Abstract
Melioidosis is caused by the gram-negative saprophytic bacterium Burkholderia pseudomallei, which is endemic to southeast Asia and northern Australia. We have previously found evidence of geographic localization of strains based on multilocus sequence typing (MLST). In this study, we examined the diversity of 277 isolates from northern Australia, which were resolved into 159 different sequence types. No sequence types were common to both Queensland and the Northern Territory, and there was significant differentiation between the alleles present in the two regions. The considerable diversity in sequence types contrasts with the limited diversity of alleles at MLST loci, supporting previous work suggesting a high rate of recombination relative to mutation in B. pseudomallei, where new sequence types are primarily generated by reassortment of existing alleles.
Melioidosis, the disease caused by the saprophytic bacterium Burkholderia pseudomallei, is endemic to southeast Asia and northern Australia (19). There is a marked heterogeneity in clinical presentation and disease severity between patients. Regional variations in disease presentations have also been noted, with prostatic abscesses and encephalomyelitis described in Australia and parotid abscesses and hepatosplenic suppuration being common presentations in Thailand (2).
We have previously demonstrated a lack of correlation between disease presentation and strain type using both restriction enzyme analysis and multilocus sequence typing (MLST) (3, 4). This suggests that host factors and the mode of acquisition (and possibly the magnitude of the infecting dose) may be more important than strain tropism or virulence. However, we did note apparent clustering of strain types, suggesting geographical localization. These differences were most marked when we compared isolates from the regions of Australia where this organism is endemic to those from countries in the southeast Asian region (4, 18). Other data have suggested some localization of strain types within one region in northern Australia, with strain differences between Darwin and regions in the Top End greater than those within the city of Darwin (3).
In the present study, we examined the diversity of B. pseudomallei isolates across the region of northern Australia where melioidosis is endemic, looking specifically for geographical localization of strain types in Queensland and the Northern Territory.
MATERIALS AND METHODS
Northern Territory isolates were drawn from the Darwin Prospective Melioidosis Study encompassing a B. pseudomallei collection maintained at the Menzies School of Health Research from consecutive Northern Territory patients with culture-confirmed melioidosis since 1989. Strains from northern Queensland were predominantly from the Townsville General Hospital. The population of these regions is irregularly distributed; in the Northern Territory, major population centers are found in Darwin, Katherine, and Gove (Nhulumbuy), with small communities of <2,000 inhabitants distributed elsewhere in the Northern Territory. In Queensland, the major population centers within the areas of endemicity are the cities of Cairns and Townsville on the east coast. There are relatively few communities and transport routes connecting the major population centers of both regions.
Isolates were selected to represent the full diversity of clinical presentation and regional variation within these areas. Since previous studies suggested that restriction enzyme analysis based on pulsed-field gel electrophoresis (PFGE) has equivalent or greater discrimination compared to MLST (4), we selected Queensland isolates for typing by MLST that represented the full diversity of strains based on the differences in PFGE profiles. Isolates had been stored at −70°C in Todd-Hewitt broth (Oxoid Australia, Melbourne, Victoria, Australia) with 20% glycerol. Bacteria were cultured on chocolate agar (Oxoid Australia) and subcultured in Todd-Hewitt broth, and DNA was extracted by using a DNeasy tissue kit (Qiagen, Hilden, Germany).
MLST was performed as previously described at Imperial College London (9). The alleles at each of the seven loci were assigned by comparing the sequences to those at the B. pseudomallei MLST website (http://bpseudomallei.mlst.net/). Novel sequences were assigned new allele numbers and were deposited in the MLST allele database. The allele numbers at each locus provide the allelic profile of each strain, and each distinct allelic profile is assigned as a sequence type (ST). All novel STs from the present study have been submitted to this database.
