relatedness of S. aureus (1, 36). Our results show that spa types alone are able to infer eBURST CC (W = 0.96 for MRSA and W = 0.94 for MSSA). The overall agreement between eBURST and BURP CC was also low (0.53 for MRSA strains and 0.56 for MSSA), although it was the highest found for any two methods in the MRSA subset. This suggests that the BURP algorithm retrieves a phylogenetic signal similar to that retrieved by eBURST, but since the latter interrogates housekeeping genes and BURP is based on the alignment of sequence repeats found in the polymorphic region of the spa gene, BURP is likely to reflect a faster evolutionary clock. This is supported by the fact that if two strains belonged to the same BURP group, they had a 74% probability of belonging to the same eBURST CC if the strains were MSSA and 76% if the strains were MRSA, but the reverse (strains belonging to the same eBURST CC also having the same BURP group) was only 50% for MSSA and 56% for MRSA, indicating that some eBURST groups have strains that belong to more than one BURP group. However, one should bear in mind that the typeability of spa typing is not 100%, as shown in Table 1, which could negatively influence an epidemiological study.
Our results also demonstrate that PFGE, while a labor-intensive and time-consuming technique, shows high levels of agreement with other methods. For instance, if any two strains share the same PFGE type, they have over 80% probability of belonging to the same eBURST CC (W = 0.85 for MSSA, 0.83 for MRSA, and 0.86 for the whole data set), whereas for PFGE subtypes, there is absolute agreement (W = 1.00, an expected fact given the high discriminatory power of PFGE at the subtype level: SID = 99.69%), which is higher than the values found for agreement between PFGE subtypes and spa typing (W = 0.97 for MRSA, 0.94 for MSSA, and 0.82 for the entire collection) or ST (W = 0.82 for MRSA, 0.79 for MSSA, and 0.77 for the entire collection).
We also found that the PFGE type-spa type combination, for the MRSA subset, improved the predictive power of each single technique for determining the SCCmec type (W = 0.71). For the MSSA subset, if two strains shared the same PFGE type-spa type, they had 100% probability of belonging to the same eBURST CC; for MRSA strains, this value was slightly lower (W = 0.94), and there was no gain of predictive power over the spa type alone (W = 0.96). In terms of costs, and for MRSA, the PFGE-SCCmec combination is possibly the best option: while less expensive than spa typing alone (data not shown), it was found that if two strains shared the same PFGE type-SCCmec type, they had 94% probability of sharing the same BURP group and 85% probability of sharing the same eBURST CC (in the last case, it was only marginally better than the PFGE type alone). However, since SCCmec characterization can only be applied to MRSA strains, the PFGE-spa type combination remains a more broadly applicable technique for the study of S. aureus. Also, SCCmec characterization does not provides 100% typeability, which can hamper the correct characterization of some MRSA strains even when combined with PFGE types, similar to spa typing.
Based on the data presented here, we can provide a rationale for the informed choice of the most appropriate typing scheme for the characterization of S. aureus isolates. Since the MRSA and MSSA subsets were shown to differ in terms of diversity and stability, they could be characterized by different typing schemes, although a unified methodology could be desirable. In our data set, PFGE subtypes were shown to predict the CC at 100% and were also the best method for ST and SCCmec prediction. Nevertheless, they have the disadvantage of requiring a normalized and curated database of patterns, the complexity of which increases dramatically with the number of isolates. Therefore, we suggest that the most suitable method to infer clonal relationships between isolates, taking MLST and eBURST as reference methods, is spa typing. Moreover, for the MSSA data set, spa types also predicted the PFGE type with 92% confidence, rendering PFGE analysis redundant. Therefore, for MSSA strains, our data support the sole use of spa typing, whereas for MRSA, a combination of PFGE and spa typing would offer additional discriminatory power. Overall, the most cost-effective combination of techniques for a detailed characterization of S. aureus isolates, irrespective of resistance to methicillin, would be a combination of PFGE and spa typing, which allows a very accurate (W = 0.995) assignment of strains to eBURST CC without performing MLST. This combination also provides the necessary discriminatory power and typeability for local epidemiological studies, as well as the possibility of defining more distant relationships between isolates for long-term epidemiological studies.
Supplementary Material
Acknowledgments
Partial support for this study was provided by project POCTI/BIA-MIC/58416/2004 from Fundação para a Ciência e Tecnologia (FCT), Lisbon, Portugal, to H. de Lencastre. N.A.F. and D.C.O. were supported by grants SFRH/BD/19208/2004 and SFRH/BPD/9374/2002, respectively, from FCT.
