Abstract
Dengue fever is an important tropical illness for which there is currently no virus-specific treatment. To shed light on mechanisms involved in the cellular response to dengue virus (DV), we assessed gene expression changes, using Affymetrix GeneChips (HG-U133A), of infected primary human cells and identified changes common to all cells. The common response genes included a set of 23 genes significantly induced upon DV infection of human umbilical vein endothelial cells (HUVECs), dendritic cells (DCs), monocytes, and B cells (analysis of variance, P < 0.05). Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), one of the common response genes, was identified as a key link between type I and type II interferon response genes. We found that DV induces TRAIL expression in immune cells and HUVECs at the mRNA and protein levels. The induction of TRAIL expression by DV was found to be dependent on an intact type I interferon signaling pathway. A significant increase in DV RNA accumulation was observed in anti-TRAIL antibody-treated monocytes, B cells, and HUVECs, and, conversely, a decrease in DV RNA was seen in recombinant TRAIL-treated monocytes. Furthermore, recombinant TRAIL inhibited DV titers in DV-infected DCs by an apoptosis-independent mechanism. These data suggest that TRAIL plays an important role in the antiviral response to DV infection and is a candidate for antiviral interventions against DV.
Dengue virus (DV) has reemerged as a major global health problem in the tropics, particularly among children (9, 26). This mosquito-borne flavivirus, for which there is no vaccine or antiviral treatment, causes an estimated 50 million infections annually (32, 34). Most DV infections result in a self-limited febrile illness (dengue fever). Less frequently, infections can cause dengue hemorrhagic fever, a potentially fatal plasma leakage syndrome.
DV replication can be effectively controlled after a short period of viremia in most individuals. It is unclear, however, what host factors induced by DV infection are involved in regulating the virus. Increases in serum levels of type I and type II interferons (IFNs) have been observed during DV infection (21, 22). Pretreatment of cells with type I IFN was shown to block DV infection of cells by a protein kinase receptor and 2′-5′ oligoadenylate synthase (OAS)-independent mechanism (5), although it has been shown that DV infection inhibits type I IFN signaling within infected cells (31).
The in vivo tropism and cellular response to DV has only been partially understood. Macrophages (10), B cells (18, 23), and dendritic cells (DCs) (27, 47) are known sites for DV replication in vivo. Primary endothelial cells and hepatocytes are infected in vitro (13, 16, 42, 46). The response to DV in these cells may be critical to control DV replication. Previous in vitro studies analyzed changes in gene expression induced by DV in human umbilical vein endothelial cells (HUVECs) (46) and monocytes (30) but reported up-regulation of different sets of genes by DV infection. Interestingly, gene expression analysis of whole blood cells in dengue patients found that the IFN-inducible gene response was attenuated in dengue shock syndrome patients compared to the response in dengue hemorrhagic fever patients (41).
In this study, we have identified a common response profile of 23 induced genes in primary human cells including HUVECs, monocytes, DCs, and B cells infected in vitro with DV. Signaling pathway analysis identified TRAIL (tumor necrosis factor [TNF]-related apoptosis-inducing) as a potential common linker between the IFN-α and IFN-γ pathways. TRAIL is a member of the TNF family that specifically promotes apoptosis in cancer cells by binding to and activating the death receptors DR4 and DR5 (12), resulting in recruitment of adaptor protein FADD (Fas-associated death domain). FADD recruits procaspase-8 into the death receptor complex, thereby causing autoproteolytic cleavage of procaspase-8, which in turn leads to activation of the apoptosis signaling pathway (43). TRAIL has also been shown to negatively regulate innate immune responses independent of apoptosis (6). Previous studies indicated that TRAIL can function as an antiviral and antitumor protein (2, 17, 25, 35, 37-39, 44, 45, 48) by inducing cell death.
We found that TRAIL regulates viral replication in DV-infected monocytes at a concentration which is higher than that used to induce cell death in tumor cell lines in vitro (1, 2). Additionally, we show that recombinant TRAIL (rTRAIL)-mediated inhibition of DV titers is not mediated through apoptosis of DV-infected DCs. These data describe an apoptosis-independent mechanism by which TRAIL might mediate antiviral activity.
MATERIALS AND METHODS
Blood sample preparation and cell culture.
Blood samples were obtained from healthy U.S. volunteers at The University of Massachusetts Medical School. Monocytes and B cells were negatively selected from heparin-anticoagulated blood using a rosetting antibody precipitation kit (StemCell) and maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) and antibiotics. Sample purity was determined by cell surface staining of freshly isolated monocytes and B cells. Primary HUVECs were obtained from pooled umbilical cords (two to five donor pools per culture). Human subject protocols were approved by Harvard University Medical School (Institutional Review Board #92-05383 and #85-01323). HUVECs were maintained by the Core Facility of the Center for Excellence in Vascular Research at Harvard Medical School in M199 medium supplemented with 10% FCS, 1 mM glutamine, endothelial cell growth stimulant, porcine intestinal heparin, and antibiotics. HUVEC cultures were split at a ratio of 1:3 or 1:4 for up to two passages. DV (DV type 2, strain New Guinea C) was cultured using standard methods in the C6/36 insect cell line. HUVECs, monocytes, and B cells were infected in vitro with DV as previously described (46). Cells were treated with uninfected C6/36 cell supernatant as a negative control (mock infection). Cells were collected for gene expression analysis at 48 h postinfection (46).
CD14 microbeads (catalog number 120-000-305; Miltenyi Biotec) were used to positively select CD14-positive cells (monocytes) from Ficoll-isolated peripheral blood mononuclear cells from healthy donors. Monocytes were cultured for 7 days in RPMI 1640 medium supplemented with 800 U/ml of granulocyte-macrophage colony-stimulating factor, 500 U/ml of interleukin-4, and 10% FCS. Cells were stained for CD1a, CD14, HLA-DR, and CD83 on day 6 to determine the monocyte to immature DC conversion.
Wild-type (2fTGH) and IFN-α pathway mutant (U1A, U3A, U4A, and U5A) fibroblast cells (24, 29, 33) were obtained from Paul Fisher (Columbia University, New York, NY) and cultured in Dulbecco's modified Eagle's medium supplemented with 10% FCS. The fibroblast cells were infected with DV for 48 h.
Affymetrix GeneChip hybridization and analysis.