The relatedness among isolates was estimated based both on allelic profiles (using eBURST v3 [8, 15]) and on differences in the concatenated sequence of alleles at all loci. The sequences of the seven loci from the isolates were joined in-frame to produce a concatenated sequence of 3,399 bp. A dendrogram was constructed from the concatenated sequences by the neighbor-joining method with the Kimura two-parameter model of pairwise genetic distances using the MEGA 3.1 program (12). The significance of the nodes on the tree was evaluated by using the bootstrap technique with 1,000 resamplings from the data set. The average pairwise diversity at each locus and of the concatenated sequence of all loci was assessed by using MEGA 3.1. The ratio of synonymous to nonsynonymous nucleotide substitutions at each locus was assessed by using START 2 (10). This parameter cannot be estimated at the ndh locus since this fragment includes parts of two other overlapping genes, with a change in reading frame across the junction sequence.
Comparisons were made between isolates from Queensland and those from the Northern Territory. We also examined the distribution of alleles at each of the loci to determine whether there were region-specific alleles. A classification index was calculated, which represents the probability of correctly classifying a ST or allele into one population where classification is based on the allele frequency between the populations (11). A value of 1 is obtained when the alleles (STs) in the two populations are completely distinct and a value of 0 when there are no differences in the allele (ST) frequencies. Statistical significance was estimated by 10,000 resamplings from the observed allele and ST frequency of the populations, to assess the probability of departure from the null hypothesis that there were no differences in the distributions of alleles and STs between populations. Comparisons were also made between isolates from offshore islands off the northern coast of Australia.
Ethical approval for the present study was obtained from the Human Research Ethics Committee of the Department of Health and Community Services and the Menzies School of Health Research. A condition of our ethical approval is that we not identify smaller communities specifically.
(The findings in this study were presented at the Fifth World Melioidosis Congress in Khon Kaen, Thailand, in November 2007.)
RESULTS
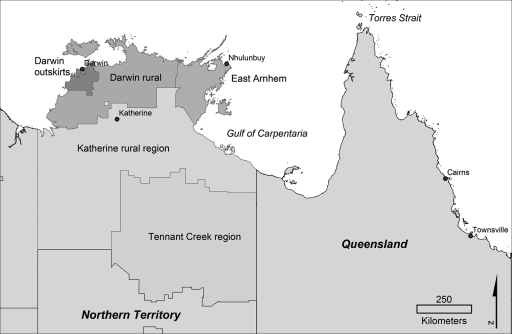
MLST was performed on 277 isolates from northern Australia; 202 were from humans, 24 from animals, and 51 were environmental. These included 230 isolates from the Top End of the Northern Territory and 47 isolates from Queensland (Fig. 1). The isolates from the Northern Territory included 153 isolates from Darwin and the surrounding region, 21 isolates from the Darwin rural region, 18 isolates from Katherine and the Katherine region, 31 isolates from Gove and the East Arnhem region (which borders Queensland), and 7 from elsewhere in the Northern Territory. Of these 230 isolates, 39 were from islands off the coast of the Top End. In Queensland, 7 isolates were from an island community in the Gulf of Carpentaria, 1 was from the Torres Strait islands, 18 were from Townsville, and the remaining 21 were from a range of locations within the state.
FIG. 1.
Map of northern Australia, showing locations of major towns and regions in the Northern Territory and Queensland.
Australian isolates demonstrate considerable diversity.
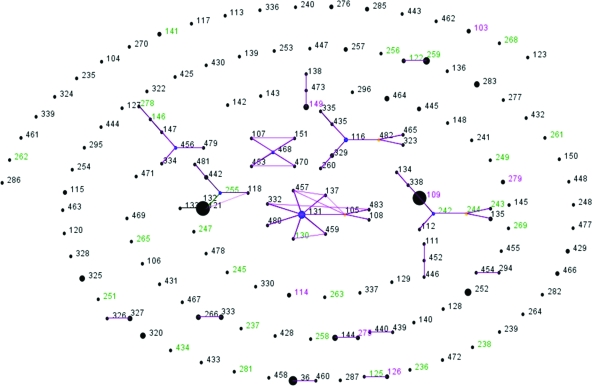
Overall, 159 STs were documented among 277 isolates (0.57 ST/isolate). The diversity of isolates found in the environment (0.67 ST/isolate) was similar to the diversities found in animals (0.67 ST/isolate) and humans (0.65 ST/isolate). The majority of STs in the present study were singletons (isolates that differed from all other isolates in this data set at more than one allele), and eBURST only identified six clonal complexes that included ≥5 STs (Fig. 2). The number of alleles at each locus ranged from 13 (ace and ndh) to 24 (narK). The synonymous/nonsynonymous mutation ratio (dS/dN) was >6 at all loci where this was able to be estimated (Table 1).