Footnotes
Published ahead of print on 7 November 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Aires de Sousa, M., T. Conceicao, C. Simas, and H. de Lencastre. 2005. Comparison of genetic backgrounds of methicillin-resistant and susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J. Clin. Microbiol. 435150-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., and H. de Lencastre. 2004. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol. Med. Microbiol. 40101-111. [DOI] [PubMed] [Google Scholar]
- 3.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck, W. D., B. Berger-Bachi, and F. H. Kayser. 1986. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J. Bacteriol. 165373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrico, J. A., F. R. Pinto, C. Simas, S. Nunes, N. G. Sousa, N. Frazao, H. de Lencastre, and J. S. Almeida. 2005. Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J. Clin. Microbiol. 435483-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carriço, J. A., C. Silva-Costa, J. Melo-Cristino, F. R. Pinto, H. de Lencastre, J. S. Almeida, and M. Ramirez. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 442524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leao, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6189-198. [DOI] [PubMed] [Google Scholar]
- 8.Cookson, B. D., D. A. Robinson, A. B. Monk, S. Murchan, A. Deplano, R. de Ryck, M. J. Struelens, C. Scheel, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, C. Cuny, W. Witte, P. T. Tassios, N. J. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, M. Muller-Premru, W. Hryniewicz, A. Rossney, B. O'Connell, B. D. Short, J. Thomas, S. O'Hanlon, and M. C. Enright. 2007. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J. Clin. Microbiol. 451830-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crisostomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 989865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 381008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 997687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.European Antimicrobial Resistance Surveillance System. 23 August 2006. posting date. European Antimicrobial Resistance Surveillance System 2005, 2006. http://www.earss.rivm.nl.
- 13.Faria, N. A., D. C. Oliveira, H. Westh, D. L. Monnet, A. R. Larsen, R. Skov, and H. de Lencastre. 2005. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. J. Clin. Microbiol. 431836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 1853307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 394190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallin, M., A. Deplano, O. Denis, R. De Mendonca, R. De Ryck, and M. J. Struelens. 2007. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 45127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho, P. L., C. Cheung, G. C. Mak, C. W. Tse, T. K. Ng, C. H. Cheung, T. L. Que, R. Lam, R. W. Lai, R. W. Yung, and K. Y. Yuen. 2007. Molecular epidemiology and household transmission of community-associated methicillin-resistant Staphylococcus aureus in Hong Kong. Diagn. Microbiol. Infect. Dis. 57145-151. [DOI] [PubMed] [Google Scholar]
- 17.Hubert, L., and P. Arabie. 1985. Comparing partitions. J. Classification 2193-218. [Google Scholar]
- 18.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 262465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 431449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuehnert, M. J., D. Kruszon-Moran, H. A. Hill, G. McQuillan, S. K. McAllister, G. Fosheim, L. K. McDougal, J. Chaitram, B. Jensen, S. K. Fridkin, G. Killgore, and F. C. Tenover. 2006. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. J. Infect. Dis. 193172-179. [DOI] [PubMed] [Google Scholar]
- 23.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 953140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 415113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milheirico, C., D. C. Oliveira, and H. de Lencastre. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: SCCmec IV multiplex. J. Antimicrob. Chemother. 6042-48. [DOI] [PubMed] [Google Scholar]
- 26.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 411574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 404289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 462155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira, D. C., C. Milheirico, S. Vinga, and H. de Lencastre. 2006. Assessment of allelic variation in the ccrAB locus in methicillin-resistant Staphylococcus aureus clones. J. Antimicrob. Chemother. 5823-30. [DOI] [PubMed] [Google Scholar]
- 30.Ridom GmbH. 2006. Ridom StaphType user guide. Ridom GmbH, Wurzburg, Germany.
- 31.Robinson, D. A., S. K. Hollingshead, J. M. Musser, A. J. Parkinson, D. E. Briles, and M. J. Crain. 1998. The IS1167 insertion sequence is a phylogenetically informative marker among isolates of serotype 6B Streptococcus pneumoniae. J. Mol. Evol. 47222-229. [DOI] [PubMed] [Google Scholar]
- 32.Sa-Leao, R., A. Tomasz, and H. de Lencastre. 2001. Multilocus sequence typing of Streptococcus pneumoniae clones with unusual drug resistance patterns: genetic backgrounds and relatedness to other epidemic clones. J. Infect. Dis. 1841206-1210. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz, D. C., and C. R. Cantor. 1984. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 3767-75. [DOI] [PubMed] [Google Scholar]
- 34.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 373556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson, E. H. 1949. Measurement of species diversity. Nature 163688. [Google Scholar]
- 36.Strommenger, B., C. Kettlitz, T. Weniger, D. Harmsen, A. W. Friedrich, and W. Witte. 2006. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 442533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tiemersma, E. W., S. L. Bronzwaer, O. Lyytikainen, J. E. Degener, P. Schrijnemakers, N. Bruinsma, J. Monen, W. Witte, and H. Grundman. 2004. Methicillin-resistant Staphylococcus aureus in Europe, 1999-2002. Emerg. Infect. Dis. 101627-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Belkum, A., M. Struelens, A. de Visser, H. Verbrugh, and M. Tibayrenc. 2001. Role of genomic typing in taxonomy, evolutionary genetics, and microbial epidemiology. Clin. Microbiol. Rev. 14547-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallace, D. L. 1983. A method for comparing two hierarchical clusterings. J. Am. Stat. Assoc. 78569-576. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.