Total cellular RNA isolated using an RNeasy kit (Qiagen) was biotin labeled and hybridized to human oligonucleotide microarrays (Affymetrix HG-U133A) as previously described (46). Experiments displaying Affymetrix present call rates of >30% were included in the analysis. Signal values from each of the 22,283 probe sets were calculated using robust multiarray analysis (14). Signal values were transformed using the inverse natural log. For repeated experiments, inverse natural log-transformed robust multiarray analysis results were normalized based on the median and geometric means and were calculated prior to importing data into GeneSpring software (Agilent). Each GeneChip was independently normalized to the median expression level of all genes on the chip. Each gene was then normalized to the median expression levels of that gene. Signals with low expression levels were excluded, and expression of statistical filters was applied as indicated. We analyzed our data by hierarchical cluster analysis using a Pearson correlation. Reproducibility was assessed as previously described (46). For the analysis of the common response, the following analysis was performed: expression signals for 22,283 genes were filtered to exclude those with very low signals (∼2,000 genes), and the remaining genes were analyzed by one-way analysis of variance (ANOVA) to identify genes with statistically significant differences between DV-infected samples (five samples) and uninfected samples (five samples). Variances were calculated using the cross-gene error model, parametric test, a P value cutoff 0.05, and the Benjamin and Hochberg false discovery rate multiple testing correction. For hierarchical cluster analysis a Pearson correlation was used.
Quantitative reverse transcription-PCR (qRT-PCR).
TaqMan quantitation of DV RNA was performed as previously described (46). TRAIL mRNA was quantified using the same reaction conditions with TaqMan primer and probes obtained from Applied Biosystems. Results were calculated using the standard curve method or relative quantification method using quantitative PCR software (Applied Biosystems).
For microfluidic card analysis or qRT-PCR, total RNA was extracted from cells using a Qiagen RNeasy kit. RNA was subjected to 384-well microfluidic card analysis as described by the manufacturer (Applied Biosystems). One hundred nanograms of total cellular RNA was reverse transcribed using TaqMan reverse transcription reagents in the presence of random hexamers as primers. Reverse transcription was performed at 25°C for 10 min and 48°C for 30 min, followed by 95°C for 5 min. For PCR, a 100-μl reaction mixture through a single port provided 2 μl of total reaction mixture per sample. The 100-μl PCR reaction mixture included 5 μl of cDNA, 45 μl of RNase/DNase-free water, and 50 μl of TaqMan Universal PCR Master Mix (2×). PCRs were cycled at 50°C for 2 min and 94.5°C for 10 min, followed by 40 cycles of 97°C for 30 s and 59.7°C for 1 min in the PCR signal detection system 7900 (Applied Biosystems). β-Actin was used as an endogenous control to equalize loading of total RNA between samples. Each data point was measured in quadruplicate, and the standard error was determined.
TRAIL protein detection.
For TRAIL enzyme-linked immunosorbent assay (ELISA) and TRAIL intracellular cytokine staining, TRAIL protein levels in culture supernatants and cell lysates were measured by ELISA (R&D Systems). The minimum detectable amount of TRAIL for this ELISA is 2.86 pg/ml. CD14+ CD4− cells (monocytes) were stained for surface TRAIL protein and CD1a+ CD14− cells (DCs) were stained for surface and intracellular TRAIL protein by flow cytometry using monoclonal anti-TRAIL-phycoerythrin (PE) antibody (BD Biosciences). Both monocytes and DCs were infected with DV at a multiplicity of infection (MOI) of 0.1 for either 48 h (monocytes) or 12, 24, and 48 h (DCs). DCs were treated with brefeldin A for the last 8 h of each time point.
TRAIL antibody treatment.
Monocytes, B cells, and HUVECs were pretreated for 24 h with TRAIL blocking monoclonal antibody (50 μg/ml; R&D Systems) or purified immunoglobulin G1 isotype control antibody followed by infection with DV for 48 h at an MOI of 1. The antibodies were left in the culture during the 48 h DV infection.
The cells were collected by centrifugation at 500 × g, washed twice in phosphate-buffered saline (PBS), and cell pellets were stored at −70°C until analysis. RNA was extracted from the cell pellet using RNeasy (Qiagen) and subjected to TaqMan qRT-PCR for detection of TRAIL and DV RNA using β-actin as an endogenous control. A standard curve was run using a precalibrated DV sample for absolute quantification of gene expression.
rTRAIL treatment.
Monocytes were treated for 24 h with rTRAIL (Merck, Germany) at concentrations from 5 to 20 μg/ml and infected with DV at an MOI of 0.1. At the end of 24 and 48 h, cells were washed with PBS (twice), and the cell pellets were stored at −70°C. RNA was extracted, and DV copy number was determined using TaqMan qRT-PCR. β-Actin was used as endogenous control.
DCs were treated for 24 h with rTRAIL (Biomol, PA) and infected with DV at an MOI of 0.1 for 12, 24, or 48 h. DCs were washed at 2 h postinfection to remove residual virus used for infection. Cells were maintained in fresh RPMI 1640 medium containing 20 μg/ml rTRAIL, 10% FCS, interleukin-4 (500 U/ml), and granulocyte-macrophage colony-stimulating factor (800 U/ml) for 12, 24 or 48 h. At each time point, cells were stained for DV antigen (DV-fluorescein isothiocyanate), CD1a-allophycocyanin, HLA-DR-peridinin chlorophyll protein-Cy5.5, and CD83-PE. Plaque assay was also performed on supernatant collected at the 48-h time point.
Detection of apoptosis in DCs. (i) Poly(ADP-ribose) polymerase 1 (PARP-1) protein cleavage detection.
Cells undergoing apoptosis cleave PARP-1 protein (116-kDa) into two smaller fragments (85 and 25 kDa) (19). PARP-1 protein cleavage is the last known stage in cell death to differentiate between apoptosis and necrosis. DCs infected with DV for 48 h were washed with PBS (two washes) and stained with PARP-1-PE antibody (BD Biosciences) followed by flow cytometry analysis.
(ii) Caspase-3 and Live/Dead Aqua stain.
Live/Dead Aqua stains dying cells while active caspase-3 is an early marker for cells undergoing apoptosis (8). DV-infected DCs either untreated or treated with TRAIL were stained for apoptosis using caspase-3 and Live/Dead Aqua dye at 12, 24, and 48 h postinfection. DCs incubated in a 65°C water bath for 20 min and camptothecin B (2 mM)-treated THP-1 cells were used as positive controls for Live/Dead Aqua stain at each time point. DCs treated with camptothecin B (2 mM/ml) for 4 h were used as a positive control for caspase-3 stain at each time point.
Plaque assay.
Culture supernatants were incubated with a monolayer of LLCMK2 cells for 2 h. Cell monolayers were washed with PBS and overlaid with medium viscosity (mix of high and low) carboxy methylcellulose (Sigma-Aldrich, St. Louis, MO). Carboxy methylcellulose was removed 6 days later, and cells were fixed and stained with 0.2% crystal violet in ethanol. DV plaques were counted visually (36).
Statistical analysis.
Means, standard deviations, and a Student's t test were calculated by the Excel program (Microsoft). The ANOVA was used to analyze gene expression profiles by GeneSpring (Agilent).
Microarray data accession numbers.
Microarray gene expression data are available at GEO under accession number GSE9378.