FIG. 2.
eBURST diagram of ST from isolates in the present study, demonstrating STs found only on islands (green) only on the mainland (black) or both islands and the mainland (cyan). The size of the dots represents the relative frequency of the ST.
TABLE 1.
Characteristics of alleles at MLST loci in Australian isolates in this study
| Locus | No. of alleles | Avg pairwise diversity | dS/dNa |
|---|---|---|---|
| ace | 13 | 2.1 | ∞ |
| gltB | 20 | 2.8 | 37.3 |
| gmhD | 17 | 2.6 | 8.4 |
| lepA | 19 | 2.7 | 38.3 |
| lipA | 17 | 2.6 | 6.6 |
| narK | 24 | 2.8 | 22.7 |
| ndh | 13 | 2.3 | NE |
NE, not estimated. The ndh locus includes parts of two other overlapping genes, with a change in reading frame across the junction sequence. dS/dN refers to the ratio of synonymous to nonsynonymous changes at each locus.
There is weak evidence of localization of alleles at each locus, but evidence of regional localization of STs within Australia.
Among the 230 NT isolates 125 STs were represented (0.54 ST/isolate), and in the 47 Queensland isolates 34 STs were described (0.72 ST/isolate). No ST was found in either Queensland or the Northern Territory. The classification index based on ST was therefore 1.00 (P < 0.001).
There were few region-specific alleles. Queensland-specific alleles were infrequently encountered and ranged from one at the gltB locus (present in 2% of Queensland isolates) to six at the lipA and narK loci (present in 15 and 13% of Queensland isolates, respectively). Northern Territory-specific alleles were seen more frequently and were present in from 3% of the Northern Territory isolates at the lipA locus to 37% at the narK locus (Table 2). The classification index by allele was ≤0.25 at all loci, but the null hypothesis that a uniform distribution of alleles was present was rejected (P < 0.001 at all loci except for ndh [P = 0.005]) (Table 2).
TABLE 2.
Alleles at MLST loci in the Northern Territory and Queensland
| Locus | No. of alleles | No. of alleles, no of isolates (proportion of isolates [%])
|
No. of common NT-Queensland alleles | Adjusted classification index (P) | |
|---|---|---|---|---|---|
| NTa specific | Queensland specific | ||||
| ace | 13 | 6, 16 (7) | 3, 6 (13) | 4 | 0.16 (<0.001) |
| gltB | 20 | 11, 24 (10) | 1, 1 (2) | 8 | 0.21 (<0.001) |
| gmhD | 17 | 8, 12 (5) | 2, 2 (4) | 7 | 0.18 (<0.001) |
| lepA | 19 | 12, 35 (15) | 1, 1 (2) | 6 | 0.15 (<0.001) |
| lipA | 17 | 6, 8 (3) | 6, 7 (15) | 5 | 0.16 (<0.001) |
| narK | 24 | 8, 84 (37) | 6, 6 (13) | 10 | 0.25 (<0.001) |
| ndh | 13 | 8, 12 (5) | 2, 4 (10) | 3 | 0.10 (0.005) |
NT, Northern Territory.