RESULTS
Identification of common gene expression response to DV infection.
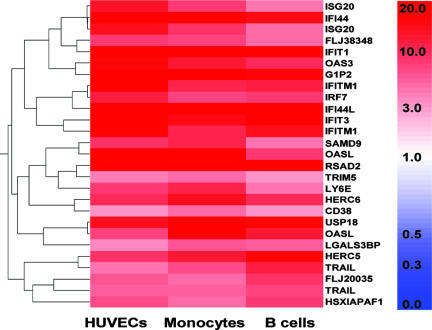
To identify common gene expression changes in host cells in response to DV infection, we analyzed the global gene expression profile in HUVECs, monocytes, DCs, and B cells using Affymetrix GeneChip arrays. Monocytes and B cells isolated from healthy donor blood were ∼80% CD14+ and ∼90% CD19+, respectively (data not shown). Monocyte-derived DCs were ∼91% CD1a+. The CD1a+ cells were stained for CD83 to confirm that the monocyte-derived DCs were immature. Very few (<1%) CD1a+ cells stained positive for CD83 surface antigen (data not shown). All the cells tested were infected with DV propagated in C6/36 mosquito cells. Control samples were mock infected with uninfected C6/36 supernatants. ANOVA identified a set of 23 transcripts that demonstrated a significant change in expression (P of <0.05) in all cell types studied (Fig. 1 and Table 1). To confirm the gene expression changes detected by Affymetrix GeneChip, 11 genes for which primers and probes were available were selected for validation by qRT-PCR. qRT-PCR confirmed up-regulation (greater than threefold) of all the 11 genes in HUVECs (Table 2).
FIG. 1.
Gene expression analysis using HG-U133A Affymetrix GeneChips. Expression levels for the 23 DV common response genes in cells exposed to DV. HUVECs (n = 2), monocytes (n = 2), and B cells (n = 1) represent cells exposed in vitro to DV for 48 h. In vitro infection profiles were normalized to C6/36 insect cell supernatant-treated samples for each cell type independently. Hierarchical cluster analysis used a Pearson correlation. Color indicates relative changes in induction (n-fold). On the color scale, dark red represents 20-fold up-regulation, and white indicates no change. Affymetrix microarray analysis was performed using GeneSpring software (Agilent).
TABLE 1.
Biological function of the 23 DV response genes
| Gene name | Functional category | Description | GenBank accession no. | Affymetrix identifier |
|---|---|---|---|---|
| G1P2 | Antiviral | IFNα-inducible protein (clone IF1-15K) | NM_005101 | 205483_s_at |
| IRF7 | Antiviral | IFN regulatory factor 7 | NM_004030 | 208436_s_at |
| ISG20 | Antiviral | IFN-stimulated gene (20 kDa) | NM_002201 | 204698_at |
| OAS3 | Antiviral | 2′-5′-OAS3 (100 kDa) | NM_006187 | 218400_at |
| OASL | Antiviral | 2′-5′-OAS3-like | NM_003733 | 205660_at |
| RSAD2 | Antiviral | Radical S-adenosyl methionine domain-containing 2 | AI337069 | 213797_at |
| TRIM5 | Antiviral | Tripartite motif-containing 5 | AF220028 | 210705_s_at |
| HSXIAPAF1 | Apoptosis | XIAP associated factor-1 | NM_017523 | 206133_at |
| TRAIL | Apoptosis | TNF (ligand) superfamily, member 10 | U57059 | 202687_s_at |
| CD38 | Cyclic ADP-ribose metabolism | CD38 antigen (p45) ecto enzyme | NM_001775 | 205692_s_at |
| HERC5 | HECT E3 ubiquitin ligase | Hect domain and RLD 5 | NM_016323 | 219863_at |
| IFI44 | IFN inducible | IFN-induced protein 44 | NM_006417 | 214453_s_at |
| IFI44L | IFN inducible | IFN-induced protein 44 | NM_006820 | 204439_at |
| IFITM1 | Immune suppression | IFN-induced transmembrane protein 1 (residues 9-27) | AA749101 | 214022_s_at |
| LGALS3BP | Immune activation | Lectin, galactoside-binding, soluble, 3 binding protein | NM_005567 | 200923_at |
| USP18 | Ubiquitination | Ubiquitin-specific protease 18 | NM_017414 | 219211_at |
| FLJ20035 | Unknown | RNA Hellicase/DEAD/DEXD protein Q6PK35 | NM_017631 | 218986_s_at |
| FLJ38348 | Unknown | Coiled-coil domain-containing 75 protein CCDC75 | AV755522 | 213294_at |
| HERC6 | Unknown | Hect domain and RLD6 | NM_017912 | 219352_at |
| IFIT1 | Unknown | IFN-induced protein with tetratricopeptide repeat 1 | NM_001548 | 203153_at |
| IFIT3 | Unknown | IFN-induced protein with tetratricopeptide repeat 3 | NM_001549 | 204747_at |
| LY6E | Unknown | Lymphocyte antigen 6 complex, locus E | NM_002346 | 202145_at |
| SAMD9 | Unknown | Sterile alpha motif domain-containing 9 | NM_017654 | 219691_at |
TABLE 2.
qRT-PCR validation of the 11 DV response genes
| Gene name | Relative increase in induction (n-fold) determined by:a
|
|
|---|---|---|
| GeneChip analysis | PCR | |
| ISG20 | 15 | 3 |
| LYE6E | 8 | 4 |
| USP18 | 14 | 5 |
| HERC5 | 9 | 6 |
| HSXIAPAF1 | 7 | 8 |
| IF144 | 27 | 10 |
| G1P2 | 28 | 15 |
| LGALS3BP | 3 | 15 |
| IFI44L | 58 | 31 |
| IFIT1 | 126 | 32 |
| IRF7 | 12 | >100 |
qRT-PCR validation of the 11 DV response genes. In vitro DV infections in HUVECs were analyzed by qRT-PCR using microfluidic cards. Relative induction was calculated with qRT-PCR software (Applied Biosystems). The results of qRT-PCR and the Affymetrix GeneChip HG-U133A were consistent for HUVECs and monocytes, with 8 (>70%) of the 11 genes induced within fourfold of each other.
To identify signaling pathways associated with the host cell response to DV infection, we analyzed our common response gene list using Pathway Architect software (Stratagene). This analysis found the commonly regulated genes to be predominantly regulated by IFN-α and IFN-γ signaling pathways. In addition, TRAIL/TNFSF10, one of the 23 common response genes, was identified as a potential linker of these two signaling pathways (data not shown).
DV infection induces TRAIL/TNFSF10 mRNA and protein production.