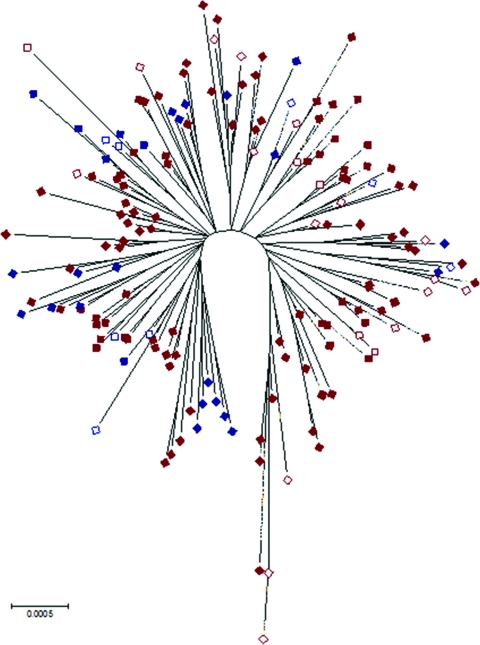
Although there were no STs common to both the Northern Territory and Queensland, the isolates from Queensland were not clearly delineated from those from the Northern Territory on a dendrogram based on differences in allelic profiles with isolates distributed throughout the major clades (data not shown). Similarly, Queensland isolates were distributed through Northern Territory isolates on a neighbor-joining tree constructed by using the concatenated sequence, but the statistical support for the nodes was generally poor, reflecting low levels of sequence diversity among housekeeping genes in B. pseudomallei and a history of recombination (Fig. 3).
FIG. 3.
Bootstrap consensus tree based on the concatenated sequence at all loci demonstrating relatedness of Queensland (blue), Northern Territory (red), and island isolates (hollow).
There is limited evidence of localization of STs in offshore islands that are geologically separate from the Australian mainland.
On the offshore islands off the northern coast of Australia 35 STs were identified compared to 131 STs from the Australian mainland. Seven STs were found in both isolates from islands, as well as on the mainland (Fig. 2); the adjusted classification index based on STs was 0.47 (P < 0.001). The classification indices based on alleles were all ≤0.15 (data not shown); however, the null hypothesis that a uniform distribution of alleles was present was rejected at the glhD, lepA, lipA, and narK loci.
No isolate in the present study is related to isolates from southeast Asia, but the predominant alleles are found in both regions.
No STs from isolates in the present study have been described in studies from other countries. ST60 was described as a ST common to both Australia and Thailand in a previous study comparing historic Thai isolates to those in the MLST database (13) but was not encountered in the present study (see below for a complete discussion). However, 12 STs from the present study were single-locus variants (i.e., differing at one MLST locus) of STs from other countries, with nucleotide differences ranging from 1 to 12. There were 72 STs that were double-locus variants of isolates from other countries. The unadjusted classification index for STs between Australian isolates in the present study and Thai isolates in the MLST database (n = 460) was 1.0 (P < 0.001); after we adjusted for the expectation under the null hypothesis, the adjusted classification index was 0.58. At the ace locus, the adjusted classification index was 0.38, at gltB it was 0.44, at gmhD it was 0.58, at lepA it was 0.54, at lipA it was 0.27, at narK it was 0.80, and at ndh it was 0.25 (all P < 0.001).
DISCUSSION
A number of studies have examined the genetic profiles of B. pseudomallei isolates by using MLST. All have noted low genetic diversity within alleles, which hampers analysis of phylogeny, but alleles seem to recombine frequently to form new STs. Godoy et al. found that B. mallei appeared to be a clone or subspecies of B. pseudomallei, being placed phylogenetically within the species, but both B. pseudomallei and B. mallei were distinct from B. thailandensis (9). We have previously found by using both PFGE and MLST that there was no evidence of clustering by virulence or clinical presentation (3, 4), and this finding was supported by a similar finding that isolates from patients with parotiditis in Thailand were genetically diverse (18). Vesaratchavest et al. also found that there was less genetic diversity in clinical isolates compared to environmental isolates, suggesting that strains may differ in their ability to cause human disease (18).
Recent evidence suggests that eBURST, the conventional algorithm used to analyze allelic profile data, may be unreliable for bacteria such as B. pseudomallei with high recombination and/or mutation ratios (16). Similarly, significant rates of recombination would be expected to impact on the reliability of inferring phylogeny from differences based on concatenated sequences. However, both our previous study and that of Vesaratchavest et al. found broad evidence of separate evolution of B. pseudomallei strains in Australia compared to that occurring in southeast Asia (4, 18). From this analysis, the population snapshot of Australian B. pseudomallei isolates using eBURST (Fig. 2) does not meet Turner's proposed criteria where eBURST is felt to be unreliable (single large straggly group, the presence of long-range single-locus variant links, and >25% of isolates in the largest group) (16). However, ongoing typing of additional Australian isolates since these data were analyzed has linked the clonal complexes, confirming the high rates of recombination across this large geographic area (data not shown).