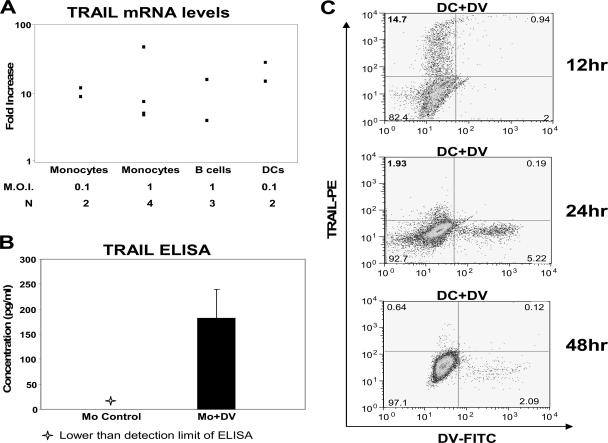
Using real-time PCR, we tested TRAIL/TNFSF10 induction in vitro during DV infection at the mRNA level. We found that TRAIL mRNA was induced in DV-infected monocytes, B cells, and DCs (Fig. 2A). We did not detect the protein TRAIL on the surface of DV-infected monocytes by flow cytometry or in the supernatant of DV-infected monocytes by ELISA, most likely due to the small amounts of TRAIL present. We detected larger amounts of TRAIL protein, 180.93 ± 101.9 pg/ml (mean ± standard deviation [SD]; n = 3), in DV-infected monocyte cell lysates compared to uninfected monocyte lysate at 48 h after infection with DV in vitro (Fig. 2B). These results represent the accumulation/utilization of TRAIL inside the cell at 48 h postinfection. In DV-infected DCs, higher intracellular TRAIL protein levels were detected at 12 h (at 14.7%) but low levels were found at the 24- or 48-h time points after infection, with less than 2% of the cells expressing TRAIL (Fig. 2C). Also, the early expression of TRAIL at the surface or intracellularly at 12 h was observed in the uninfected DC compartment. The lack of TRAIL detection after 12 h postinfection indicates that TRAIL expression is transient in DCs. Both monocyte and DC experiments corroborate the increase in TRAIL mRNA expression originally observed after DV infections by gene expression analysis.
FIG. 2.
TRAIL mRNA and protein induction in DV-infected cells. (A) TRAIL mRNA levels were measured by qRT-PCR. Monocytes were infected with DV at an MOI of 0.1 and 1 PFU/ml, DCs were infected with DV at an MOI of 0.1 PFU/ml, and B cells were infected with DV at an MOI of 1 PFU/ml for 48 h. TRAIL mRNA expression was quantified by qRT-PCR analysis on total RNA extracts. β-Actin mRNA, a constitutively expressed protein, was used as a control probe. Data shown are representative of multiple (N) experiments. (B) TRAIL protein levels were measured in cell lysates. Monocytes (Mo) were infected with DV at an MOI of 1 PFU/cell and then cultured for 48 h. Levels of TRAIL protein were quantified in cell lysates using a TRAIL ELISA (R&D Systems). Data shown are representative of three experiments. (C) Intracellular TRAIL protein levels were determined in DCs infected with DV for 12, 24, and 48 h at an MOI of 0.1. Cells were treated with brefeldin A for the last 8 h of each time point. Levels of TRAIL protein were quantified by flow cytometry using TRAIL-PE (BD Biosciences). Data shown are a representation of two experiments. FITC, fluorescein isothiocyanate.
TRAIL inhibits DV replication.
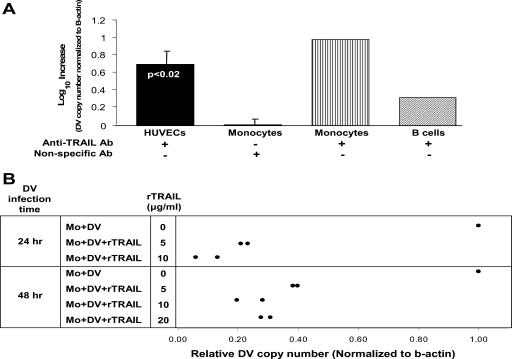
TRAIL neutralizing antibodies were used to block TRAIL function in DV-infected monocytes, B cells, and HUVECs. The concentration of anti-TRAIL antibody (R&D Systems) was titrated (data not shown) to block cell surface TRAIL protein in primary monocytes treated with IFN-β (100 U/ml) as indicated by previously published data (7). At 48 h postinfection, monocytes showed a 0.7 ± 0.3 (mean ± SD) log increase in DV RNA amount (P < 0.02, unpaired Student's t test; n = 5). In duplicate experiments, an average 0.99 and 0.32 log increase in DV RNA was observed in B cells and HUVECs, respectively (Fig. 3A). Irrelevant antibody treatment did not affect DV copy number in monocytes (0.008 ± 0.09 log increase in DV RNA).
FIG. 3.
TRAIL regulates DV replication. (A) TRAIL blocking monoclonal antibody increases DV progeny. Monocytes, B cells, HUVECs were infected with DV at an MOI of 1 PFU/cell and then cultured for 48 h. TRAIL blocking antibody (50 ng/ml) was added 24 h prior to infection with DV. DV copy number was quantified by qRT-PCR analysis. β-Actin mRNA, a constitutively expressed protein, was used as a control probe. The numbers of DV-infected cells are the means of five experiments for monocytes and two experiments each for B cells and HUVECs. Results for nonspecific antibody-treated monocytes are the means of three experiments (± SD). (B) rTRAIL treatment inhibits DV copy number. Monocytes were infected with DV at an MOI of 0.1 PFU/cell and then cultured for 24 and 48 h. Monocytes were pretreated with rTRAIL 6 h prior to infection with DV. DV copy number was quantified by qRT-PCR analysis. β-Actin mRNA, a constitutively expressed protein, was used as a control probe. Results are the means of two experiments. Mo, monocyte.