In contrast, the present study found less robust evidence of geographic localization at a significantly smaller scale than the Australian-Thai comparison. Although no STs were common to both regions within Australia, isolates were not distinct on analysis of the concatenated sequences and allelic profile. Further, an examination of isolates from small offshore islands showed even more limited evidence that geographic localization is present at this level. The diversity of isolates should be noted with caution, however, since isolates were not chosen randomly and thus may not be representative of the population frequency distribution of STs. For example, many isolates from Townsville have previously been shown to be similar by PFGE (17), and we generally limited MLST in the present study to one strain of each of the Townsville PFGE patterns. This approach would be expected to reduce the number of STs represented by more than one isolate, but even using this sampling approach 21% of the STs from both the Northern Territory and Queensland were represented by multiple isolates, and no ST was represented by isolates from both of these regions.
More recently, McCombie et al., using a historical collection of B. pseudomallei isolates collected in Thailand in the 1960s, suggested that one ST (ST60) found in Thailand was common to a group of environmental isolates from Australia (13). However, we have reason to believe that these isolates may not have been from Australia (5). This emphasizes the need to establish provenance and verify typing for all studies where clinical or geographical correlates are being examined.
Melioidosis is acquired by exposure to the organisms in the local environment, and genetic differentiation between strains from widely separate regions is not unexpected since long-range wind dispersal is unlikely for a non-spore-forming soil saprophyte. However, the finding that there was a clear lack of separation by geographic region within Australia, compared to between Australia and southeast Asia, may mirror such a distinction that is seen in other flora and fauna. We have previously suggested that this may have represented dissemination during transient land bridges between these regions in the Miocene epoch (4). The descriptions of B. pseudomallei in South America (1, 14), the Indian subcontinent (6), and regions of Africa and Madagascar (7) suggest that B. pseudomallei originated prior to the separation of the Gondwana supercontinent. However, the striking genetic homogeneity in alleles globally argues against this. Further work is required to define the true global epidemiology of this pathogen, as well as to examine the global population structure outside of the classic areas of endemicity in this highly recombinant species.
One finding that is yet to be explained is that the predominant alleles are common to all regions. Within Australia, we did not find STs that were common to both regions, but the majority of STs were comprised of alleles common to both regions. This is reflected in the high classification index by ST but lower classification index by allele. However, we found that the predominant alleles at each locus are often common to both areas but recombine in unique configurations to form the regional STs observed. There are several possible reasons for the sharing of prevalent alleles, but the most likely hypothesis is that B. pseudomallei is a relatively young species and, since their original distribution over southeast Asia and northern Australia, sufficient time has not elapsed for all of the ancestral alleles to accumulate mutations that would distinguish the alleles present in these two regions. Because of this the broad differentiation between Australian and Thai isolates, and within Australian regions, is apparent at the level of STs, due to a high level of recombination, but not yet at the level of alleles.
Acknowledgments
We thank Sharon Peacock, Ed Feil, Bill Hanage, and David Aanensen for helpful discussions regarding the analysis of MLST data; Jeffrey Hanna from the Queensland Tropical Public Health Unit and Helen Smith from Queensland Health Scientific Services for providing isolates and data; and Gary Lum and the Royal Darwin Hospital microbiology laboratory staff for expertise in isolate identification.
This study was supported by a project grant from the Australian National Health and Medical Research Council (NHMRC). A.C.C. is supported by an NHMRC Health Professional Research Fellowship.
Footnotes
Published ahead of print on 14 November 2007.