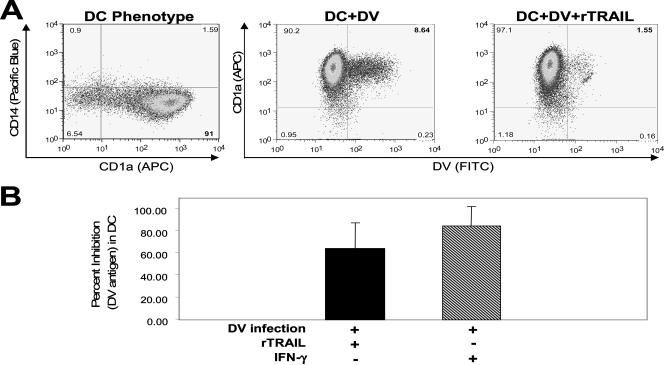
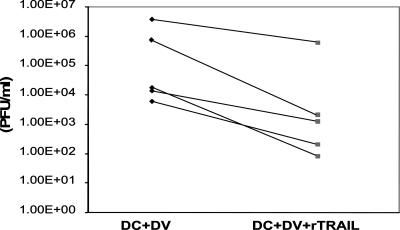
Monocytes pretreated with 5 to 20 μg/ml of rTRAIL (Merck) showed a 0.5 to 1 log decrease in DV RNA copy number after 24 and 48 h (Fig. 3B), respectively. We also determined inhibition of DV infection in monocyte-derived DCs 48 h postinfection. Since DCs are more efficiently infected by DV than the other cells used, we were able to show by flow cytometry the percent infected cells and to use a plaque assay of the culture supernatant to measure progeny virus. DCs treated with rTRAIL (20 μg/ml) showed a decrease of 63.63% ± 23.90% (mean ± SD) in DV antigen (Fig. 4A and B). IFN-γ (500 U/ml), previously shown to inhibit DV in DCs (11), inhibited DV antigen by 84.68% ± 17.44% (mean ± SD) (Fig. 4B). Further, DV infectious progeny was significantly reduced (P < 0.007, paired Student's t test; n = 5) in the supernatant of DV-infected DCs treated with rTRAIL (Fig. 5). The range of inhibition was 0.9 to 2.8 logs.
FIG. 4.
TRAIL inhibits DV antigen in monocyte-derived DCs. (A) rTRAIL treatment inhibits DV infection in DV-infected DCs. DCs were pretreated with rTRAIL for 24 h followed by infection with DV at an MO of 0.1. At the 48-h time point the cells were stained intracellularly for DV antigen (2H2 monoclonal antibody). Data presented from one experiment are representative of seven experiments (± SD). (B) Data from all seven experiments are presented with SD analysis for both the rTRAIL (20 μg/ml) (n = 7) and IFN-γ (500 U/ml) (n = 6) treatments. Data are presented as percent inhibition of DV infection following rTRAIL or IFN-γ treatment. FITC, fluorescein isothiocyanate; APC, allophycocyanin.
FIG. 5.
rTRAIL reduces levels of DV progeny in DV-infected DCs. DCs pretreated with rTRAIL for 24 h were infected with DV for 48 h at an MOI of 0.1. At the end of the experiment culture supernatants were collected and stored at −70°C. LLCMK2 cell monolayers were exposed to the cell culture supernatants to determine the DV titers (PFU/ml). Inhibition of DV titers was observed in rTRAIL-treated DV-infected DC supernatants. Results for five experiments are shown.
Type I IFN regulates TRAIL mRNA induction.
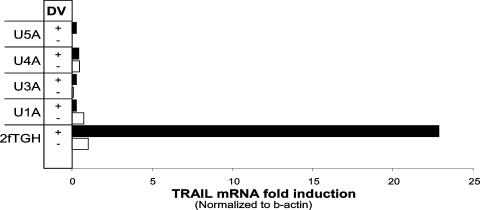
Tyk2, STAT1, JAK1, and IFNARα 2c mutants (U1A, U3A, U4A, and U5A) derived from wild-type fibroblast cells (2fTGH) were used to determine whether the IFN-α signaling pathway regulates TRAIL mRNA induction during DV infection. TaqMan qRT-PCR was used to quantify TRAIL mRNA levels. Unlike the wild-type cells, none of the mutant cell lines showed up-regulation of TRAIL in response to DV infection (Fig. 6). These data show that type I IFN signaling is necessary for TRAIL mRNA induction.
FIG. 6.
TRAIL induction in response to DV infection is IFN-α dependent. 2fTGH (wild type) and U1A, U3A, U4A, and U5A (IFN-α mutant) fibroblast cells were infected with DV at an MOI of 1 PFU/cell and then cultured for 48 h. TRAIL mRNA levels were quantified by qRT-PCR analysis. β-Actin mRNA, a constitutively expressed protein, was used as a control probe. Results are representative of two experiments.
TRAIL inhibition of DV is apoptosis independent.
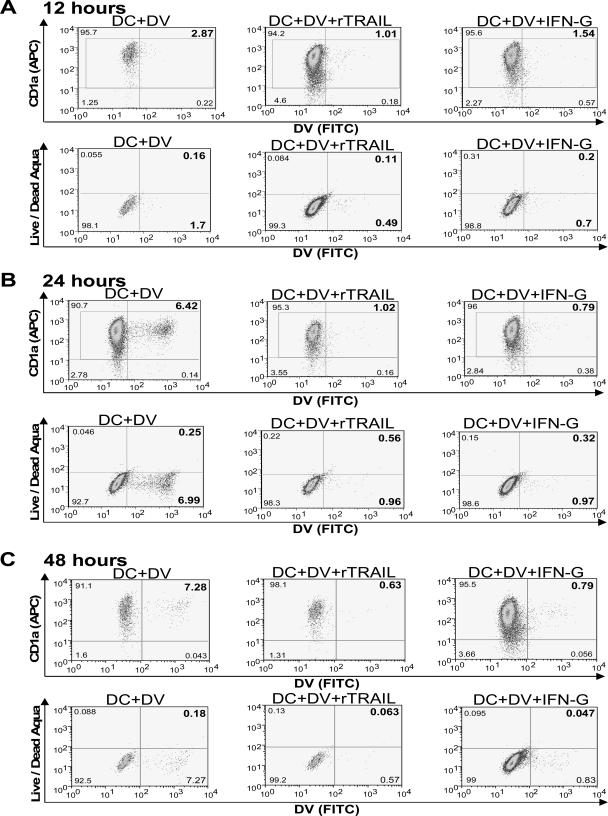
Previously published reports have found that TRAIL functions by inducing apoptosis of cells (35). Hence, we investigated whether rTRAIL-mediated inhibition of DV was dependent on apoptosis of DV-infected cells. DV-infected DCs treated with or without rTRAIL were stained for DV antigen and Live/Dead Aqua dye or early (caspase-3) and late (PARP-1) markers of apoptosis. The decrease in DV infection of rTRAIL-treated DCs did not correlate with an increase in staining with Live Dead Aqua, a dye which detects a cell in its early stage of apoptosis (Fig. 7). Similar results for absence of apoptosis were observed when DCs were stained for active caspase-3 and cleaved PARP-1 proteins (data not shown).
FIG. 7.
Cell death flow cytometry-based assay of CD1a+ cells. Absence of apoptosis in rTRAIL-treated DV-infected DCs. Monocytes were pretreated overnight with rTRAIL as described in Materials and Methods, followed by infection with DV for 48 h at an MOI of 1. Live/Dead Aqua fluorescence was used to identify apoptotic DCs at 12 (A), 24 (B), and 48 (C) h after rTRAIL treatment and DV infection. Data from a representative experiment are shown for each time point from a total of two (12 h and 24 h) and five (48 h) experiments. FITC, fluorescein isothiocyanate; APC, allophycocyanin.
DISCUSSION
We initially set out to identify the cellular immune responses to DV infection. Using global gene expression profiling, we found that IFN-α and IFN-γ signaling pathway-inducible genes were commonly regulated in several DV-susceptible cell types. TRAIL, one of the common response genes, was identified as a novel antiviral molecule against DV. Furthermore, our data suggest that the rTRAIL-mediated decrease in DV infection is mediated by a novel apoptosis-independent mechanism.
DV infection is an acute infection cleared within approximately 1 week (22). We hypothesized that there is a set of cellular genes which is necessary to clear the virus in susceptible cells. We identified a common gene expression profile of 23 differentially regulated genes from GeneChip analysis of DV-infected HUVECs and peripheral blood mononuclear cell subsets (B cells and monocytes). These findings extend previous studies of gene expression in DV-infected HUVECs (46) and macrophages (30) that were limited to global gene expression analysis in one cell type. All of the 23 common response genes were among the list of differentially expressed genes detected in HUVECs using differential display and GeneChip microarrays (46). These genes were not reported as up-regulated by DV in the cytokine gene array study of Moreno-Altamirano et al. (30). However, their study did not investigate these specific genes.
Of the 23 genes that comprised the common DV response profile, functions and/or inducers of 19 genes have been described in the literature (Table 2). These include classical antiviral response genes (OAS3 and IRF7), more recently identified antiviral genes (ISG15, HERC5, RSAD2, TRIM5, TRAIL, OASL, and ISG20), genes regulating ubiquitination (USP18), cell adhesion and cyclic ADP-ribose metabolism (CD38), apoptosis (XAF1), immune suppression (IFITM1), immune activation (LGALs3BP), and nine other genes (FLJ20035, FLJ38348, HERC6, IFI44, IFI44L, IFIT1, IFIT3, LY6E, and SAMD9) with unknown function. Functions of the genes identified in this study may be pivotal in understanding the role of the cellular responses to DV infection. Of the common response genes, G1p2, Mx1, and OAS3 were also associated with dengue shock in patients with severe DV infection (41).
One of the common response genes, TRAIL, was identified as a potential link between IFN-α and IFN-γ signaling pathways. We selected TRAIL for further study based on its previously documented antiviral and antitumor functions (2, 17, 25, 35, 37-39, 44, 45, 48). This member of the TNF family of ligands is capable of initiating apoptosis through the engagement of its death receptors, TRAIL-R1 (DR4) and TRAIL-R2 (DR5) (4). In vitro and in vivo studies have demonstrated tumoricidal activity of TRAIL without significant toxicity toward normal cells or tissues (35, 38). TRAIL-mediated killing by activated CD4+ T cells, NK cells, and B cells has been shown to occur with influenza virus and others (17, 37, 45). IFNs enhance expression of TRAIL while, on the other hand, TRAIL treatment can enhance expression of IFN-inducible genes like IFITM1, IFIT1, STAT1, LGal3BP, and PRKR as well as IFN-β itself (20). The molecular cross-talk and functional synergy observed between the TRAIL and IFN signaling pathways is not limited to the genes involved in apoptosis and may have implications for the physiologic role and mechanism of action of TRAIL protein. Findings in this study (Fig. 3 and 6) support the idea that type I IFN, which is known to inhibit DV, might induce TRAIL as a part of the type I IFN response against DV.
Primary monocytes, DCs, B cells, and HUVECs infected with DV induced TRAIL mRNA expression. TRAIL protein levels were also found to be highly induced in DV-infected monocyte cell lysates, but using fluorescence-activated cell sorting, we were unable to detect TRAIL on monocyte and DC surfaces after DV infection or secreted TRAIL protein in the supernatant from DV-infected primary monocytes. Recently, Matsuda et al. reported that TRAIL protein secreted by HepG2 cells after DV infection was partly responsible for apoptosis of uninfected HepG2 cells (28). Global gene expression analysis in DV-infected HepG2 cells and primary cells like monocytes, B cells, and HUVECs performed in our laboratory has shown a distinct set of differentially regulated genes in HepG2 versus primary human cells (data not shown). Unlike primary cells, TRAIL mRNA levels were not found to be up-regulated in DV-infected HepG2 cells by GeneChip analysis (data not shown). DV infection of primary cells may better reflect a physiological response. In the previous study of Matsuda et al., 80% of DV-infected HepG2 cells stained positive for cell death (28), raising the possibility that the TRAIL protein detected in the supernatant of DV-infected cells was not secreted but resulted from cell death. DCs infected with DV stained minimally for apoptosis detection dye (Live/Dead Aqua) and markers (PARP-1 and caspase-3) (Fig. 7). Our results indicate that under the conditions tested, DV infection does not induce apoptosis in monocytes and DCs.
The concentration of rTRAIL (5 to 20 μg/ml) in this study that was able to inhibit DV replication was similar to the one used in studies demonstrating TRAIL-induced apoptosis of tumor cells (1, 2). Hence, we hypothesized that TRAIL might be inhibiting DV copy number by acting as an antiviral agent and not by inducing apoptosis. An apoptosis-independent TRAIL antiviral function is a novel finding which should be further studied. One possible mechanism of TRAIL antiviral function could be TRAIL-mediated increased expression of known or novel antiviral cellular protein(s) which are secreted by cells.
TRAIL has been shown to mediate antiviral functions in vivo in mouse models of influenza and encephalomyocarditis virus infection (15). Influenza viral clearance was prolonged in mice injected with anti-TRAIL antibody. TRAIL expressed by NK cells was crucial to limit encephalomyocarditis virus replication in vivo (38). In this study, we observed TRAIL-dependent inhibition of DV replication. Previous studies have shown that both type I and type II IFNs are critical in controlling different stages of DV infection in mice (40), although the precise mechanism(s) by which IFNs mediate an antiviral response is unknown. We postulated that TRAIL protein was induced by and contributed to the type I IFN-mediated antiviral function. Using wild-type (2fTGH) and type I IFN mutant (U1A, U3A, U4A, and U5A) fibroblast cells (24, 29, 33), we found that induction of TRAIL gene expression by DV was type I IFN dependent. Furthermore, TRAIL inhibited DV in a novel apoptosis-independent manner. This is the first study to show that TRAIL might regulate a pathogen in an apoptosis-independent manner. Activation of the TRAIL signaling pathway might therefore be used as an antiviral therapy in the future.
Acknowledgments
This work was supported by grants P01-AI34533 and U19-AI057319 from the National Institutes of Health.
We gratefully acknowledge Kay Case at the Vascular Research Division, Brigham and Women's Hospital, for HUVEC preparations; Phyllis Spatrick for processing the Affymetrix chips; Jurand Janus for technical assistance with plaque assays; and Marcia Woda for flow cytometry analysis. We acknowledge Paul B. Fisher for sharing of cell line reagents and Fundasangre (Caracas, Venezuela) for providing anti-dengue virus antibodies.