REFERENCES
- 1.Biegeleisen, J. Z., R. Mosquera, and W. B. Cherry. 1964. A case of human melioidosis: clinical, epidemiological and laboratory findings. Am. J. Trop. Med. Hyg. 1389-99. [DOI] [PubMed] [Google Scholar]
- 2.Cheng, A. C., and B. J. Currie. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18383-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, A. C., N. P. Day, M. J. Mayo, D. Gal, and B. J. Currie. 2004. Burkholderia pseudomallei strain type, based on pulsed-field gel electrophoresis, does not determine disease presentation in melioidosis. Microbes Infect. 7104-109. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, A. C., D. Godoy, M. Mayo, D. Gal, B. G. Spratt, and B. J. Currie. 2004. Isolates of Burkholderia pseudomallei from Northern Australia are distinct by multilocus sequence typing, but strain types do not correlate with clinical presentation. J. Clin. Microbiol. 425477-5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Currie, B. J., A. D. Thomas, D. Godoy, D. A. Dance, A. C. Cheng, L. Ward, M. Mayo, T. L. Pitt, and B. G. Spratt. 2007. Australian and Thai isolates of Burkholderia pseudomallei are distinct by multilocus sequence typing: revision of a case of mistaken identity. J. Clin. Microbiol. -3829.453828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dance, D. A., M. D. Smith, H. M. Aucken, and T. L. Pitt. 1999. Imported melioidosis in England and Wales. Lancet 353208. [DOI] [PubMed] [Google Scholar]
- 7.Dance, D. A. B. 1991. Melioidosis: the tip of the iceberg? Clin. Microbiol. Rev. 452-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godoy, D., G. Randle, A. J. Simpson, D. M. Aanensen, T. L. Pitt, R. Kinoshita, and B. G. Spratt. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 412068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 171230-1231. [DOI] [PubMed] [Google Scholar]
- 11.Jolley, K. A., D. J. Wilson, P. Kriz, G. McVean, and M. C. Maiden. 2005. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol. Biol. Evol. 22562-569. [DOI] [PubMed] [Google Scholar]
- 12.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 13.McCombie, R. L., R. A. Finkelstein, and D. E. Woods. 2006. Multilocus sequence typing of historical Burkholderia pseudomallei isolates collected in Southeast Asia from 1964 to 1967 provides insight into the epidemiology of melioidosis. J. Clin. Microbiol. 442951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miralles, I. S., C. Maciel Mdo, M. R. Angelo, M. M. Gondini, L. H. Frota, C. M. dos Reis, and E. Hofer. 2004. Burkholderia pseudomallei: a case report of a human infection in Ceara, Brazil. Rev. Inst. Med. Trop. Sao Paulo 4651-54. [DOI] [PubMed] [Google Scholar]
- 15.Spratt, B. G., W. P. Hanage, B. Li, D. M. Aanensen, and E. J. Feil. 2004. Displaying the relatedness among isolates of bacterial species: the eBURST approach. FEMS Microbiol. Lett. 241129-134. [DOI] [PubMed] [Google Scholar]
- 16.Turner, K. M., W. P. Hanage, C. Fraser, T. R. Connor, and B. G. Spratt. 2007. Assessing the reliability of eBURST using simulated populations with known ancestry. BMC Microbiol. 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulett, G. C., B. J. Currie, T. W. Clair, M. Mayo, N. Ketheesan, J. Labrooy, D. Gal, R. Norton, C. A. Smith, J. Barnes, J. Warner, and R. G. Hirst. 2001. Burkholderia pseudomallei virulence: definition, stability, and association with clonality. Microbes Infect. 3621-631. [DOI] [PubMed] [Google Scholar]
- 18.Vesaratchavest, M., S. Tumapa, N. P. Day, V. Wuthiekanun, W. Chierakul, M. T. Holden, N. J. White, B. J. Currie, B. G. Spratt, E. J. Feil, and S. J. Peacock. 2006. Nonrandom distribution of Burkholderia pseudomallei clones in relation to geographical location and virulence. J. Clin. Microbiol. 442553-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White, N. J. 2003. Melioidosis. Lancet 3611715-1722. [DOI] [PubMed] [Google Scholar]