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Abdollahi, T., N. M. Robertson, A. Abdollahi, and G. Litwack. 2003. Identification of interleukin 8 as an inhibitor of tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in the ovarian carcinoma cell line OVCAR3. Cancer Res. 634521-4526. [PubMed] [Google Scholar]
- 2.Bouralexis, S., D. M. Findlay, and A. Evdokiou. 2005. Death to the bad guys: targeting cancer via Apo2L/TRAIL. Apoptosis 1035-51. [DOI] [PubMed] [Google Scholar]
- 3.Clarke, P., S. M. Meintzer, S. Gibson, C. Widmann, T. P. Garrington, G. L. Johnson, and K. L. Tyler. 2000. Reovirus-induced apoptosis is mediated by TRAIL. J. Virol. 748135-8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cretney, E., J. L. McQualter, N. Kayagaki, H. Yagita, C. C. Bernard, I. S. Grewal, A. Ashkenazi, and M. J. Smyth. 2005. TNF-related apoptosis-inducing ligand (TRAIL)/Apo2L suppresses experimental autoimmune encephalomyelitis in mice. Immunol. Cell Biol. 83511-519. [DOI] [PubMed] [Google Scholar]
- 5.Diamond, M. S., and E. Harris. 2001. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 289297-311. [DOI] [PubMed] [Google Scholar]
- 6.Diehl, G. E., H. H. Yue, K. Hsieh, A. A. Kuang, M. Ho, L. A. Morici, L. L. Lenz, D. Cado, L. W. Riley, and A. Winoto. 2004. TRAIL-R as a negative regulator of innate immune cell responses. Immunity 21877-889. [DOI] [PubMed] [Google Scholar]
- 7.Ehrlich, S., C. Infante-Duarte, B. Seeger, and F. Zipp. 2003. Regulation of soluble and surface-bound TRAIL in human T cells, B cells, and monocytes. Cytokine 24244-253. [DOI] [PubMed] [Google Scholar]
- 8.Greidinger, E. L., D. K. Miller, T. T. Yamin, L. Casciola-Rosen, and A. Rosen. 1996. Sequential activation of three distinct ICE-like activities in Fas-ligated Jurkat cells. FEBS Lett. 390299-303. [DOI] [PubMed] [Google Scholar]
- 9.Gubler, D. J. 2001. Human arbovirus infections worldwide. Ann. N. Y. Acad. Sci. 95113-24. [DOI] [PubMed] [Google Scholar]
- 10.Halstead, S. B. 1989. Antibody, macrophages, dengue virus infection, shock, and hemorrhage: a pathogenetic cascade. Rev. Infect. Dis. 11(Suppl. 4)S830-S839. [DOI] [PubMed] [Google Scholar]
- 11.Ho, L. J., L. F. Hung, C. Y. Weng, W. L. Wu, P. Chou, Y. L. Lin, D. M. Chang, T. Y. Tai, and J. H. Lai. 2005. Dengue virus type 2 antagonizes IFN-alpha but not IFN-gamma antiviral effect via down-regulating Tyk2-STAT signaling in the human dendritic cell. J. Immunol. 1748163-8172. [DOI] [PubMed] [Google Scholar]
- 12.Huang, Y., and M. S. Sheikh. 2006. TRAIL death receptors and cancer therapeutics. Toxicol. Appl. Pharmacol. 224284-289. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Y. H., H. Y. Lei, H. S. Liu, Y. S. Lin, C. C. Liu, and T. M. Yeh. 2000. Dengue virus infects human endothelial cells and induces IL-6 and IL-8 production. Am. J. Trop. Med. Hyg. 6371-75. [DOI] [PubMed] [Google Scholar]
- 14.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4249-264. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa, E., M. Nakazawa, M. Yoshinari, and M. Minami. 2005. Role of tumor necrosis factor-related apoptosis-inducing ligand in immune response to influenza virus infection in mice. J. Virol. 797658-7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jessie, K., M. Y. Fong, S. Devi, S. K. Lam, and K. T. Wong. 2004. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J. Infect. Dis. 1891411-1418. [DOI] [PubMed] [Google Scholar]
- 17.Kemp, T. J., B. D. Elzey, and T. S. Griffith. 2003. Plasmacytoid dendritic cell-derived IFN-alpha induces TNF-related apoptosis-inducing ligand/Apo-2L-mediated antitumor activity by human monocytes following CpG oligodeoxynucleotide stimulation. J. Immunol. 171212-218. [DOI] [PubMed] [Google Scholar]
- 18.King, A. D., A. Nisalak, S. Kalayanrooj, K. S. Myint, K. Pattanapanyasat, S. Nimmannitya, and B. L. Innis. 1999. B cells are the principal circulating mononuclear cells infected by dengue virus. Southeast Asian J. Trop. Med. Public Health 30718-728. [PubMed] [Google Scholar]
- 19.Kumari, S. R., H. Mendoza-Alvarez, and R. Alvarez-Gonzalez. 1998. Functional interactions of p53 with poly(ADP-ribose) polymerase (PARP) during apoptosis following DNA damage: covalent poly(ADP-ribosyl)ation of p53 by exogenous PARP and noncovalent binding of p53 to the M(r) 85,000 proteolytic fragment. Cancer Res. 585075-5078. [PubMed] [Google Scholar]
- 20.Kumar-Sinha, C., S. Varambally, A. Sreekumar, and A. M. Chinnaiyan. 2002. Molecular cross-talk between the TRAIL and interferon signaling pathways. J. Biol. Chem. 277575-585. [DOI] [PubMed] [Google Scholar]
- 21.Kurane, I., B. L. Innis, S. Nimmannitya, A. Nisalak, A. Meager, and F. A. Ennis. 1993. High levels of interferon alpha in the sera of children with dengue virus infection. Am. J. Trop. Med. Hyg. 48222-229. [DOI] [PubMed] [Google Scholar]
- 22.Libraty, D. H., T. P. Endy, H. S. Houng, S. Green, S. Kalayanarooj, S. Suntayakorn, W. Chansiriwongs, D. W. Vaughn, A. Nisalak, F. A. Ennis, and A. L. Rothman. 2002. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J. Infect. Dis. 1851213-1221. [DOI] [PubMed] [Google Scholar]
- 23.Lin, Y. W., K. J. Wang, H. Y. Lei, Y. S. Lin, T. M. Yeh, H. S. Liu, C. C. Liu, and S. H. Chen. 2002. Virus replication and cytokine production in dengue virus-infected human B lymphocytes. J. Virol. 7612242-12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutfalla, G., S. J. Holland, E. Cinato, D. Monneron, J. Reboul, N. C. Rogers, J. M. Smith, G. R. Stark, K. Gardiner, K. E. Mogensen, et al. 1995. Mutant U5A cells are complemented by an interferon-alpha beta receptor subunit generated by alternative processing of a new member of a cytokine receptor gene cluster. EMBO J. 145100-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma, H., Y. Liu, S. Liu, H. F. Kung, X. Sun, D. Zheng, and R. Xu. 2005. Recombinant adeno-associated virus-mediated TRAIL gene therapy suppresses liver metastatic tumors. Int. J. Cancer 116314-321. [DOI] [PubMed] [Google Scholar]
- 26.Mairuhu, A. T., J. Wagenaar, D. P. Brandjes, and E. C. van Gorp. 2004. Dengue: an arthropod-borne disease of global importance. Eur. J. Clin. Microbiol. Infect. Dis. 23425-433. [DOI] [PubMed] [Google Scholar]
- 27.Marovich, M., G. Grouard-Vogel, M. Louder, M. Eller, W. Sun, S. J. Wu, R. Putvatana, G. Murphy, B. Tassaneetrithep, T. Burgess, D. Birx, C. Hayes, S. Schlesinger-Frankel, and J. Mascola. 2001. Human dendritic cells as targets of dengue virus infection. J. Investig. Dermatol. Symp. Proc. 6219-224. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda, T., A. Almasan, M. Tomita, K. Tamaki, M. Saito, M. Tadano, H. Yagita, T. Ohta, and N. Mori. 2005. Dengue virus-induced apoptosis in hepatic cells is partly mediated by Apo2 ligand/tumour necrosis factor-related apoptosis-inducing ligand. J. Gen. Virol. 861055-1065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.McKendry, R., J. John, D. Flavell, M. Muller, I. M. Kerr, and G. R. Stark. 1991. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl. Acad. Sci. USA 8811455-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno-Altamirano, M. M., M. Romano, M. Legorreta-Herrera, F. J. Sanchez-Garcia, and M. J. Colston. 2004. Gene expression in human macrophages infected with dengue virus serotype-2. Scand. J. Immunol. 60631-638. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 10014333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nathan, M. B., and R. Dayal-Drager. 2006. Recent Epidemiological trends, the global strategy and public health advances in dengue. Report of the Scientific Working Group on Dengue, 2006. WHO TDR/SWG/08. World Health Organization, Geneva, Switzerland.
- 33.Pellegrini, S., J. John, M. Shearer, I. M. Kerr, and G. R. Stark. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 94605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen, L. R., and A. A. Marfin. 2005. Shifting epidemiology of Flaviviridae. J. Travel Med. 12(Suppl. 1)S3-S11. [DOI] [PubMed] [Google Scholar]
- 35.Pitti, R. M., S. A. Marsters, S. Ruppert, C. J. Donahue, A. Moore, and A. Ashkenazi. 1996. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J. Biol. Chem. 27112687-12690. [DOI] [PubMed] [Google Scholar]
- 36.Russell, P. K., A. Nisalak, P. Sukhavachana, and S. Vivona. 1967. A plaque reduction test for dengue virus neutralizing antibodies. J. Immunol. 99285-290. [PubMed] [Google Scholar]
- 37.Sato, K., S. Hida, H. Takayanagi, T. Yokochi, N. Kayagaki, K. Takeda, H. Yagita, K. Okumura, N. Tanaka, T. Taniguchi, and K. Ogasawara. 2001. Antiviral response by natural killer cells through TRAIL gene induction by IFN-alpha/beta. Eur. J. Immunol. 313138-3146. [DOI] [PubMed] [Google Scholar]
- 38.Sato, K., T. Nakaoka, N. Yamashita, H. Yagita, H. Kawasaki, C. Morimoto, M. Baba, and T. Matsuyama. 2005. TRAIL-transduced dendritic cells protect mice from acute graft-versus-host disease and leukemia relapse. J. Immunol. 1744025-4033. [DOI] [PubMed] [Google Scholar]
- 39.Secchiero, P., M. Vaccarezza, A. Gonelli, and G. Zauli. 2004. TNF-related apoptosis-inducing ligand (TRAIL): a potential candidate for combined treatment of hematological malignancies. Curr. Pharm. Des. 103673-3681. [DOI] [PubMed] [Google Scholar]
- 40.Shresta, S., K. L. Sharar, D. M. Prigozhin, H. M. Snider, P. R. Beatty, and E. Harris. 2005. Critical roles for both STAT1-dependent and STAT1-independent pathways in the control of primary dengue virus infection in mice. J. Immunol. 1753946-3954. [DOI] [PubMed] [Google Scholar]
- 41.Simmons, C. P., S. Popper, C. Dolocek, T. N. Chau, M. Griffiths, N. T. Dung, T. H. Long, D. M. Hoang, N. V. Chau, T. T. Thao le, T. T. Hien, D. A. Relman, and J. Farrar. 2007. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J. Infect. Dis. 1951097-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suksanpaisan, L., A. Cabrera-Hernandez, and D. R. Smith. 2007. Infection of human primary hepatocytes with dengue virus serotype 2. J. Med. Virol. 79300-307. [DOI] [PubMed] [Google Scholar]
- 43.Thorburn, J., L. M. Bender, M. J. Morgan, and A. Thorburn. 2003. Caspase- and serine protease-dependent apoptosis by the death domain of FADD in normal epithelial cells. Mol. Biol. Cell 1467-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wajant, H., J. Gerspach, and K. Pfizenmaier. 2005. Tumor therapeutics by design: targeting and activation of death receptors. Cytokine Growth Factor Rev. 1655-76. [DOI] [PubMed] [Google Scholar]
- 45.Wang, M., Y. Liu, S. Liu, and D. Zheng. 2004. 8-Chloro-adenosine sensitizes a human hepatoma cell line to TRAIL-induced apoptosis by caspase-dependent and -independent pathways. Oncol. Rep. 12193-199. [PubMed] [Google Scholar]
- 46.Warke, R. V., K. Xhaja, K. J. Martin, M. F. Fournier, S. K. Shaw, N. Brizuela, N. de Bosch, D. Lapointe, F. A. Ennis, A. L. Rothman, and I. Bosch. 2003. Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J. Virol. 7711822-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, S. J., G. Grouard-Vogel, W. Sun, J. R. Mascola, E. Brachtel, R. Putvatana, M. K. Louder, L. Filgueira, M. A. Marovich, H. K. Wong, A. Blauvelt, G. S. Murphy, M. L. Robb, B. L. Innes, D. L. Birx, C. G. Hayes, and S. S. Frankel. 2000. Human skin Langerhans cells are targets of dengue virus infection. Nat. Med. 6816-820. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, X., R. M. Cheung, R. Komaki, B. Fang, and J. Y. Chang. 2005. Radiotherapy sensitization by tumor-specific TRAIL gene targeting improves survival of mice bearing human non-small cell lung cancer. Clin. Cancer Res. 116657-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]